Maternal Under- and Over-Nutrition during Gestation Causes Islet Hypertrophy and Sex-Specific Changes to Pancreas DNA Methylation in Fetal Sheep
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Histological Analyses
2.3. DNA Isolation and Sequencing
2.4. Data Analyses
3. Results
3.1. Fetal Islet Size and Number
3.2. β-Cell Proliferation and Apoptosis
3.3. Glucose and Somatostatin Positive Area
3.4. Sequencing Information
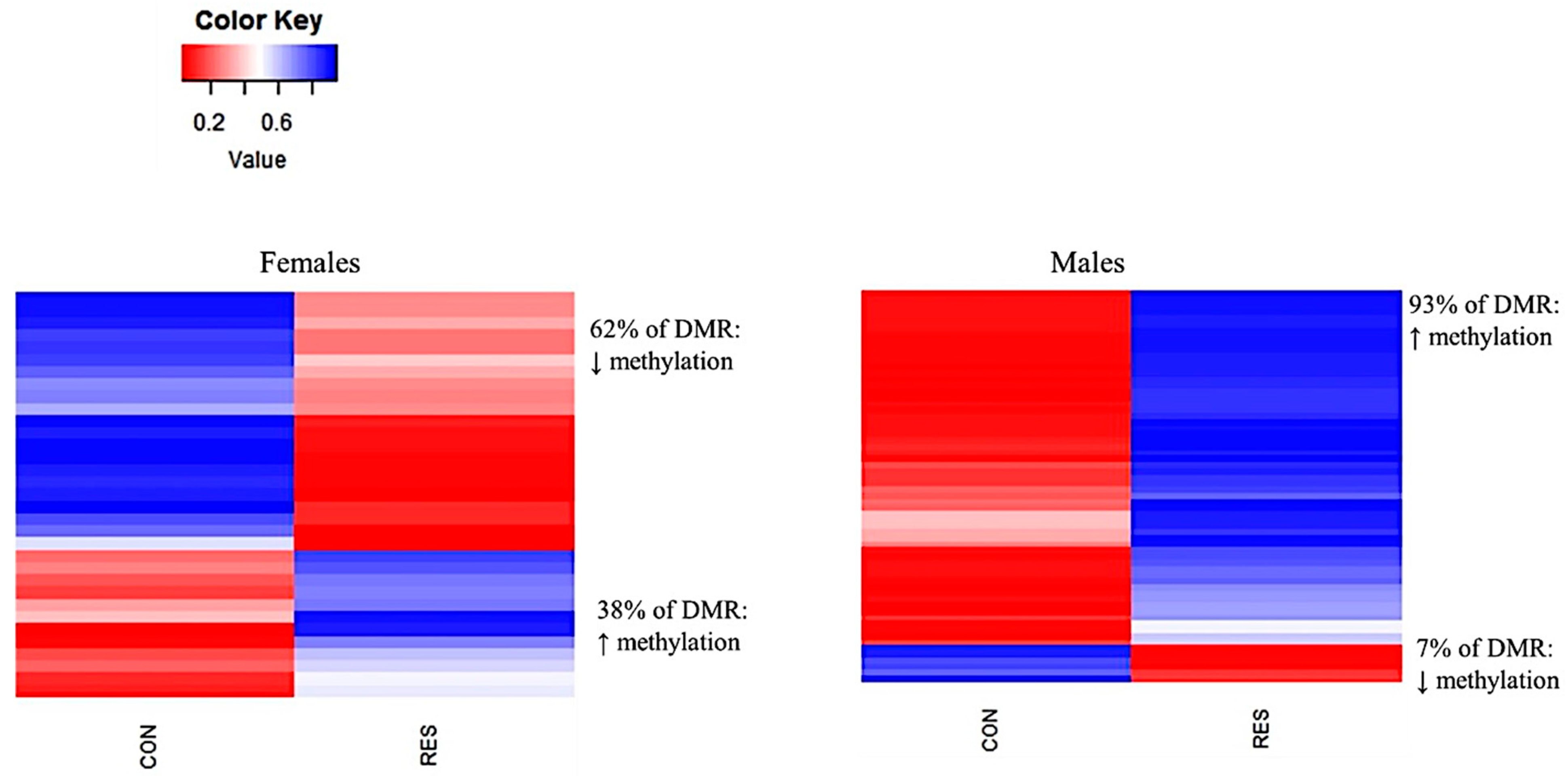
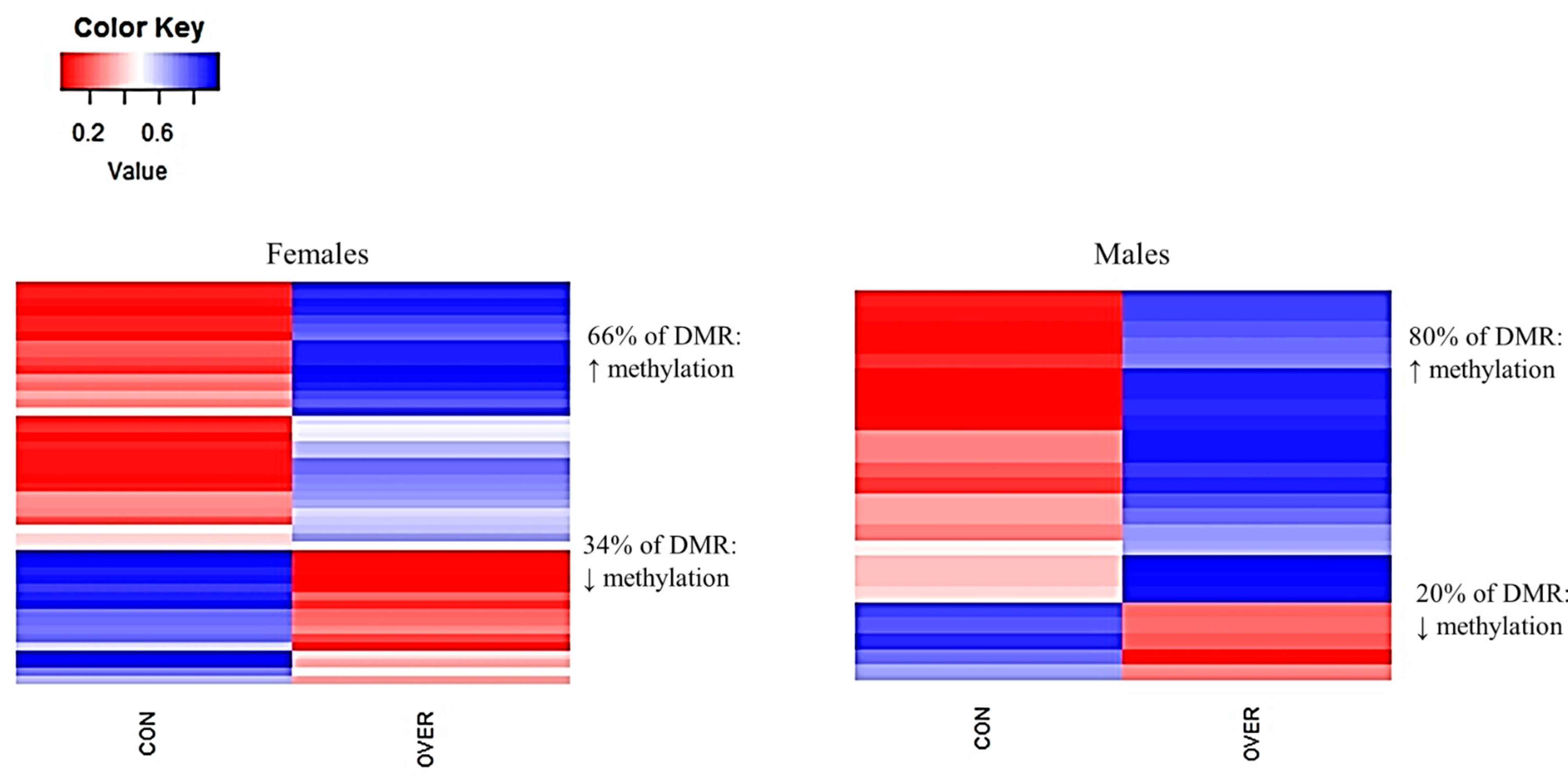
3.5. Differentally Methylated Loci (DML) and Regions (DMRs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cerf, M.E.; Williams, K.; Chapman, C.S.; Louw, J. Compromised-Cell Development and Cell Dysfunction in Weanling Offspring from Dams Maintained on a High-Fat Diet during Gestation. Pancreas 2007, 34, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Gittes, G.K. Developmental biology of the pancreas: A comprehensive review. Dev. Biol. 2009, 326, 4–35. [Google Scholar] [CrossRef] [Green Version]
- Cole, L.; Anderson, M.; Antin, P.B.; Limesand, S.W. One process for pancreatic beta-cell coalescence into islets involves an epithelial-mesenchymal transition. J. Endocrinol. 2009, 203, 19–31. [Google Scholar] [CrossRef]
- Wilson, M.E.; Scheel, D.; German, M.S. Gene expression cascades in pancreatic development. Mech. Dev. 2003, 120, 65–80. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Wallace, J.M.; Spencer, T.E. Board-invited review: Intrauterine growth retardation: Implications for the animal sciences. J. Anim. Sci. 2006, 84, 2316–2337. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.L.; Reed, S.A.; Pillai, S.M.; Jones, A.K.; McFadden, K.K.; Zinn, S.A.; Govoni, K.E. Physiology and endocrinology symposium: The effects of poor maternal nutrition during gestation on offspring postnatal growth and metabolism. J. Anim. Sci. 2017, 95, 2222–2232. [Google Scholar] [CrossRef] [Green Version]
- Mastorci, F.; Vassalle, C.; Chatzianagnostou, K.; Marabotti, C.; Siddiqui, K.; Eba, A.O.; Mhamed, S.A.S.; Bandopadhyay, A.; Nazzaro, M.S.; Passera, M.; et al. Undernutrition and Overnutrition Burden for Diseases in Developing Countries: The Role of Oxidative Stress Biomarkers to Assess Disease Risk and Interventional Strategies. Antioxidants 2017, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Fowden, A.L.; Hill, D.J. Intra-uterine programming of the endocrine pancreas. Br. Med. Bull. 2001, 60, 123–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.J.; Ford, S.P.; Means, W.J.; Hess, B.W.; Nathanielsz, P.W.; Du, M. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J. Physiol. 2006, 575, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Ford, S.P.; Zhang, L.; Zhu, M.; Miller, M.M.; Smith, D.T.; Hess, B.W.; Moss, G.E.; Nathanielsz, P.W.; Nijland, M.J. Maternal obesity accelerates fetal pancreatic β-cell but not α-cell development in sheep: Prenatal consequences. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R835–R843. [Google Scholar] [CrossRef]
- Ford, S.P.; Long, N.M. Evidence for similar changes in offspring phenotype following either maternal undernutrition or overnutrition: Potential impact on fetal epigenetic mechanisms. Reprod. Fertil. Dev. 2011, 24, 105–111. [Google Scholar] [CrossRef]
- Phillips, T. The role of methylation in gene expression. Nat. Educ. 2008, 1, 116. [Google Scholar]
- Hochberg, Z.; Feil, R.; Constancia, M.; Fraga, M.; Junien, C.; Carel, J.-C.; Boileau, P.; Le Bouc, Y.; Deal, C.L.; Lillycrop, K.; et al. Child health, developmental plasticity, and epigenetic programming. Endocr. Rev. 2011, 32, 159–224. [Google Scholar] [CrossRef]
- Sosa-Larios, T.C.; Cerbón, M.A.; Morimoto, S. Epigenetic alterations caused by nutritional stress during fetal programming of the endocrine pancreas. Arch. Med. Res. 2015, 46, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Namous, H.; Peñagaricano, F.; Del Corvo, M.; Capra, E.; Thomas, D.L.; Stella, A.; Williams, J.L.; Marsan, P.A.; Khatib, H. Integrative analysis of methylomic and transcriptomic data in fetal sheep muscle tissues in response to maternal diet during pregnancy. BMC Genomics 2018, 19, 123. [Google Scholar] [CrossRef] [Green Version]
- Pillai, S.M.; Jones, A.K.; Hoffman, M.L.; McFadden, K.K.; Reed, S.A.; Zinn, S.A.; Govoni, K.E. Fetal and organ development at gestational days 45, 90, 135 and at birth of lambs exposed to under- or over-nutrition during gestation. Transl. Anim. Sci. 2017, 1, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Research Council, N. Nutrient Requirements of Sheep; National Academies Press: Washington, DC, USA, 1985; ISBN 9780309035965. [Google Scholar]
- Limesand, S.W.; Rozance, P.J.; Macko, A.R.; Anderson, M.J.; Kelly, A.C.; Hay, W.W., Jr. Reductions in insulin concentrations and β-cell mass precede growth restriction in sheep fetuses with placental insufficiency. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E516–E523. [Google Scholar] [CrossRef] [Green Version]
- Blackwood, E.M.; Kadonaga, J.T. Going the distance: A current view of enhancer action. Science 1998, 281, 60–63. [Google Scholar] [CrossRef] [Green Version]
- Mora, A.; Sandve, G.K.; Gabrielsen, O.S.; Eskeland, R. In the loop: Promoter-enhancer interactions and bioinformatics. Brief. Bioinform. 2016, 17, 980–995. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Zheng, X.; Yang, J.; Imamichi, T.; Stephens, R.; Lempicki, R.A. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinform. 2009, 27, 13.11.1–13.11.13. [Google Scholar] [CrossRef]
- Bouwens, L.; Rooman, I. Regulation of pancreatic beta-cell mass. Physiol. Rev. 2005, 85, 1255–1270. [Google Scholar] [CrossRef]
- Holemans, K.; Aerts, L.; Van Assche, F.A. Lifetime consequences of abnormal fetal pancreatic development. J. Physiol. 2003, 547, 11–20. [Google Scholar] [CrossRef]
- Weir, G.C.; Bonner-Weir, S. Five stages of evolving beta-cell dysfunction during progression to diabetes. Diabetes 2004, 53 (Suppl. 3), S16–S21. [Google Scholar] [CrossRef] [Green Version]
- Limesand, S.W.; Jensen, J.; Hutton, J.C.; Hay, W.W., Jr. Diminished beta-cell replication contributes to reduced beta-cell mass in fetal sheep with intrauterine growth restriction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R1297–R1305. [Google Scholar] [CrossRef]
- Keomanivong, F.E.; Camacho, L.E.; Lemley, C.O.; Kuemper, E.A.; Yunusova, R.D.; Borowicz, P.P.; Kirsch, J.D.; Vonnahme, K.A.; Caton, J.S.; Swanson, K.C. Effects of realimentation after nutrient restriction during mid- to late gestation on pancreatic digestive enzymes, serum insulin and glucose levels, and insulin-containing cell cluster morphology. J. Anim. Physiol. Anim. Nutr. 2017, 101, 589–604. [Google Scholar] [CrossRef]
- Weir, G.C.; Bonner-Weir, S. Islet β cell mass in diabetes and how it relates to function, birth, and death. Ann. N. Y. Acad. Sci. 2013, 1281, 92–105. [Google Scholar] [CrossRef]
- Titlbach, M.; Fält, K.; Falkmer, S. Postnatal maturation of the islets of Langerhans in sheep. Light microscopic, immunohistochemical, morphometric, and ultrastructural investigations with particular reference to the transient appearance of argyrophil insulin immunoreactive cells. Diabetes Res. 1985, 2, 5–15. [Google Scholar]
- Hyatt, M.A.; Keisler, D.H.; Budge, H.; Symonds, M.E. Maternal parity and its effect on adipose tissue deposition and endocrine sensitivity in the postnatal sheep. J. Endocrinol. 2010, 204, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Molitch, M.E. Neuroendocrinology and the neuroendocrine system. In Goldman’s Cecil Medicine, 24th ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1425–1431. [Google Scholar]
- Hoffman, M.L.; Peck, K.N.; Wegrzyn, J.L.; Reed, S.A.; Zinn, S.A.; Govoni, K.E. Poor maternal nutrition during gestation alters the expression of genes involved in muscle development and metabolism in lambs. J. Anim. Sci. 2016, 94, 3093–3099. [Google Scholar] [CrossRef]
- Lillycrop, K.A.; Phillips, E.S.; Jackson, A.A.; Hanson, M.A.; Burdge, G.C. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 2005, 135, 1382–1386. [Google Scholar] [CrossRef]
- Park, J.H.; Stoffers, D.A.; Nicholls, R.D.; Simmons, R.A. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J. Clin. Investig. 2008, 118, 2316–2324. [Google Scholar] [CrossRef] [Green Version]
- Ong, T.P.; Ozanne, S.E. Developmental programming of type 2 diabetes: Early nutrition and epigenetic mechanisms. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 354–360. [Google Scholar] [CrossRef]
- Lillycrop, K.A.; Slater-Jefferies, J.L.; Hanson, M.A.; Godfrey, K.M.; Jackson, A.A.; Burdge, G.C. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br. J. Nutr. 2007, 97, 1064–1073. [Google Scholar]
- Hall, E.; Volkov, P.; Dayeh, T.; Esguerra, J.L.S.; Salö, S.; Eliasson, L.; Rönn, T.; Bacos, K.; Ling, C. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 2014, 15, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, Y.; Shirane, M.; Nakayama, K.I. TMEM55B contributes to lysosomal homeostasis and amino acid-induced mTORC1 activation. Genes Cells 2018, 23, 418–434. [Google Scholar] [CrossRef] [Green Version]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Chen, X.; Xiao, Y.; Wen, J.; Chen, J.; Wang, K.; Chen, G. Gestational diabetes mellitus alters DNA methylation profiles in pancreas of the offspring mice. J. Diabetes Complicat. 2019, 33, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Doush, Y.; Surani, A.A.; Navarro-Corcuera, A.; McArdle, S.; Ellen Billett, E.; Montiel-Duarte, C. SP1 and RARα regulate AGAP2 expression in cancer. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parveen, N.; Dhawan, S. DNA Methylation Patterning and the Regulation of Beta Cell Homeostasis. Front. Endocrinol. 2021, 12, 651258. [Google Scholar] [CrossRef] [PubMed]
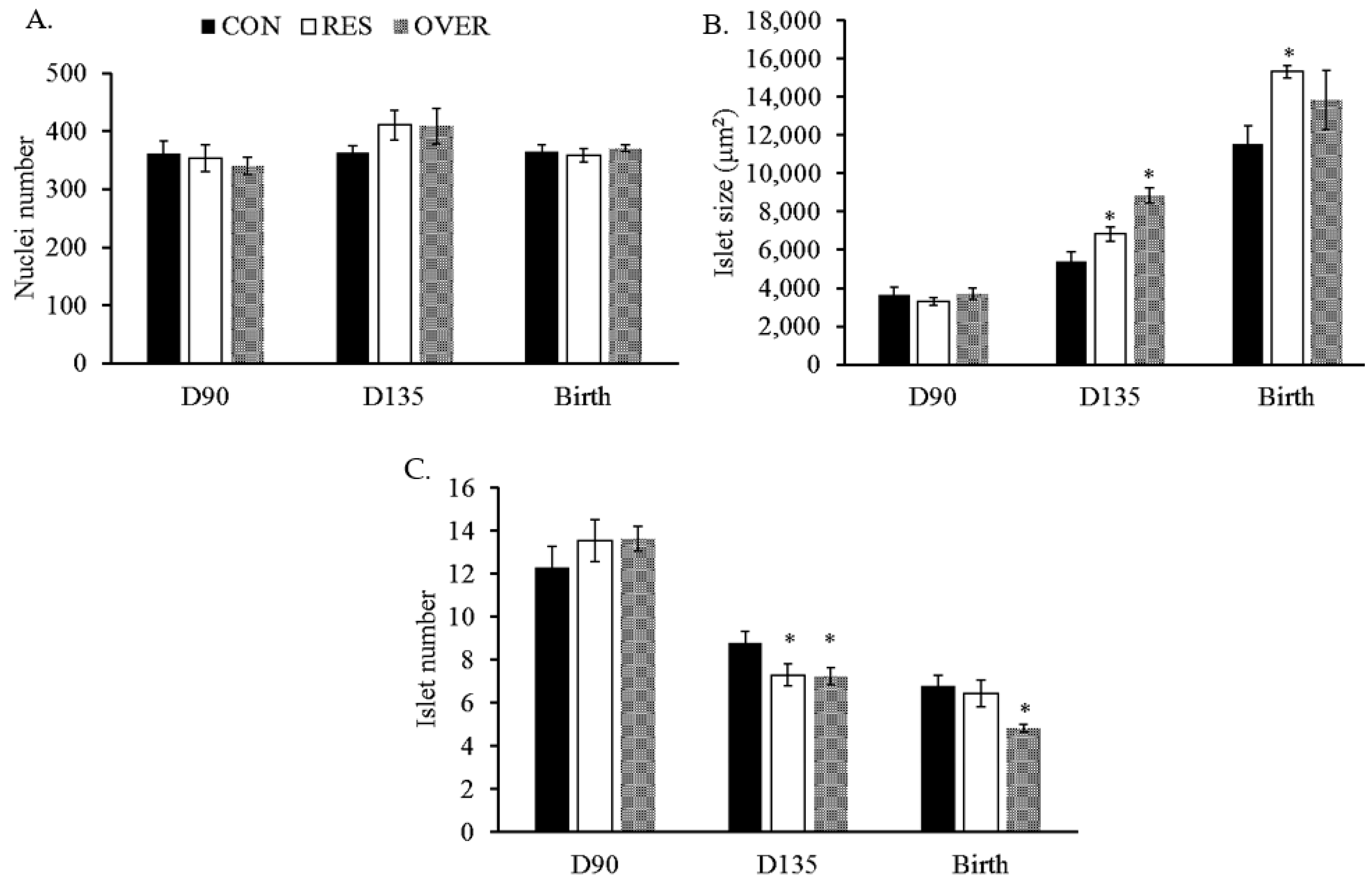
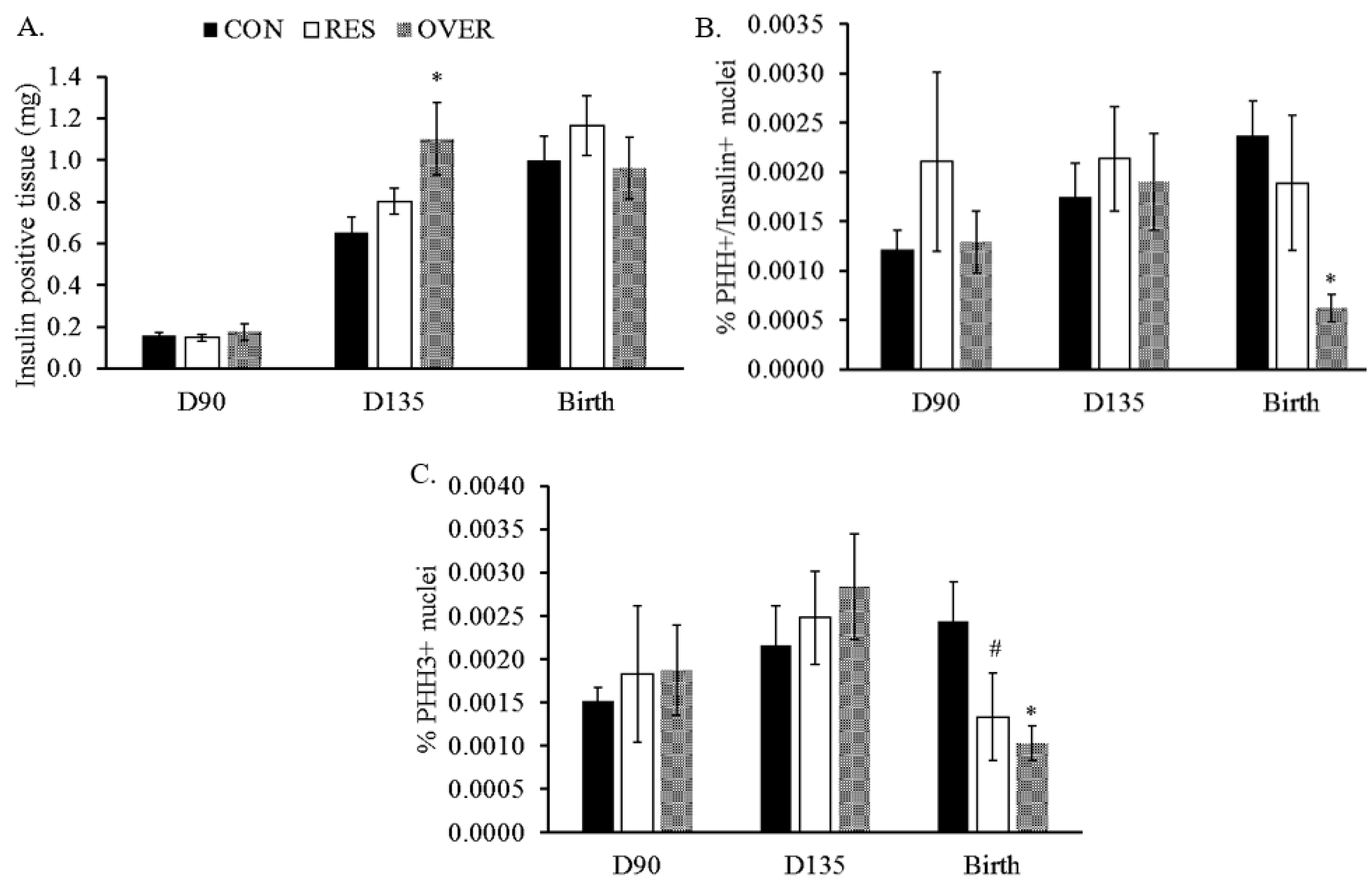
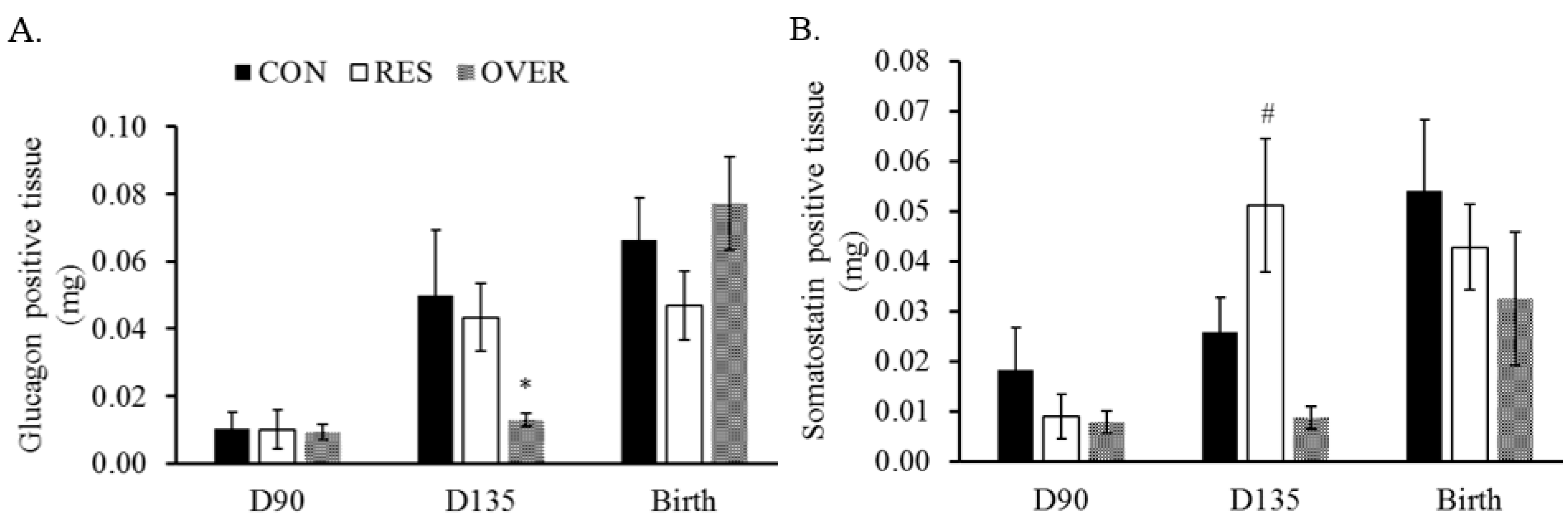
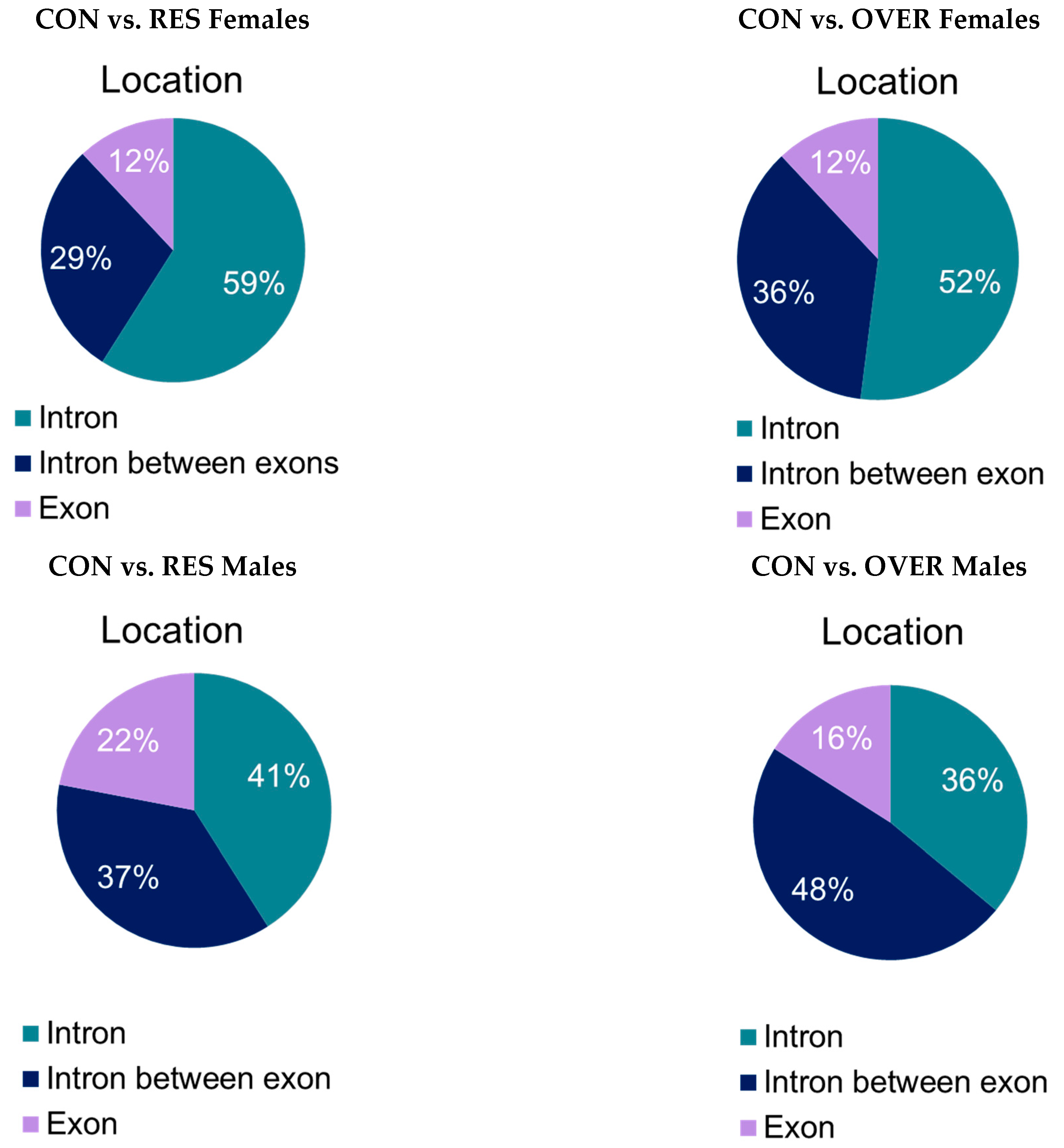
| Treatment | |||
|---|---|---|---|
| CON | RES | OVER | |
| Total # Reads 1 | |||
| Males | 4,999,063 | 5,261,852 | 4,830,722 |
| Females | 7,810,231 | 4,225,227 | 6,023,562 |
| # Reads Post Trimming 2 | |||
| Males | 4,903,981 | 4,174,833 | 3,310,469 |
| Females | 6,844,615 | 4,252,496 | 5,386,296 |
| % Mapping 3 | |||
| Males | 24 | 21 | 26 |
| Females | 44 | 41 | 43 |
| Treatment Comparison | Sex | DML | DMR |
|---|---|---|---|
| CON vs. RES | Female | 2416 | 34 |
| CON vs. RES | Male | 1514 | 64 |
| CON vs. OVER | Female | 1874 | 48 |
| CON vs. OVER | Male | 608 | 25 |
| Gene Name | Molecular Function | Cellular Component |
|---|---|---|
| EFCAB6 | GO:0005509: calcium ion binding | N/A |
| CACNG5 | GO:0005245: voltage-gated calcium channel activity; GO:0016247: channel regulator activity | GO:0032281: AMPA glutamate receptor complex |
| HDLBP | GO:0003723: RNA binding | N/A |
| KBTBD12 | N/A | GO:0031463: Cul3-RING ubiquitin ligase complex |
| MAP3K15 | GO:0004709: MAP kinase kinase kinase activity; GO:0005524: ATP binding | GO:0005622: Intracellular |
| SLC10A5 | GO:0008508: bile acid: sodium symporter activity | GO:0005887: Integral component of plasma membrane |
| TXNDC11 | GO:0003756: protein disulfide isomerase activity | GO:0005623: Cell; GO:0005783: endoplasmic reticulum; GO:0005789: Endoplasmic reticulum membrane; GO:0016021: Integral component of membrane |
| TCN1 | GO:0031419: cobalamin binding | GO:0005615: Extracellular space |
| TMEM55A | GO:0034597: phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity | GO:0005765: Lysosomal membrane; GO:0016021: integral component of membrane; GO:0031902: late endosome membrane |
| WDR48 | N/A | GO:0005634: Nucleus; GO:0005764: lysosome; GO:0005770: Late endosome |
| Gene Name | Molecular Function | Cellular Component |
|---|---|---|
| HMGCL | GO:0000287: magnesium ion binding; GO:0004419: hydroxymethylglutaryl-CoA lyase activity; GO:0005102: receptor binding; GO:0030145: manganese ion binding; GO:0042803: protein homodimerization activity; GO:0046872: metal ion binding | GO:0005743: mitochondrial inner membrane; GO:0005759: mitochondrial matrix; GO:0005777: peroxisome |
| NAPA | GO:0005483: soluble NSF attachment protein activity; GO:0019905: syntaxin binding | GO:0005774: vacuolar membrane; GO:0031201: SNARE complex; GO:0043209: myelin sheath; GO:0070044: synaptobrevin 2-SNAP-25-syntaxin-1a complex; GO:0070062: extracellular exosome |
| SETD3 | GO:0003713: transcription coactivator activity; GO:0042800: histone methyltransferase activity (H3-K4 specific); GO:0046975: histone methyltransferase activity (H3-K36 specific) | GO:0000790: nuclear chromatin |
| TNFRSF19 | GO:0004872: receptor activity | GO:0005886: plasma membrane; GO:0016021: integral component of membrane |
| CHMP4C | N/A | GO:0070062: extracellular exosome; GO:0090543: Flemming body |
| CLDN15 | GO:0005198: structural molecule activity | GO:0005886: plasma membrane; GO:0005923: bicellular tight junction; GO:0016021: integral component of membrane; GO:0016328: lateral plasma membrane |
| HOXC13 | GO:0001077: transcriptional activator activity; RNA polymerase II core promoter proximal region sequence-specific binding; GO:0003682: chromatin binding; GO:004356: sequence-specific DNA binding | GO:0005634: nucleus |
| KMT2C | GO:0008270: zinc ion binding; GO:0042800: histone methyltransferase activity (H3-K4 specific); GO:0044822: poly(A) RNA binding | GO:0044666: MLL3/4 complex |
| LPGAT1 | N/A | GO:0005737: cytoplasm; GO:0016021: integral component of membrane |
| MYOM3 | GO:0016746: transferase activity, transferring acyl groups | GO:0031430: M band |
| MYH11 | GO:0003774: motor activity; GO:0005524: ATP binding; GO:0008307: structural constituent of muscle | GO:0016459: myosin complex; GO:0070062: extracellular exosome |
| PAX5 | GO:0000978: RNA polymerase II core promoter proximal region sequence-specific DNA binding; GO:0001077: transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | GO:0005634: nucleus |
| Gene Name | Molecular Function | Cellular Component |
|---|---|---|
| OPLAH | GO:0005524: ATP binding; GO:0016787: hydrolase activity; GO:0017168: 5-oxoprolinase (ATP-hydrolyzing) activity | GO:0005829: cytosol |
| AGAP2 | GO:0005096: GTPase activator activity; GO:0005525: GTP binding | GO:0005622: intracellular |
| EVL | N/A | GO:0005737: cytoplasm; GO:0005856: cytoskeleton; GO:0005925: focal adhesion; GO:0016020: membrane; GO:0030027: lamellipodium |
| FBXL2 | GO:0004842: ubiquitin-protein transferase activity; GO:0005516~calmodulin binding, GO:0019903~protein phosphatase binding, GO:0036312~phosphatidylinositol 3-kinase regulatory subunit binding, | GO:0016020: membrane; GO:0019005: SCF ubiquitin ligase complex |
| PPARGC1B | GO:0000166: nucleotide binding; GO:0001104:RNA polymerase II transcription cofactor activity, GO:0003676: nucleic acid binding; GO:0008134:transcription factor binding; GO:0030374:ligand-dependent nuclear receptor transcription coactivator activity; GO:0030546: receptor activator activity | GO:0005634: nucleus; GO:0005739: mitochondrion; GO:0016592: mediator complex |
| SEC23B | GO:0008270: zinc ion binding | GO:0000139: Golgi membrane; GO:0005783: endoplasmic reticulum; GO:0005789: endoplasmic reticulum membrane; GO:0005794: Golgi apparatus; GO:0030127: COPII vesicle coat; GO:0033116: endoplasmic reticulum-Golgi intermediate compartment membrane; GO:0048471: perinuclear region of cytoplasm |
| XPC | GO:0003684: damaged DNA binding; GO:0003697: single-stranded DNA binding | GO:0000111: nucleotide-excision repair factor 2 complex; GO:0005730: nucleolus; GO:0005737: cytoplasm; GO:0005886: plasma membrane; GO:0070062: extracellular exosome; GO:0071942: XPC complex |
| CNTNAP5 | N/A | GO:0016021:integral component of membrane |
| DAGLB | GO:0016787: hydrolase activity | GO:0005765: lysosomal membrane; GO:0005886: plasma membrane; GO:0016021: integral component of membrane |
| DUSP16 | GO:0004725: protein tyrosine phosphatase activity; GO:0017017: MAP kinase tyrosine/serine/threonine phosphatase activity | GO:0005654: nucleoplasm; GO:0005737: cytoplasm |
| HOXD3 | GO:0003700: transcription factor activity, sequence-specific DNA binding; GO:0043565: sequence-specific DNA binding | GO:0005654: nucleoplasm; GO:0016235: aggresome |
| IGSF3 | N/A | GO:0005886: plasma membrane; GO:0009986: cell surface; GO:0016021: integral component of membrane |
| ITGB2 | GO:0001948: glycoprotein binding; GO:0005515: protein binding; GO:0019901: protein kinase binding; GO:0030369: ICAM-3 receptor activity; GO:0046872: metal ion binding; GO:0050839: cell adhesion molecule binding | GO:0009986: cell surface; GO:0016020: membrane; GO:0034687: integrin alphaL-beta2 complex; GO:0070062: extracellular exosome |
| ITIH4 | GO:0004867: serine-type endopeptidase inhibitor activity | GO:0005576: extracellular region; GO:0005737: cytoplasm; GO:0005886: plasma membrane; GO:0070062: extracellular exosome; GO:0072562: blood microparticle |
| KAT6A | GO:0003677: DNA binding; GO:0003713: transcription coactivator activity; GO:0004402: histone acetyltransferase activity; GO:0008270: zinc ion binding | GO:0000786: nucleosome; GO:0005794: Golgi apparatus; GO:0016605: PML body; GO:0070776: MOZ/MORF histone acetyltransferase complex |
| PALLD | N/A | GO:0005634: nucleus; GO:0005739: mitochondrion; GO:0005884: actin filament; GO:0005886: plasma membrane; GO:0005925: focal adhesion |
| PLXNA2 | GO:0017154: semaphorin receptor activity | GO:0002116: semaphorin receptor complex; GO:0005887: integral component of plasma membrane |
| SEMA3B | GO:0030215: semaphorin receptor binding; GO:0038191: neuropilin binding; GO:0045499: chemorepellent activity | GO:0005615: extracellular space |
| SEMA5A | GO:0030215: semaphorin receptor binding; GO:0045499: chemorepellent activity | GO:0016021: integral component of membrane; GO:0070062: extracellular exosome |
| TACC2 | N/A | GO:0005730: nucleolus; GO:0005737: cytoplasm; GO:0015630: microtubule cytoskeleton |
| ZDHHC5 | GO:0008270: zinc ion binding; GO:0016409: palmitoyltransferase activity; GO:0019706: protein-cysteine S-palmitoyltransferase activity | GO:0005737: cytoplasm; GO:0005886: plasma membrane; GO:0016021: integral component of membrane; GO:0030425: dendrite |
| ZNF469 | N/A | GO:0003676: nucleic acid binding; GO:0046872: metal ion binding |
| Gene Name | Molecular Function | Cellular Component |
|---|---|---|
| FBXL2 | GO:0004842: ubiquitin-protein transferase activity; GO:0005516: calmodulin binding; GO:0019903: protein phosphatase binding; GO:0036312: phosphatidylinositol 3-kinase regulatory subunit binding | GO:0016020: membrane; GO:0019005: SCF ubiquitin ligase complex |
| MAP7D1 | GO:0005198: structural molecule activity | GO:0015630: microtubule cytoskeleton |
| B3GNT8 | GO:0008378: galactosyltransferase activity; GO:0016262: protein N-acetylglucosaminyltransferase activity | GO:0000139: Golgi membrane; GO:0016021: integral component of membrane; GO:0070062: extracellular exosome |
| ABLIM2 | GO:0008270: zinc ion binding | GO:0015629: actin cytoskeleton |
| EVPL | GO:0005198: structural molecule activity | GO:0001533: cornified envelope; GO:0005737: cytoplasm; GO:0045111: intermediate filament cytoskeleton; GO:0070062: extracellular exosome |
| HAGH | GO:0004416: hydroxyacylglutathione hydrolase activity; GO:0046872: metal ion binding | GO:0005759: mitochondrial matrix; GO:0070062: extracellular exosome |
| ING5 | GO:0003682: chromatin binding; GO:0008270: zinc ion binding | GO:0070776: MOZ/MORF histone acetyltransferase complex |
| OPRL1 | GO:0001626: nociceptin receptor activity; GO:0042923: neuropeptide binding | GO:0005887: integral component of plasma membrane; GO:0043005: neuron projection |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peterson, M.; Gauvin, M.; Pillai, S.; Jones, A.; McFadden, K.; Cameron, K.; Reed, S.; Zinn, S.; Govoni, K. Maternal Under- and Over-Nutrition during Gestation Causes Islet Hypertrophy and Sex-Specific Changes to Pancreas DNA Methylation in Fetal Sheep. Animals 2021, 11, 2531. https://doi.org/10.3390/ani11092531
Peterson M, Gauvin M, Pillai S, Jones A, McFadden K, Cameron K, Reed S, Zinn S, Govoni K. Maternal Under- and Over-Nutrition during Gestation Causes Islet Hypertrophy and Sex-Specific Changes to Pancreas DNA Methylation in Fetal Sheep. Animals. 2021; 11(9):2531. https://doi.org/10.3390/ani11092531
Chicago/Turabian StylePeterson, Maria, Mary Gauvin, Sambhu Pillai, Amanda Jones, Katelyn McFadden, Katelynn Cameron, Sarah Reed, Steven Zinn, and Kristen Govoni. 2021. "Maternal Under- and Over-Nutrition during Gestation Causes Islet Hypertrophy and Sex-Specific Changes to Pancreas DNA Methylation in Fetal Sheep" Animals 11, no. 9: 2531. https://doi.org/10.3390/ani11092531
APA StylePeterson, M., Gauvin, M., Pillai, S., Jones, A., McFadden, K., Cameron, K., Reed, S., Zinn, S., & Govoni, K. (2021). Maternal Under- and Over-Nutrition during Gestation Causes Islet Hypertrophy and Sex-Specific Changes to Pancreas DNA Methylation in Fetal Sheep. Animals, 11(9), 2531. https://doi.org/10.3390/ani11092531





