Simple Summary
Skeletal muscle is the most abundant tissue in fish and the main product of the Chilean salmonid aquaculture industry. Intensive farming conditions generate stress and increased susceptibility to infectious diseases, such as salmonid rickettsial septicemia (SRS), that directly affect this tissue. However, the immunocompetence of skeletal muscle during infection is poorly understood. To further explore the interplay between pathogen infection and stress on this tissue, we analyze the transcriptional profile of isolated rainbow trout (Oncorhynchus mykiss) muscle cells pretreated with 3 h of the stress hormone cortisol, and then infected with the SRS etiologic agent Piscirickettsia salmonis for 8 h, using RNA sequencing technology. For the first time, the obtained data reveals the biological processes related to programmed cell death, negative regulation of cell proliferation, and innate immune response. These results are validated by real-time qPCR. Furthermore, cortisol pretreatment significantly stimulated bacterial gene expression compared to infected cells. These data demonstrated that fish skeletal muscle can activate an intrinsic immune-like response against P. salmonis that is differentially regulated by cortisol. The information provided here will help us to understand the molecular mechanisms of fish muscle cells respond to infection, which could prevent P. salmonis outbreaks in skeletal muscle under stress conditions.
Abstract
Salmonid rickettsial septicemia (SRS) is the major infectious disease of the Chilean salmonid aquaculture industry caused by Piscirickettsia salmonis. Intensive farming conditions generate stress and increased susceptibility to diseases, being skeletal muscle mainly affected. However, the interplay between pathogen infection and stress in muscle is poorly understood. In this study, we perform an RNA-seq analysis on rainbow trout myotubes that are pretreated for 3 h with cortisol (100 ng/mL) and then infected with P. salmonis strain LF-89 for 8 h (MOI 50). Twelve libraries are constructed from RNA samples (n = 3 per group) and sequenced on Illumina HiSeq 4000. A total of 704,979,454 high-quality reads are obtained, with 70.25% mapped against the reference genome. In silico DETs include 175 total genes—124 are upregulated and 51 are downregulated. GO enrichment analysis reveals highly impacted biological processes related to apoptosis, negative regulation of cell proliferation, and innate immune response. These results are validated by RT-qPCR of nine candidate transcripts. Furthermore, cortisol pretreatment significantly stimulated bacterial gene expression of ahpC and 23s compared to infection. In conclusion, for the first time, we describe a transcriptomic response of trout myotubes infected with P. salmonis by inducing apoptosis, downregulating cell proliferation, and intrinsic immune-like response that is differentially regulated by cortisol.
1. Introduction
Piscirickettsiosis, also called salmonid rickettsial septicemia (SRS), is one of the most important infectious diseases affecting the Chilean salmonid aquaculture industry [1]. SRS is caused by Piscirickettsia salmonis, which is a gram-negative bacterium that is aerobic, non-encapsulated, generally non-motile, pleomorphic, but predominately coccoid, and facultative intracellular [2]. Previous studies have revealed that P. salmonis enters immune cells (macrophages) of rainbow trout (Oncorhynchus mykiss) and regulates apoptotic processes [3,4]. Among the mechanisms that P. salmonis uses to infect are secretion system type 4 deficient in organelle trafficking/intracellular multiplication (Dot/Icm), adhesion molecules (type 4 pili), and the inhibition of reactive oxygen species [5]. This septicemic disease affects a variety of salmonid species, such as Atlantic salmon (Salmo salar), Coho salmon (Oncorhynchus kisutch), and rainbow trout [6]. Globally, rainbow trout is a salmonid of high commercial interest for the industry, with its skeletal muscle being the most important tissue in terms of aquaculture products [7].
Currently, the aquaculture industry mainly uses intensive farming conditions that generate stress [8]. Stress initially produces and releases hydrocortisone hormone, better known as “cortisol”, the main neuroendocrine regulator of the stress response [9]. Cortisol triggers metabolic and physiological changes in several processes of fishes, particularly in the immune system [10]. The mechanism of cortisol action is related to its interaction with corticosteroid receptors and regulation of stress-sensitive genes [11]. In fish muscle, this signaling pathway promotes an imbalance between protein synthesis and degradation, leading to skeletal muscle atrophy [12].
Previously, in vivo studies have found evidence of the immune response of skeletal muscle during an infectious process [13,14]. Although muscle was recently suggested to be an immunocompetent organ by itself in fish [15], these data could be masked by infiltration of immune cells in skeletal muscle. To demonstrate this, few studies have performed stimulations with lipopolysaccharide [16] or proinflammatory cytokines [17] in salmonid muscle cells. A recent study demonstrated the in vitro ability of rainbow trout myotubes to implement an inflammatory and antimicrobial response against P. salmonis [18]. In this sense, isolated and differentiated muscle cells, or myotubes, could be better considered to regulate fish muscle physiology against the systemic effects of infection through in vivo studies [19]. This inflammatory response of skeletal muscle to P. salmonis has been further investigated by RNA-seq in an in vivo challenge, revealing interesting details [20]. Moreover, high-throughput mRNA sequencing (RNA-seq) has emerged as an optimal strategy for analyzing global gene expression [21].
In the present work, we performed an RNA-seq analysis in vitro to evaluate whether cortisol-mediated stress produces an effect in rainbow trout myotubes infected with P. salmonis. The data demonstrate that cortisol and/or infection induce transcriptomic changes in fish muscle cells and potentially mediate apoptosis, cell proliferation, and the innate immune response. Finally, cortisol pretreatment differentially stimulated bacterial gene expression in infected myotubes.
2. Materials and Methods
2.1. Primary Culture of Rainbow Trout Myotubes
Healthy P. salmonis free-juvenile rainbow trout (weight 9.4 ± 1.8 g; length 10 ± 2 cm) were obtained from Río Blanco “Federico Albert Taupp” Pisciculture (Los Andes, Valparaíso, Chile). Fishes were reared and used according to a protocol approved by the bioethical committee for animal experiments of Universidad Andrés Bello. Muscle cells were obtained from four rainbow trout (5–8 g). The dorsal white skeletal muscle that was disintegrated in Dulbecco’s modified Eagle’s medium (DMEM, #D7777, Sigma-Aldrich, San Luis, MO, USA) containing 9 mM NaHCO3, 20 mM HEPES, and 10% donor horse serum (#04-004-1A, Biological Industries, Beit HaEmek, Israel) at pH 7.4, with 100 U/mL penicillin and 100 μg/mL streptomycin (#30-004-CI, Corning Inc., Corning, NY, USA). After mechanical dissociation, the muscle was digested with 0.2% collagenase type II (#17101-015, Thermo Fisher, Waltham, MA, USA) in DMEM for 1 h at 18 °C. The suspension was centrifuged at 300× g for 5 min and then digested with 0.1% trypsin-EDTA (#25-053-CI, Corning Inc.) in DMEM for 30 min at 18 °C. After trypsin was deactivated, the suspension was filtered, centrifuged at 500× g for 10 min, and finally resuspended in 5 mL of DMEM. The cell suspension was collected to perform cell counts and to validate viability with Trypan Blue 0.2% solution (#T8164, Sigma-Aldrich).
Muscle cells were seeded in 12-well plates (cell density of 8 × 105 per well) previously treated with poly-l-lysine (2 µg/cm2, #P5899, Sigma-Aldrich) and laminin (20 µg/mL, #L2020, Sigma-Aldrich). First, cells were incubated at 18 °C for 7 days in DMEM, 9 mM NaHCO3, 20 mM HEPES, and 10% fetal bovine serum (#2442, Sigma-Aldrich) at pH 7.4, 100 U/mL penicillin, and 100 μg/mL streptomycin and then differentiated into myotubes by an additional seven days of culture in DMEM, 9 mM NaHCO3, 20 mM HEPES, 2% fetal bovine serum, with 100 U/mL penicillin and 10 mg/mL streptomycin. The procedure to obtain rainbow trout myotubes was repeated three times independently (n = 3).
2.2. Piscirickettsia salmonis Culture
The bacteria P. salmonis strain LF-89 (ATCC VR-1361) were grown as previously reported [22]. Briefly, P. salmonis was cultivated in basal media composed of yeast extract 2.0 g/L, peptone from meat digested 2.0 g/L, (NH4)2SO4 1.32 g/L, MgSO4·7H2O 0.1 g/L, K2HPO4 6.3 g/L, NaCl 9.0 g/L, CaCl2·2H2O 0.08 g/L, and FeSO4·7H2O 0.02 g/L. Then, bacteria were grown with orbital shaking (100 rpm) at 23 °C and monitored using turbidimetry at an optical density (OD) of 600 nm.
2.3. Experimental Design
The concentration and time used for all treatments were previously standardized by [18]. Briefly, myotubes from each plate were divided into the following four experimental groups: (i) Control (CTRL, medium + cortisol vehicle), (ii) cortisol treatment (CORT), (iii) P. salmonis infection (INF), and (iv) cells pretreated with cortisol and then infected with bacteria (C+INF). Before treatments, myotubes were washed once with 1X PBS (phosphate-buffered saline) at pH 7.4. First, myotubes were preincubated with cortisol (100 ng/mL, #H0888, Sigma-Aldrich) or cortisol vehicle (DMSO) in DMEM, 9 mM NaHCO3, and 20 mM HEPES for 3 h at 18 °C. In parallel, P. salmonis was quantified by measuring the OD 600 nm to ensure it was between 0.3 and 0.6 (logarithmic phase) using the basal medium, and the McFarland formula was applied [23,24]. Then, bacteria were centrifuged at 5000× g for 30 min, the supernatant was discarded, and the cells were resuspended in cell medium. After 3 h of incubation with cortisol or vehicle, the medium was changed. The myotubes were infected with P. salmonis at a multiplicity of infection (MOI) of 50 or medium without bacteria for 8 h at 18 °C.
2.4. Immunostaining
Immunostaining for P. salmonis and rainbow trout myotubes was performed to detect the subcellular location in fish muscle cells. For this, bacteria were detected with a FITC-conjugated antibody (in green), and muscle cells were visualized with an actin cytoskeleton marker (in red) and nuclear staining (in blue). Briefly, myotubes were cultured with a 12 mm coverslip (8 × 105 cells/well) and treated as previously mentioned. Then, the cells were washed with 1X PBS and fixed with 4% paraformaldehyde (diluted with 1X PBS) for 10 min at room temperature (RT). After three washes, cells were permeabilized with 0.1% BSA (bovine serum albumin)–3% Triton X-100 (diluted with 1X PBS) for 30 min at RT. Fixed and permeabilized cells were incubated with anti-P. salmonis primary antibody (#D-FP-SRS-IF, Ango, Santiago, Chile) diluted 1:100 in 3% BSA-1X PBS for 1 h at RT in a humidified chamber. After another three washes, the cells were incubated with FITC-conjugated secondary antibodies against anti-P. salmonis (#D-FP-SRS-IF, Ango) and anti-phalloidin (#A22287, Thermo Fisher) conjugated with Alexa Fluor® 647. Both antibodies were diluted 1:200 in 3% BSA-1X PBS for 1 h at RT in a humidified chamber. Three more washes were performed, and incubation with DAPI solution (#62248, Thermo Fisher) was performed at a dilution of 1:1000 for 10 min at RT in a humidified chamber. Finally, the cells were washed once more with 1X PBS, and the coverslips were mounted with Vectashield Antifade Mounting Media (#H-1000, Vector lab., Burlingame, CA, USA) for confocal microscopy analysis.
2.5. Confocal Microscopy Analysis
The acquisition settings were the same in all treatments for the three independent plates. Images were taken using a TCS SP8 confocal fluorescence microscope (Leica, Wetzlar, Germany) with numerical aperture 1.4 × 100 oil objective, ×2 digital zoom (NA = 1.4; HC PL APO CS2 100×) and a z-step of 1 μm optical sections (velocity scan 600 Hz; resolution 1024 × 1024 pixels, equivalent to 58.13 μm × 58.13 μm). The following three laser wavelengths were used for DAPI (Ex 405 nm and Em 410–483 nm), FITC (Ex 488 nm and Em 502–621 nm), and Alexa Fluor® 647 (Ex 638 nm and Em 650–776 nm), and signals were detected with an ultrahigh dynamic photomultiplier (PMT) spectral detector. Maximum intensity projections of confocal z-stack images of P. salmonis in whole nuclei (containing 17–22 stacks with resolution equivalent to 4.81–6.21 μm) were analyzed. All pictures and three-dimensional (3D) reconstructions of myotubes were performed using Leica Application Suite X v.3.5.5 software (Leica).
2.6. Library Construction and Sequencing
Myotubes were cultured in 12-well plates (8 × 105 cells/well), and they were treated as previously mentioned. The RNA from myotubes was extracted using E.Z.N.A® Total RNA kit (#R6834, Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s recommendations. The RNA collected was measured by fluorometry using the Qubit RNA BR assay kit (#Q10211, Thermo Fisher). Then, a capillary electrophoresis Fragment Analyzer™ Automated CE System (Advanced Analytical Technologies, Inc., Ames, IA, USA), was used to confirm the RNA integrity. RNA samples with RNA quality numbers (RQN) ≥ 8.5 were selected. A total of 12 cDNA libraries corresponding to 1 µg of RNA from three biological replicates (n = 3) of each condition (CTRL, CORT, INF, and C+INF) were generated using TruSeq™ RNA Sample Preparation kit v2, Set A (#RS-122-2001, Illumina, San Diego, CA, USA), according to the manufacturer’s protocol. Then, libraries were quantified using the KAPA Library Quantification Kit (#KK4824, Roche, Basilea, Switzerland), and their integrity was analyzed by a fragment analyzer. All libraries were sequenced using a paired-end strategy (2 × 100 bp) with the Illumina HiSeq 4000 platform by the Macrogen sequencing company (Seoul, South Korea). Raw read sequences were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra; accessed date 31 May 2021) under the accession number PRJNA732666.
2.7. Raw Data Processing and RNA Sequencing Analysis
The transcriptional response of rainbow trout myotubes associated with CORT or C+INF was determined by RNA-seq after the previously mentioned treatments. Additionally, CTRL and INF, but without cortisol, were included as controls during the assay. All RNA-seq analyses were performed according to [25], with minor modifications. Briefly, raw reads were visualized using CLC Genomic Workbench 9.0 software. Then, the adapters, low-quality reads (Q < 30), and reads length <50 bp were discarded. High-quality reads were mapped onto the reference rainbow trout genome (Omyk_1.0, RefSeq assembly accession: GCF_002163495.1) which consists of 71,413-coding sequence (CDS) [26] using default mapping parameters: Mismatches = 2, minimum fraction length = 0.9, minimum fraction similarity = 0.8, and maximum hits per read = 5.
The unique mapped reads were used for differential expression analysis using empirical analysis of DGE incorporated in the CLC platform. Transcripts with absolute fold-change values > 2.0 and a false discovery rate (FDR)-corrected p-value < 0.05 were considered differentially expressed transcripts (DETs).
2.8. Functional Annotation and Gene Ontology Analysis
Based on the DETs, an ontological enrichment analysis was performed to obtain an overall view of the main biological processes, cellular components, molecular functions, and KEGG pathways that were affected during the trial.
Functional annotation was performed to obtain the gene ontology (GO) ID of each transcript, and a search against different fish databases, including rainbow trout, Atlantic salmon, Coho salmon, zebrafish (Danio rerio), and Atlantic cod (Gadus morhua), was performed with the BLASTx tool and the UniProt database (https://www.uniprot.org/; accessed date 6 December 2020). The enrichment analysis was performed by the DAVID-GO v.6.8 tool (https://david.ncifcrf.gov/; accessed date 23 January 2021) with the gene IDs of transcripts that were differentially expressed [27]. Standard settings for the DAVID analysis were gene count: 2 and EASE score: 1. The cut-off for the modified Fisher exact p-value was 3.30 × 10−1. Then, the resulting GO analysis was plotted in bubble charts.
2.9. Real-Time PCR Validation and Bacterial Gene Expression
The gene expression values in silico involved in enriched biological processes were validated by real-time PCR assays (RT-qPCR). Validation was performed with genes involved in the immune response (nod1, adam15, ikba), apoptosis (hip1r, s30bl, rbl1), and growth (arhgap32, mprip, mylk3).
RNA was extracted and quantified as previously mentioned, and 1 μg was reverse transcribed to cDNA using the QuantiTect® reverse transcription kit (#205313, Qiagen, Hilden, Germany). The primers used to amplify the candidate genes were designed using Primer3 (https://primer3.ut.ee/; accessed date 3 February 2021), validated with NetPrimer (http://www.premierbiosoft.com/netprimer/; accessed date 3 February 2021), and synthesized by Integrated DNA Technologies, Inc. (San José, CA, USA).
An Mx3000P qPCR system (Stratagene, San Diego, CA, USA) was used for all RT-qPCR. The reactions contained 7.3 µL of 2X Brilliant II SYBR® Green qPCR Master Mix (#600828, Agilent Technologies, Santa Clara, CA, USA), 0.2 µL of ROX reference dye (5 µM), 0.75 µL of each primer (250 nM) and 6 µL of cDNA (200 ng) in a 15 µL final volume. Furthermore, all RT-qPCR assays were performed using triplicates, no-reverse transcriptase control, and a no-template control. The list of primers used in this study is listed in Supplementary Table S1. Amplifications were performed with the following thermal cycling conditions: Initial activation at 95 °C for 10 min, followed by 40 cycles of 30 s at 95 °C, 30 s of Tm, and 30 s at 72 °C. A dissociation curve was generated to confirm the generation of a single PCR product. Gene expression analysis was performed using the geNorm program [28], combining the geometric average of two stable reference genes: 40S ribosomal protein 30S (fau) and β-actin (actb).

Table 1.
The number of differentially expressed transcripts (DETs) in rainbow trout myotubes treated with cortisol and/or infected with P. salmonis. The table showed the number of total DETs, upregulated, and downregulated between all comparisons. Abbreviations: CTRL, control; CORT, cortisol; INF, infection; C+INF, cortisol+infection.
Additionally, the expression profiles of P. salmonis genes were analyzed to determine differences between infected and cortisol pretreated rainbow trout myotubes. RT-qPCR of pathogenesis-related genes of P. salmonis, including Dot/Icm (dotA, dotB, icmE, icmK), type 4 pili (pilA, pilQ), thiol-specific peroxidase (ahpC), and bacterial load (23s) was performed with total RNA from myotubes infected with P. salmonis (INF) and myotubes preincubated with cortisol and then infected with P. salmonis (C+INF). For this, the primer sequences used were obtained as follows: Dot/Icm [29], type 4 pili [30], ahpC [31], and 23s [32]. The succinate dehydrogenase A subunit (sdhA) and DNA gyrase B subunit (gyrB) genes were used as normalizing genes, which were previously standardized by [33].
2.10. Statistical Analysis
The data are expressed as the mean ± standard error of the mean (SEM). Differences in means between groups were determined using one-way ANOVA followed by Tukey’s honestly significant difference posttest (HSD). Correlations between RNA-seq and RT-qPCR data were assessed through multiple linear regressions using Pearson correlation (r) and p-values. Data were accepted as significant at a p-value < 0.05. All statistical analyses were performed using GraphPad Prism v.9.0.
3. Results
3.1. Intracellular Location of P. salmonis between Rainbow Trout Myotubes with Cortisol and/or P. salmonis
The immunostaining for P. salmonis in rainbow trout myotubes was performed to detect its subcellular location. The images and 3D reconstructions of both INF (Figure 1a) and C+INF (Figure 1b) indicated an intracellular location of bacteria (in green) in muscle cells (in red) near the nuclei (in blue). No P. salmonis was detected in the CTRL and CORT groups (Supplementary Figure S1). The isolated images for all conditions are available in Supplementary Figure S1.
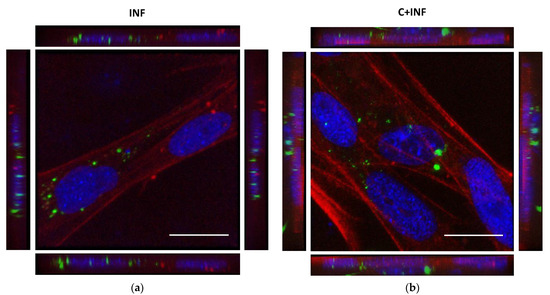
Figure 1.
Intracellular location of P. salmonis between rainbow trout myotubes infection and cortisol pretreatment. Representative confocal images of the immunofluorescence staining of P. salmonis (green) and phalloidin (actin cytoskeleton, red). (a) Primary culture of rainbow trout skeletal muscle cells infected with P. salmonis strain LF-89 (MOI 50) for 8 h (INF). (b) Rainbow trout myotubes pretreated with 3 h of cortisol (100 ng/mL) followed by infection with P. salmonis (C+INF). Nuclei were stained with DAPI (blue). The images represent a maximum projection for the total nuclear volume. Scale bar: 20 µm.
3.2. Transcriptomic Responses of Rainbow Trout Muscle Cells with Cortisol and/or P. salmonis
A total of 776,965,304 million readings were obtained from 12 cDNA libraries (Supplementary Table S2). After adapters were removed and low-quality readings were discarded, 704,979,454 high-quality readings were obtained (Supplementary Table S2). A total of 468,598,883 high-quality readings (70.25%) were specifically mapped against the 71,413 CDSs of the rainbow trout genome (Supplementary Table S3).
Differential expression analysis in silico was performed by specifically mapping the readings against a reference genome. A table was generated, indicating a total of 175 differentially expressed transcripts (DETs) for all comparisons and 124 upregulated and 51 downregulated genes (Table 1). In addition, the CTRL vs. INF presented 1 was unique, and 12 were shared DETs. For CTRL vs. CORT, 6 DETs were unique, and 40 were shared. For CTRL vs. C+INF, 11 were unique, and 27 were shared DETs. For CORT vs. INF, 6 were unique, and 33 were shared DETs. For INF vs. C+INF, 6 was unique, and 24 were shared DETs. Finally, CORT vs. C+INF no presented unique DETs, and 9 were shared.
3.3. Gene Ontology Enrichment Analysis of Treated Rainbow Trout Myotubes
In the biological process component of GO analysis (Figure 2), the CTRL vs. CORT group presented the following GO terms: Activation of cysteine-type endopeptidase activity involved in the apoptotic process (or caspase activation), negative regulation of cell migration, apoptotic process, and innate immune response.
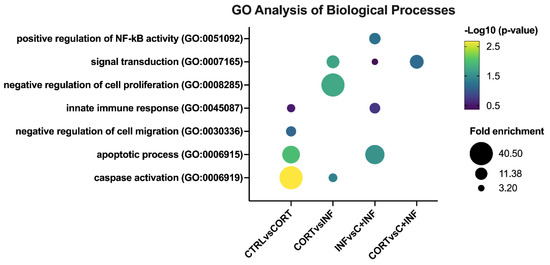
Figure 2.
GO analysis of biological processes in rainbow trout myotubes treated with cortisol and/or infected with P. salmonis. The most representative and significant biological processes are represented in bubble plots and are sorted by group. The dot size indicates the fold enrichment associated with the process, and the dot color indicates the significance of the enrichment (−log10(the modified Fisher exact p-value)). Abbreviations: CTRL, control; CORT, cortisol; INF, infection; C+INF, cortisol+infection.
When comparing the CORT vs. INF groups, signal transduction, caspase activation, and negative regulation of cell proliferation were detected. In the comparison of the INF vs. C+INF group, the main biological processes were positive regulation of NF-kappaB transcription factor activity, signal transduction, apoptotic process, and innate immune response. However, no enrichment of biological processes for the CTRL vs. INF and CTRL vs. C+INF comparisons was observed. In the CORT vs. C+INF groups, only signal transduction was detected.
Regarding the cellular component of the GO analysis (Supplementary Figure S2), intracellular membrane-bound organelle, autophagosome, and cytosol were the most enriched terms in the CTRL vs. CORT and CORT vs. INF groups. Then, in the INF vs. C+INF groups, cytosol, endoplasmic reticulum membrane, and plasma membrane were detected. In the CTRL vs. INF groups, adherens junctions and cytosol were observed. In the CTRL vs. C+INF groups, the cellular components identified were early endosome membrane, endoplasmic reticulum membrane plasma membrane, and cytosol. Cytosol was enriched when comparing the CORT vs. C+INF groups.
The molecular function GO analysis revealed that phosphatidylinositol-3,4-bisphosphate binding, phosphatidylinositol-3,4,5-triphosphate binding, protein binding, ATP binding, metal ion binding, and protein binding were the most enriched terms in the CTRL vs. CORT groups (Supplementary Figure S3). When comparing the CORT vs. INF groups, the GO terms were ATP binding and protein binding. In the INF vs. C+INF group, protein binding and heat shock protein binding were detected. In the CTRL vs. INF groups, only protein binding was identified. Phosphatidylinositol-3-3,4,5-trisphosphate, phosphatidylinositol-3,4-biphosphate binding, and protein binding were detected in the CTRL vs. C+INF groups. Then, in the CORT vs. C+INF groups, only protein binding was detected. Finally, no enriched KEGG pathways were detected for any comparison.
3.4. Validation of RNA Sequencing Data by Real-Time PCR and Bacterial Gene Expression
The DETs from enriched biological processes of immune response (innate immune response (GO:0045087) and positive regulation of NF-kappaB transcription factor (GO:0051092)), apoptosis (caspase activation (GO:0006919) and apoptosis process (GO:0006915)), and growth (negative regulation of cell migration (GO:0030336) and negative regulation of cell proliferation (GO:0008285)) were identified in RNA-seq analysis. From these DETs, the top three in fold change were selected for RT-PCR. Figure 3a indicates the results of the validation in the experimental group infected with P. salmonis. Figure 3b displays the data for the group treated with cortisol. Figure 3c shows the validation obtained after pretreatment with cortisol and then infection with P. salmonis.
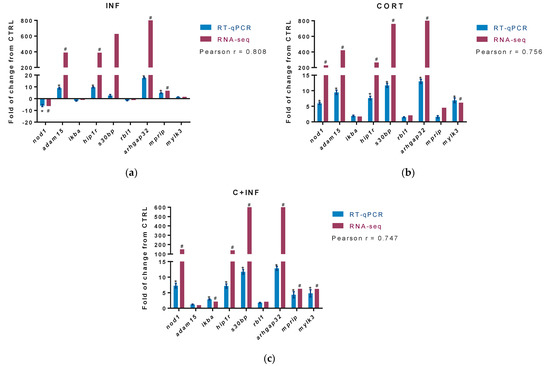
Figure 3.
RT-qPCR validation of differentially expressed transcripts. The potential target genes selected for RT-qPCR validation of RNA-seq were nod1, adam15, ikba, hip1r, s30bl, rbl1, arhgap32, mprip, and mylk3. For RNA-seq, “#” indicates fold-change (absolute values > 2.0) and FDR corrected p-value (p < 0.05). (a) Validation between CTRL vs. INF. (b) Validation between CTRL vs. CORT. (c) Validation between CTRL vs. C+INF. For RT-qPCR, relative expression was normalized against fau and actb, and “*” indicates significant differences from the control (mean ± SEM, n = 3, p < 0.05). Pearson correlation (r) between RNA-seq and qPCR are also indicated (p < 0.05). Abbreviations: CTRL, control; CORT, cortisol; INF, infection; C+INF, cortisol+infection; nod1, Nucleotide-binding oligomerization domain-containing protein 1; adam15, Disintegrin and metalloproteinase domain-containing protein 15; ikba, NF-kappa B inhibitor alpha; hip1r, Huntingtin-interacting protein 1-related protein; s30bp, SAP30-binding protein; rbl1, Retinoblastoma-like protein 1; arhgap32, Rho GTPase activating protein 32; mprip, myosin phosphatase Rho interacting protein; mylk3, myosin light chain kinase 3; fau, 40S ribosomal protein S30; actb, β-actin.
Our results present high correlations in the group infected with P. salmonis (r = 0.808), in the group treated with cortisol (r = 0.756), and in the group pretreated with cortisol and infected with P. salmonis (r = 0.747), between the expression values of those candidate genes used in the RNA-seq and RT-qPCR.
In the mRNA levels of P.salmonis genes, no expression was detected in the CTRL and CORT groups. The results indicate the significant upregulation of the ahpC and 23s genes in the C+INF group compared to the INF group (Figure 4). No changes were observed in the other evaluated genes.
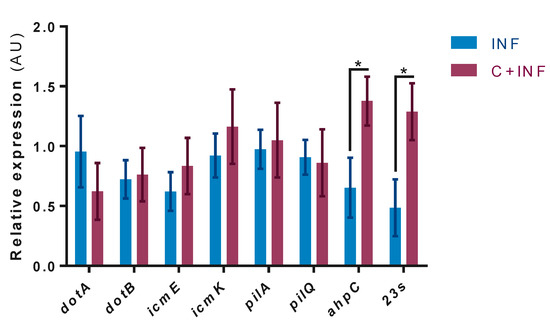
Figure 4.
Bacterial gene expression between infected myotubes with cortisol pretreatment. Pathogenesis-related gene expression by RT-qPCR of P. salmonis: Dot/Icm (dotA, dotB, icmE, icmK), type 4 pili (pilA, pilQ), thiol-specific peroxidase (ahpC), and bacterial load (23s). Data were normalized against sdhA and gyrB and are expressed in arbitrary units (AU) as the mean ± SEM (n = 3). The “*” indicates significant differences between INF vs. C+INF (p < 0.05). Abbreviations: INF, infection; C+INF, cortisol+infection; dotA, deficient in organelle trafficking A; dotB, deficient in organelle trafficking B; icmE, intracellular multiplication E; icmK, intracellular multiplication K; ahpC, alkyl hydroperoxide reductase C; sdhA, succinate dehydrogenase A subunit; gyrB, DNA gyrase B subunit.
4. Discussion
RNA-seq is widely used to evaluate the transcriptomic immune response during several fish pathogen infections in vivo [34,35], but few studies have evaluated this transcriptomic response in vitro [36,37]. Here, we performed RNA-seq analysis to evaluate the effects of cortisol on rainbow trout myotubes infected with P. salmonis. The quality obtained from RNA-seq data was as expected, and the mapping results against the rainbow trout reference genome were in line with studies using this strategy [25,38]. According to these results, other recent reports in trout presented similar results using comparable parameters of RNA-seq analysis [39,40].
In myotubes treated with cortisol and/or infected with P. salmonis, GO enrichment analysis revealed innate immune response, apoptotic process, and negative regulation of cell proliferation as the most impacted biological processes. A recent study of RNA-seq on salmonid immune cells infected with P. salmonis observed similar processes [36], which could indicate that muscle cells respond similarly to infection. This is also consistent with the observations in skeletal muscle of salmon infected with these bacteria [41], and in in vitro and in vivo infections of rainbow trout skeletal muscle with P. salmonis LF-89 strains [18,20].
For validation, the selected genes involved in the immune response were nod1, adam15, and ikba. Nucleotide-binding oligomerization domain-containing protein 1 (nod1) is a cytosolic receptor that recognizes intracellular pathogen-associated molecular patterns via nuclear factor-kappa B (NF-kappaB), promotes proinflammatory cytokine transcription, and activates apoptosis by inflammatory caspases [42,43]. Disintegrin and metalloproteinase domain-containing protein 15 (adam15) is a transmembrane glycoprotein involved in immune cell recruitment to inflammation sites, among other processes [44,45]. Interestingly, no changes were detected in myotubes infected with cortisol pretreatment compared to infection or cortisol. This could suggest immunomodulation of cortisol during infection. During infection and proinflammatory responses, NF-kappaB inhibitor alpha (ikba) becomes phosphorylated, promoting ubiquitination and degradation, enabling NF-kB to translocate to the nucleus and activate the transcription of immune-related genes [46]. The immune response could be explained in part by immune cells detected in skeletal muscle [47], but notably, in this study, there was no infiltration of immune cells. Hence, our results suggest an intrinsic, innate immunity of myotubes during infection. Stress can modulate the immune response in fish [10], and we also detected differential regulation by cortisol. This is reflected by the apoptotic process enrichment observed during trials, for which the hip1r, s30bp, and rbl1 genes were selected for validation. Huntingtin-interacting protein 1-related protein (hip1r) is a component of the endocytic machinery that interacts with the actin cytoskeleton and is involved in the intrinsic pathway of apoptosis [48]. The SAP30-binding protein (s30bp) is a member of the Sin3-associate polypeptide (SAP) complex, which regulates apoptosis by transcriptional corepressor of cell survival-related genes [49]. Retinoblastoma-like protein 1 (rbl1) is a key regulator of entry into cell division that acts as a transcription coactivator of glucocorticoid receptor-induced apoptosis [50]. These results are consistent with previous reports, revealing that cortisol-mediated stress can also regulate apoptosis and during infection. Most likely, P. salmonis could either interfere with proteins that inhibit the apoptosis process or activate intrinsic programmed cell death. Supporting these data, in rainbow trout myoblasts infected with Flavobacterium psychrophilum, apoptosis was induced through NF-κB signaling as a mechanism for nutrient acquisition and survival [51]. However, further analyses are required to comprehend the mechanisms by which this bacterium induces apoptosis.
Growth-related genes (arhgap32, mprip, mylk3) were evaluated because a negative regulation of cell proliferation was detected. Rho GTPase activating protein 32 (arhgap32) induces GTP hydrolysis of GTPases that control actin assembly of the cytoskeleton in cell growth [52]. Myosin phosphatase Rho interacting protein (mprip) binds to myosin phosphatase of actin fibers and regulates the cytoskeletal structure of muscle cells [53]. Myosin light chain kinase 3 (mylk3) encodes a protein kinase that phosphorylates myosin, promotes sarcomere formation, and increases the contractility of muscle [54]. It has been described that cortisol regulates muscle growth [55], but there is no information during infection in vitro. We found that perhaps either an imbalance of the proliferation/differentiation rates or alteration of the cytoskeleton were involved. During myogenesis in fish, a balance between proliferation and differentiation in muscle cells is necessary for optimal growth [56]. When one of these processes is affected, the other is compensated. In this case, a downregulation of proliferation occurred and was then compensated by an increase in differentiation. Previous studies have described that P. salmonis can affect the cytoskeleton during infection [57,58]. Supporting this, a decreased pattern of the actin cytoskeleton was presented following the immunostaining of infected rainbow trout myotubes compared to cortisol pretreated myotubes, indicating that P. salmonis could decrease the expression levels of genes related to the cytoskeleton structure of rainbow trout myotubes that were upregulated by cortisol.
Finally, we analyzed the expression of pathogenesis-related genes to determine whether cortisol has a synergistic effect. Surprisingly, two genes were upregulated: Large-subunit ribosomal (23s), a marker for bacterial load, and alkyl hydroperoxide reductase C (ahpC). The immunostaining of P. salmonis presented a similar pattern of bacterial abundance, which is consistent with the mRNA levels of 23s. Otherwise, thiol-specific peroxidase catalyzes the reduction of hydrogen peroxide and organic hydroperoxides to water and alcohols, respectively [59]. This mechanism protects the bacterium against oxidative stress generated by the host’s immune cells, contributing to pathogenesis [60]. This is especially interesting because it supports the immune response of muscle cells. In contrast, a transcriptomic profile of P. salmonis infecting SHK-1 cells alternated between replicative and stationary phases that activated a stringent response [37]. Additionally, dual RNA-seq between P. salmonis and Atlantic salmon spleen/head kidney indicated metabolic amino acid dependency [61]. These data suggest that this bacterium could also uptake nutrients from skeletal muscle. Together, stress-induced cortisol production could negatively affect muscle cells and promote pathogenesis during infection.
5. Conclusions
In summary, for the first time, we described a transcriptomic response of myotubes treated with cortisol and/or infected with P. salmonis by inducing an innate immune response, apoptosis, and negative regulation of cell proliferation. Additionally, cortisol pretreatment differentially stimulated the bacterial gene expression of ahpC and 23s in infected myotubes. Although few DETs were detected, these had significant changes in magnitude and were related to key processes for muscle growth and immunity. Our data suggest that fish muscle cells have an intrinsic immune response that is differentially regulated by cortisol. The information provided here will help us to understand the molecular mechanisms of fish muscle cells respond to infection, which could lead to pathogen outbreaks in skeletal muscle under stress conditions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani11082399/s1, Supplementary Figure S1: Immunofluorescence of rainbow trout myotubes pretreated with cortisol and/or infected with P. salmonis, Supplementary Figure S2: GO analysis of cellular components in rainbow trout myotubes treated with cortisol and/or infected with P. salmonis, Supplementary Figure S3: GO analysis of molecular functions in rainbow trout myotubes treated with cortisol and/or infected with P. salmonis, Supplementary Table S1: List of primers, amplicon size, and annealing temperature used in the RT-qPCR validation, Supplementary Table S2. Sequencing data obtained by RNA-Seq with an Illumina HiSeq 4000. Supplementary Table S3: Supplementary Table S3. Mapping data.
Author Contributions
Conceptualization, R.Z. and A.M.; methodology, R.Z., P.D., and M.B.-M.; software, R.Z. and P.D.; validation, R.Z., P.D., and M.B.-M.; formal analysis, R.Z.; investigation, A.M.; resources, C.M., C.A., J.A.V., and A.M.; data curation, R.Z. and P.D.; writing—original draft preparation, R.Z.; writing—review and editing, R.Z., C.M., C.A., J.A.V., and A.M.; visualization, R.Z., J.A.V., and A.M.; supervision, J.A.V. and A.M.; project administration, R.Z., J.A.V., and A.M.; funding acquisition, J.A.V. and A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agencia Nacional de Investigación y Desarrollo (ANID, Chile), grant number FONDECYT 1171307, (to Alfredo Molina), FONDECYT 1201498 (to Juan Antonio Valdés) and Fondo de Financiamiento de Centros de Investigación en Áreas Prioritarias (FONDAP) grant number INCAR 15110027.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, approved by the Agencia Nacional de Investigación y Desarrollo (ANID, Chile, 1171307, 2017), and the Ethics Committee for Animal Experiments of the Universidad Andres Bello (N° 006/2017). The hatchery of rainbow trout obtained from Río Blanco “Federico Albert Taupp” Pisciculture (Los Andes, Valparaíso, Chile) has been certified of being free of pathogens included in List 1, 2, and 3 of the Chilean sanitary regulations.
Informed Consent Statement
Not applicable.
Data Availability Statement
The commercial antibodies and reagents used in this study were listed in the Material and Methods section. The nucleotide sequences used in this study were collected from the National Center for Biotechnology Information (NCBI) GenBank repository. Raw read sequences obtained from sequencing were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra; accessed date 31 May 2021) under the accession number PRJNA732666. The datasets generated and/or analyzed during the current study are not publicly available due to privacy or ethical restrictions but are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Rozas, M.; Enríquez, R. Piscirickettsiosis and Piscirickettsia salmonis in fish: A review. J. Fish Dis. 2014, 37, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Makrinos, D.L.; Bowden, T.J. Growth characteristics of the intracellular pathogen, Piscirickettsia salmonis, in tissue culture and cell-free media. J. Fish Dis. 2017, 40, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, Ú.M.; Bron, J.E.; Brown, L.; Pourahmad, F.; Bricknell, I.R.; Thompson, K.D.; Adams, A.; Ellis, A.E. Survival and replication of Piscirickettsia salmonis in rainbow trout head kidney macrophages. Fish Shellfish. Immunol. 2008, 25, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Rojas, V.; Galanti, N.; Bols, N.C.; Jiménez, V.; Paredes, R.; Marshall, S.H. Piscirickettsia salmonis induces apoptosis in macrophages and monocyte-like cells from rainbow trout. J. Cell. Biochem. 2010, 110, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Machuca, A.; Martinez, V. Transcriptome analysis of the intracellular facultative pathogen Piscirickettsia salmonis: Expression of putative groups of genes associated with virulence and iron metabolism. PLoS ONE 2016, 11, e0168855. [Google Scholar] [CrossRef]
- Figueroa, J.; Cárcamo, J.; Yañez, A.; Olavarria, V.; Ruiz, P.; Manríquez, R.; Muñoz, C.; Romero, A.; Avendaño-Herrera, R. Addressing viral and bacterial threats to salmon farming in Chile: Historical contexts and perspectives for management and control. Rev. Aquac. 2019, 11, 299–324. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2018—Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018. [Google Scholar]
- Ashley, P.J. Fish welfare: Current issues in aquaculture. Appl. Anim. Behav. Sci. 2007, 104, 199–235. [Google Scholar] [CrossRef]
- Sadoul, B.; Geffroy, B. Measuring cortisol, the major stress hormone in fishes. J. Fish Biol. 2019, 94, 540–555. [Google Scholar] [CrossRef]
- Tort, L. Stress and immune modulation in fish. Dev. Comp. Immunol. 2011, 35, 1366–1375. [Google Scholar] [CrossRef]
- Aluru, N.; Vijayan, M.M. Stress transcriptomics in fish: A role for genomic cortisol signaling. Gen. Comp. Endocrinol. 2009, 164, 142–150. [Google Scholar] [CrossRef]
- Valenzuela, C.A.; Ponce, C.; Zuloaga, R.; González, P.; Avendaño-Herrera, R.; Valdés, J.A.; Molina, A. Effects of crowding on the three main proteolytic mechanism of skeletal muscle in rainbow trout (Oncorhynchus mykiss). BMC Vet. Res. 2020, 16, 294. [Google Scholar] [CrossRef]
- Marino, M.; Scuderi, F.; Provenzano, C.; Bartoccioni, E. Skeletal muscle cells: From local inflammatory response to active immunity. Gene Ther. 2011, 18, 109–116. [Google Scholar] [CrossRef]
- Wiendl, H.; Hohlfeld, R.; Kieseier, B.C. Immunobiology of muscle: Advances in understanding an immunological microenvironment. Trends Immunol. 2005, 26, 373–380. [Google Scholar] [CrossRef]
- Valenzuela, C.A.; Zuloaga, R.; Poblete-Morales, M.; Vera-Tobar, T.; Mercado, L.; Avendaño-Herrera, R.; Valdés, J.A.; Molina, A. Fish skeletal muscle tissue is an important focus of immune reactions during pathogen infection. Dev. Comp. Immunol. 2017, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Aedo, J.E.; Reyes, A.E.; Avendaño-Herrera, R.; Molina, A.; Valdés, J.A. Bacterial lipopolysaccharide induces rainbow trout myotube atrophy via Akt/FoxO1/Atrogin-1 signaling pathway. Acta Biochim. Biophys. Sin. 2015, 47, 932–937. [Google Scholar] [CrossRef]
- Pooley, N.J.; Tacchi, L.; Secombes, C.J.; Martin, S.A. Inflammatory responses in primary muscle cell cultures in Atlantic salmon (Salmo salar). BMC Genom. 2013, 14, 747. [Google Scholar] [CrossRef] [PubMed]
- Carrizo, V.; Valenzuela, C.A.; Zuloaga, R.; Aros, C.; Altamirano, C.; Valdés, J.A.; Molina, A. Effect of cortisol on the immune-like response of rainbow trout (Oncorhynchus mykiss) myotubes challenged with Piscirickettsia salmonis. Vet. Immunol. Immunopathol. 2021, 237, 110240. [Google Scholar] [CrossRef]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Montserrat, N.; Riera-Codina, M.; Capilla, E.; Navarro, I.; Gutiérrez, J. Contribution of in vitro myocytes studies to understanding fish muscle physiology. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 199, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Carrizo, V.; Valenzuela, C.A.; Aros, C.; Dettleff, P.; Valenzuela-Muñoz, V.; Gallardo-Escarate, C.; Altamirano, C.; Molina, A.; Valdés, J.A. Transcriptomic analysis reveals a Piscirickettsia salmonis-induced early inflammatory response in rainbow trout skeletal muscle. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 39, 100859. [Google Scholar] [CrossRef]
- Chandhini, S.; Rejish Kumar, V.J. Transcriptomics in aquaculture: Current status and applications. Rev. Aquac. 2019, 11, 1379–1397. [Google Scholar] [CrossRef]
- Henríquez, M.; González, E.; Marshall, S.H.; Henríquez, V.; Gómez, F.A.; Martínez, I.; Altamirano, C. A novel liquid medium for the efficient growth of the salmonid pathogen Piscirickettsia salmonis and optimization of culture conditions. PLoS ONE 2013, 8, e71830. [Google Scholar] [CrossRef][Green Version]
- Lagos, F.; Cartes, C.; Vera, T.; Haussmann, D.; Figueroa, J. Identification of genomic islands in Chilean Piscirickettsia salmonis strains and analysis of gene expression involved in virulence. J. Fish Dis. 2017, 40, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- McFarland, J. The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J. Am. Med. Assoc. 1907, 49, 1176–1178. [Google Scholar] [CrossRef]
- Aedo, J.E.; Zuloaga, R.; Bastías-Molina, M.; Meneses, C.; Boltaña, S.; Molina, A.; Valdés, J.A. Early transcriptomic responses associated with the membrane-initiated action of cortisol in the skeletal muscle of rainbow trout (Oncorhynchus mykiss). Physiol. Genom. 2019, 51, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Aury, J.M.; Louis, A.; Dehais, P.; Bardou, P.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis oflarge gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- Gomez, F.A.; Tobar, J.A.; Henríquez, V.; Sola, M.; Altamirano, C.; Marshall, S.H. Evidence of the presence of a functional Dot/Icm type IV-B secretion system in the fish bacterial pathogen Piscirickettsia salmonis. PLoS ONE 2013, 8, e54934. [Google Scholar] [CrossRef]
- Sánchez, P.; Oliver, C.; Hernández, M.; Cortés, M.; Cecilia Rauch, M.; Valenzuela, K.; Garduño, R.A.; Avendaño-Herrera, R.; Yáñez, A.J. In vitro genomic and proteomic evidence of a type IV pili-like structure in the fish pathogen Piscirickettsia salmonis. FEMS Microbiol. Lett. 2018, 365, 169. [Google Scholar] [CrossRef]
- Vera, T. Differential Gene Expression of Piscirickettsia salmonis Strains in Adaptive Response to Different Culture Systems: A Strategy to Identify Possible Intracellular Survival Mechanisms and Pathogenicity. Doctoral Thesis, Universität Regensburg, Regensburg, Germany, Universidad Austral de Chile, Valdivia, Chile, 2013. [Google Scholar]
- Dettleff, P.; Bravo, C.; Patel, A.; Martinez, V. Patterns of Piscirickettsia salmonis load in susceptible and resistant families of Salmo salar. Fish Shellfish. Immunol. 2015, 45, 67–71. [Google Scholar] [CrossRef]
- Flores-Herrera, P.; Arredondo-Zelada, O.; Marshall, S.H.; Gómez, F.A. Selection and validation of reliable housekeeping genes to evaluate Piscirickettsia salmonis gene expression. Infect. Genet. Evol. 2018, 63, 151–157. [Google Scholar] [CrossRef]
- Maekawa, S.; Wang, P.C.; Chen, S.C. Comparative study of immune reaction against bacterial infection from transcriptome analysis. Front. Immunol. 2019, 10, 153. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome analysis based on RNA-Seq in understanding pathogenic mechanisms of diseases and the immune system of fish: A comprehensive review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef]
- Rozas-Serri, M.; Pena, A.; Maldonado, L. Transcriptomic profiles of post-smolt Atlantic salmon challenged with Piscirickettsia salmonis reveal a strategy to evade the adaptive immune response and modify cell-autonomous immunity. Dev. Comp. Immunol. 2018, 81, 348–362. [Google Scholar] [CrossRef]
- Zúñiga, A.; Aravena, P.; Pulgar, R.; Travisany, D.; Ortiz-Severín, J.; Chávez, F.P.; Maass, A.; González, M.; Cambiazo, V. Transcriptomic changes of Piscirickettsia salmonis during intracellular growth in a salmon macrophage-like cell line. Front. Cell. Infect. Microbiol. 2020, 9, 426. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Liu, Z.; Zhou, Y.; Xia, B.; Wang, Y.; Kang, Y.; Wang, J. Transcriptome analysis provides insights into hepatic responses to moderate heat stress in the rainbow trout (Oncorhynchus mykiss). Gene 2017, 619, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Le Luyer, J.; Deschamps, M.H.; Proulx, E.; Poirier Stewart, N.; Droit, A. RNA-Seq transcriptome analysis of pronounced biconcave vertebrae: A common abnormality in rainbow trout (Oncorhynchus mykiss, Walbaum) fed a low-phosphorus diet. J. Next Gener. Seq. Appl. 2015, 2, 112. [Google Scholar] [CrossRef]
- Sudhagar, A.; Ertl, R.; Kumar, G.; El-Matbouli, M. Transcriptome profiling of posterior kidney of brown trout, Salmo trutta, during proliferative kidney disease. Parasit Vectors 2019, 12, 569. [Google Scholar] [CrossRef] [PubMed]
- Tacchi, L.; Bron, J.E.; Taggart, J.B.; Secombes, C.J.; Bickerdike, R.; Adler, M.A.; Takle, H.; Martin, S.A. Multiple tissue transcriptomic responses to Piscirickettsia salmonis in Atlantic salmon (Salmo salar). Physiol. Genom. 2011, 43, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Wang, Y.; Gao, Y.; Li, X.; Chu, Q.; Cui, J.; Xu, T. Recognition of lipopolysaccharide and activation of NF-κB by cytosolic sensor NOD1 in teleost fish. Front. Immunol. 2018, 9, 1413. [Google Scholar] [CrossRef]
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. NOD-like receptors: Versatile cytosolic sentinels. Physiol. Rev. 2015, 95, 149–178. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Maratha, A.; Butt, A.Q.; Shevlin, E.; Miggin, S.M. TRIF-mediated TLR3 and TLR4 signaling is negatively regulated by ADAM15. J. Immunol. 2013, 190, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Charrier-Hisamuddin, L.; Laboisse, C.L.; Merlin, D. ADAM-15: A metalloprotease that mediates inflammation. FASEB J. 2008, 22, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.G.; Matsui, T.; Tergaonkar, V.; Rodriguez-Esteban, C.; Izpisua-Belmonte, J.C.; Verma, I.M. Zebrafish IkappaB kinase 1 negatively regulates NF-kappaB activity. Curr. Biol. 2005, 15, 1291–1295. [Google Scholar] [CrossRef]
- Pillon, N.J.; Bilan, P.J.; Fink, L.N.; Klip, A. Cross-talk between skeletal muscle and immune cells: Muscle-derived mediators and metabolic implications. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E453–E465. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yoon, S.; Won, M.; Sim, S.H.; Ko, J.J.; Han, S.; Lee, K.; Bae, J. HIP1R interacts with a member of Bcl-2 family, BCL2L10, and induces BAK-dependent cell death. Cell. Physiol. Biochem. 2009, 23, 043–052. [Google Scholar] [CrossRef] [PubMed]
- Grzenda, A.; Lomberk, G.; Zhang, J.S.; Urrutia, R. Sin3: Master scaffold and transcriptional corepressor. Biochim. Biophys. Acta Gene Regul. Mech. 2009, 1789, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Chan, S.W.; Hong, W. Retinoblastoma protein is functionally distinct from its homologues in affecting glucocorticoid receptor-mediated transcription and apoptosis. J. Biol. Chem. 2001, 276, 13762–13770. [Google Scholar] [CrossRef]
- Iturriaga, M.; Espinoza, M.B.; Poblete-Morales, M.; Feijoo, C.G.; Reyes, A.E.; Molina, A.; Avendaño-Herrera, R.; Valdés, J.A. Cytotoxic activity of Flavobacterium psychrophilum in skeletal muscle cells of rainbow trout (Oncorhynchus mykiss). Vet. Microbiol. 2017, 210, 101–106. [Google Scholar] [CrossRef]
- Diring, J.; Mouilleron, S.; McDonald, N.Q.; Treisman, R. RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability. Nat. Cell Biol. 2019, 21, 845–855. [Google Scholar] [CrossRef]
- Riddick, N.; Ohtani, K.I.; Surks, H.K. Targeting by myosin phosphatase-RhoA interacting protein mediates RhoA/ROCK regulation of myosin phosphatase. J. Cell. Biochem. 2008, 103, 1158–1170. [Google Scholar] [CrossRef]
- Yu, H.; Chakravorty, S.; Song, W.; Ferenczi, M.A. Phosphorylation of the regulatory light chain of myosin in striated muscle: Methodological perspectives. Eur. Biophys. J. 2016, 45, 779–805. [Google Scholar] [CrossRef]
- Vainshtein, A.; Sandri, M. Signaling pathways that control muscle mass. Int. J. Mol. Sci. 2020, 21, 4759. [Google Scholar] [CrossRef]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Alvarez, C.A.; Gomez, F.A.; Mercado, L.; Ramírez, R.; Marshall, S.H. Piscirickettsia salmonis imbalances the innate immune response to succeed in a productive infection in a salmonid cell line model. PLoS ONE 2016, 11, e0163943. [Google Scholar] [CrossRef]
- Ramírez, R.; Gomez, F.A.; Marshall, S.H. The infection process of Piscirickettsia salmonis in fish macrophages is dependent upon interaction with host-cell clathrin and actin. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free. Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Kadzhaev, K.; Zingmark, C.; Golovliov, I.; Bolanowski, M.; Shen, H.; Conlan, W.; Sjöstedt, A. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 2009, 4, e5463. [Google Scholar] [CrossRef]
- Valenzuela-Miranda, D.; Gallardo-Escárate, C. Dual RNA-Seq uncovers metabolic amino acids dependency of the intracellular bacterium Piscirickettsia salmonis infecting Atlantic salmon. Front. Microbiol. 2018, 9, 2877. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).