Diet and Exercise Modulate GH-IGFs Axis, Proteolytic Markers and Myogenic Regulatory Factors in Juveniles of Gilthead Sea Bream (Sparus aurata)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
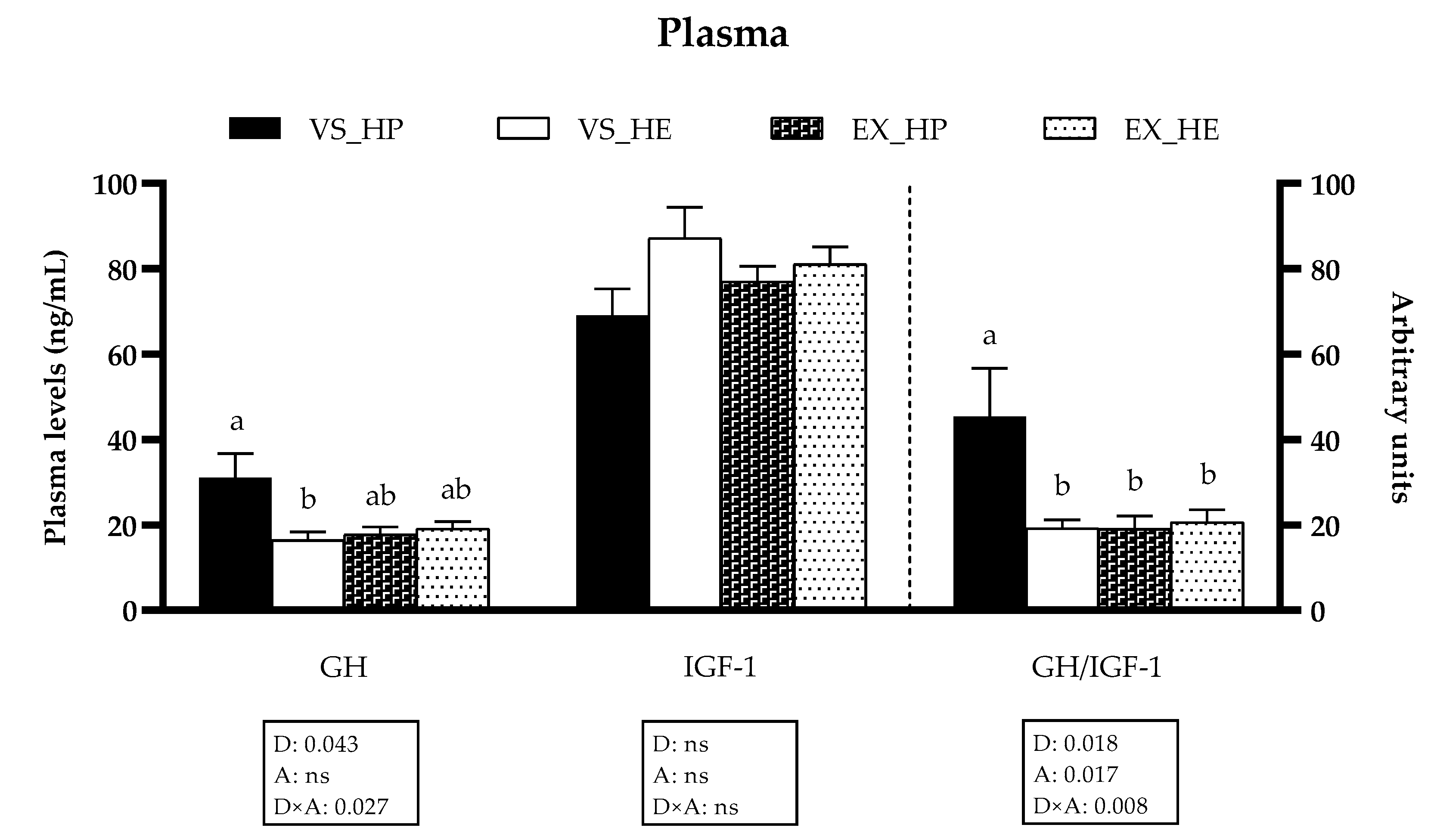
2.2. GH and IGF-1 Plasma Levels
2.3. Gene Expression
2.3.1. RNA Extraction and cDNA Synthesis
2.3.2. Quantitative Real-Time PCR (qPCR)
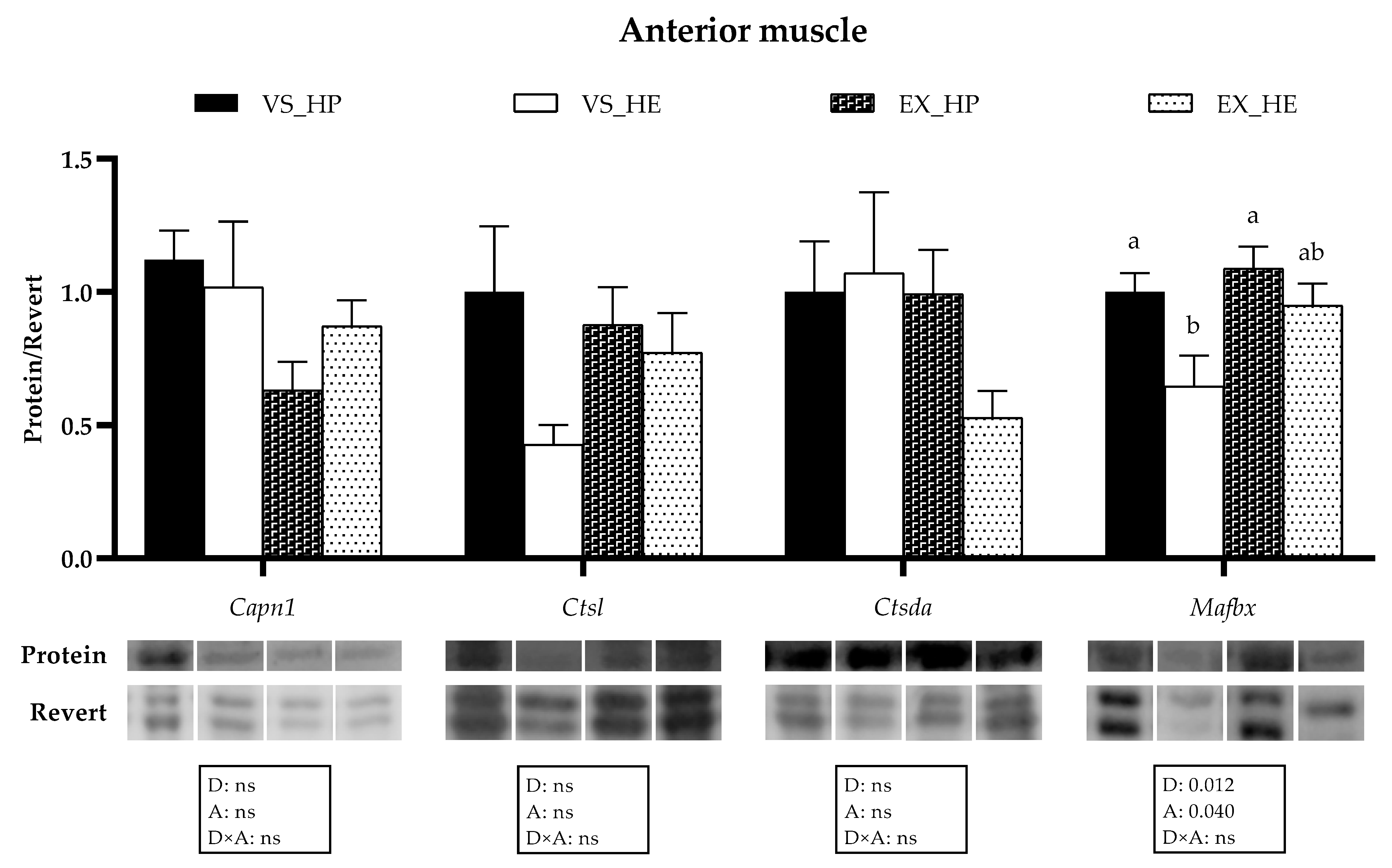
2.4. Western Blot Analysis
2.5. Muscle Texture Measurement
2.6. Statistical Analysis
3. Results
3.1. GH and IGF-1 Plasma Levels
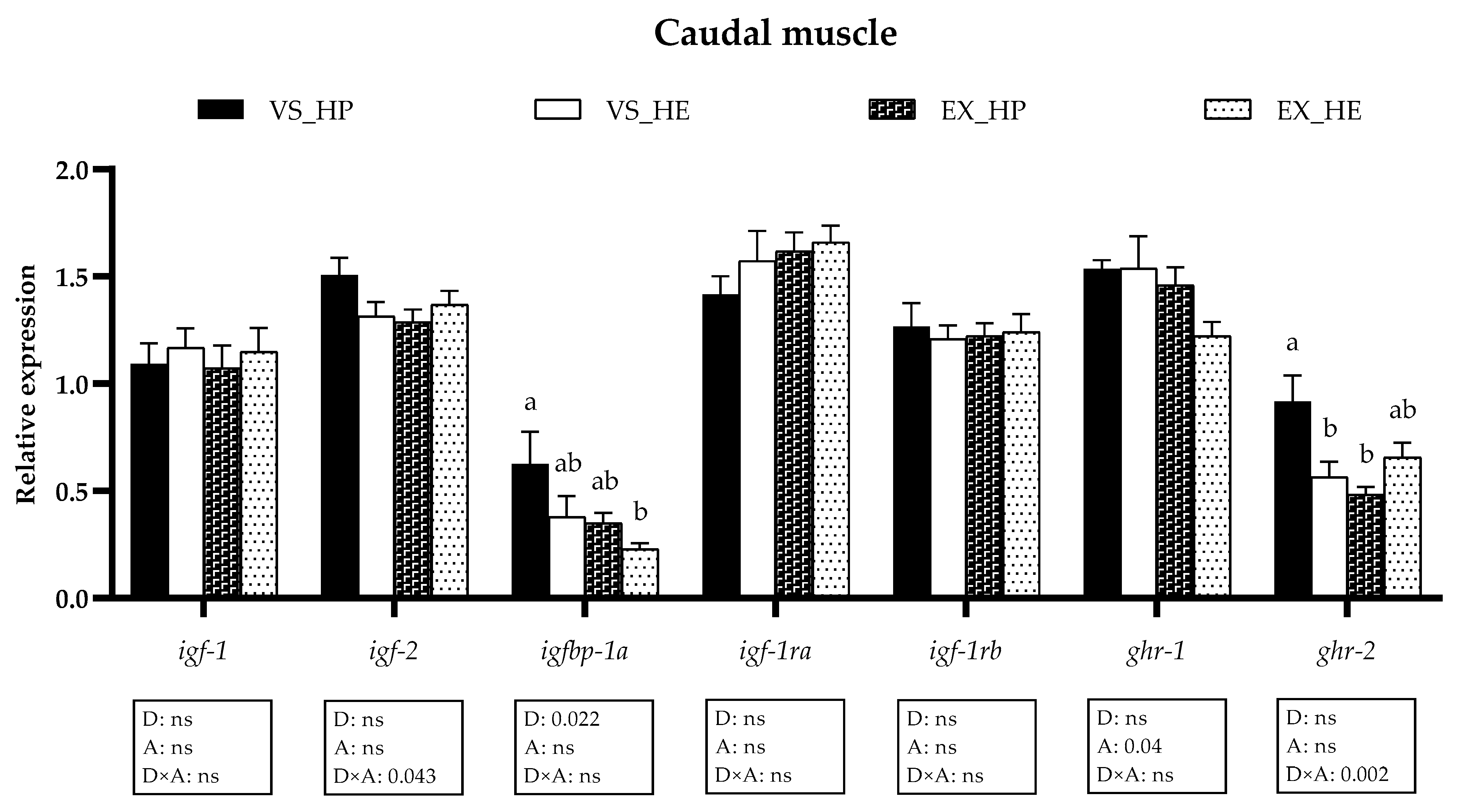
3.2. GH-IGFs Axis Components Gene Expression in Liver
3.3. GH-IGFs Axis Components Gene Expression in Anterior and Caudal Muscle
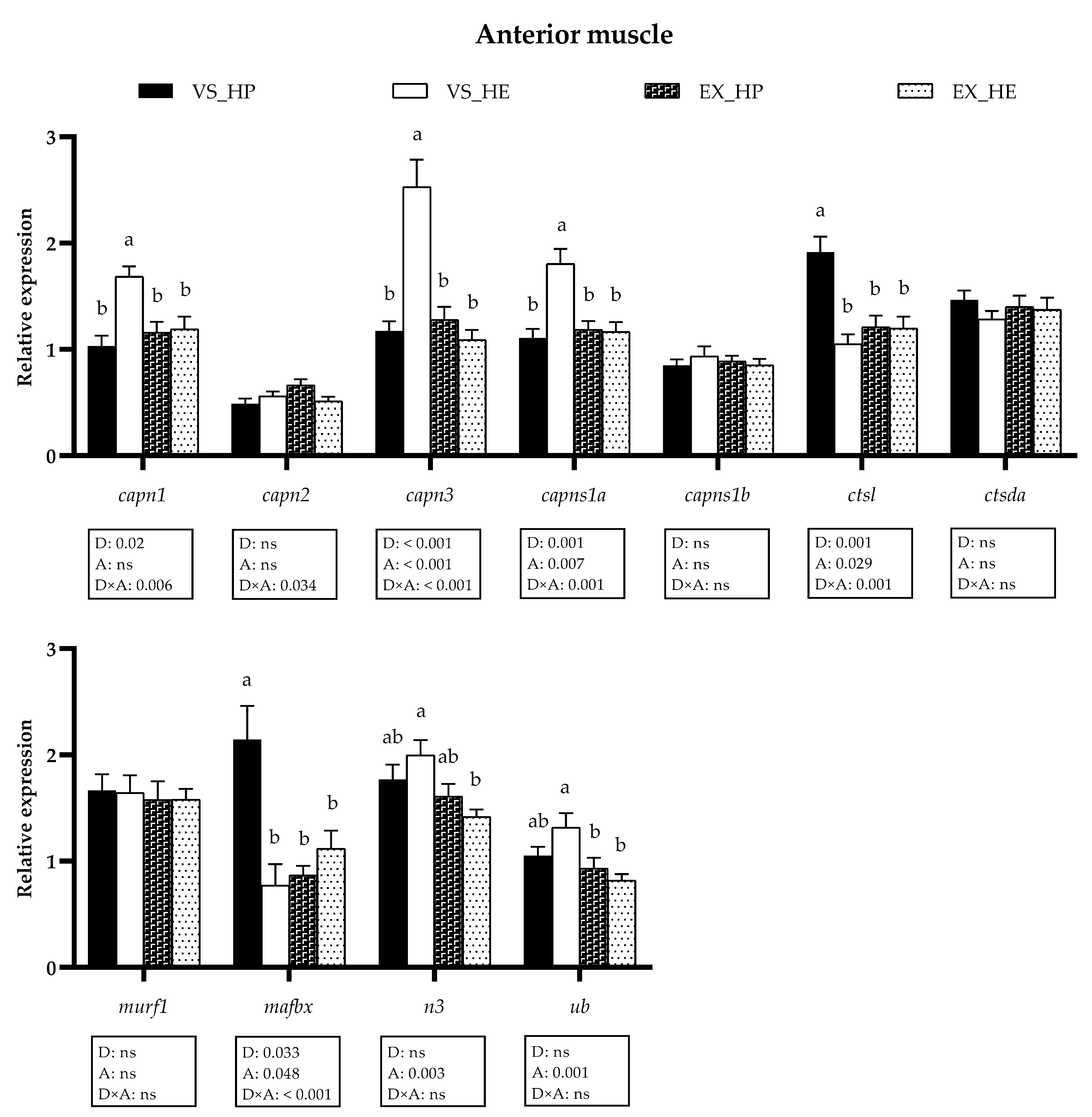
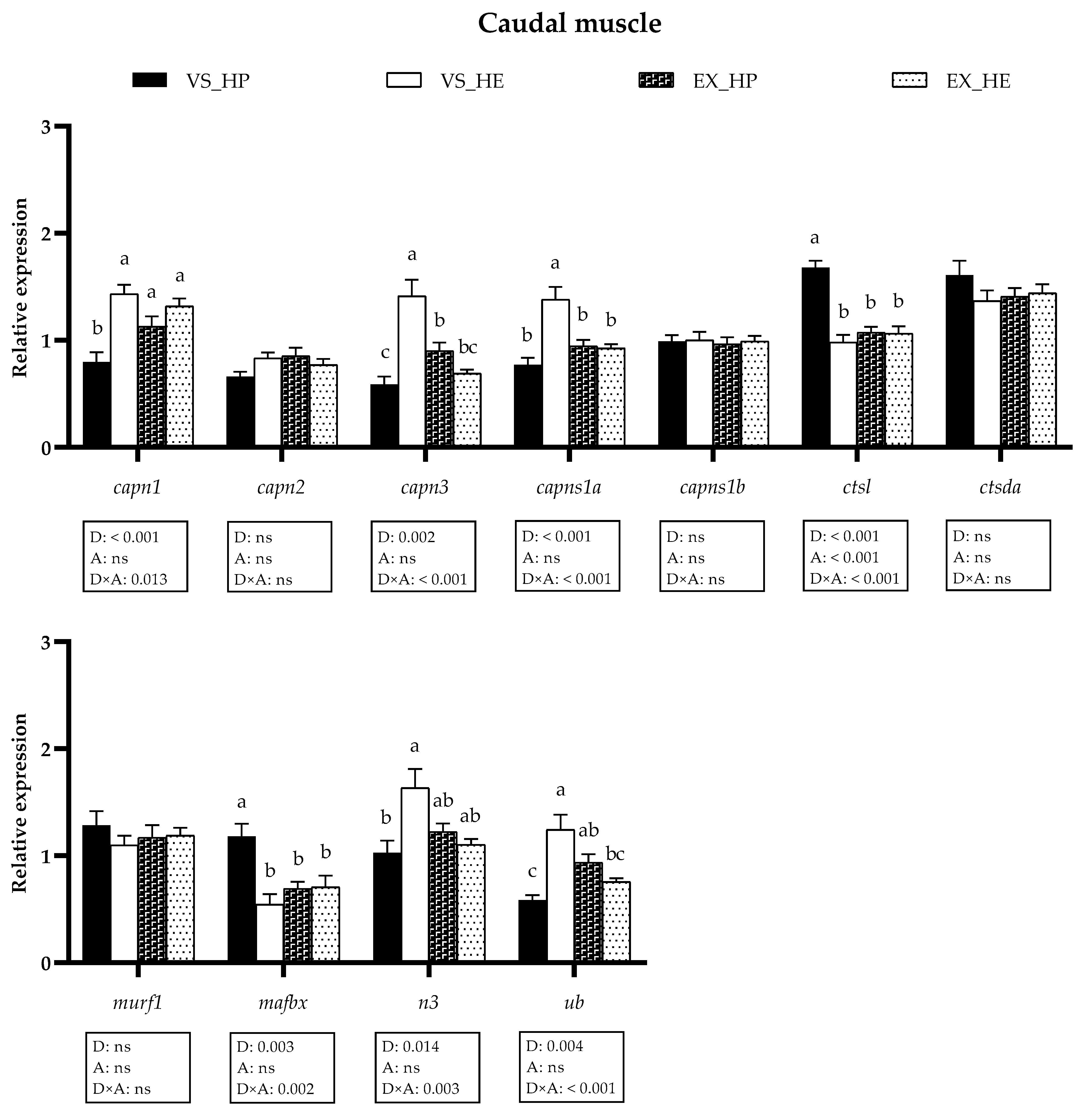
3.4. Proteolytic Markers Gene and Protein Expression
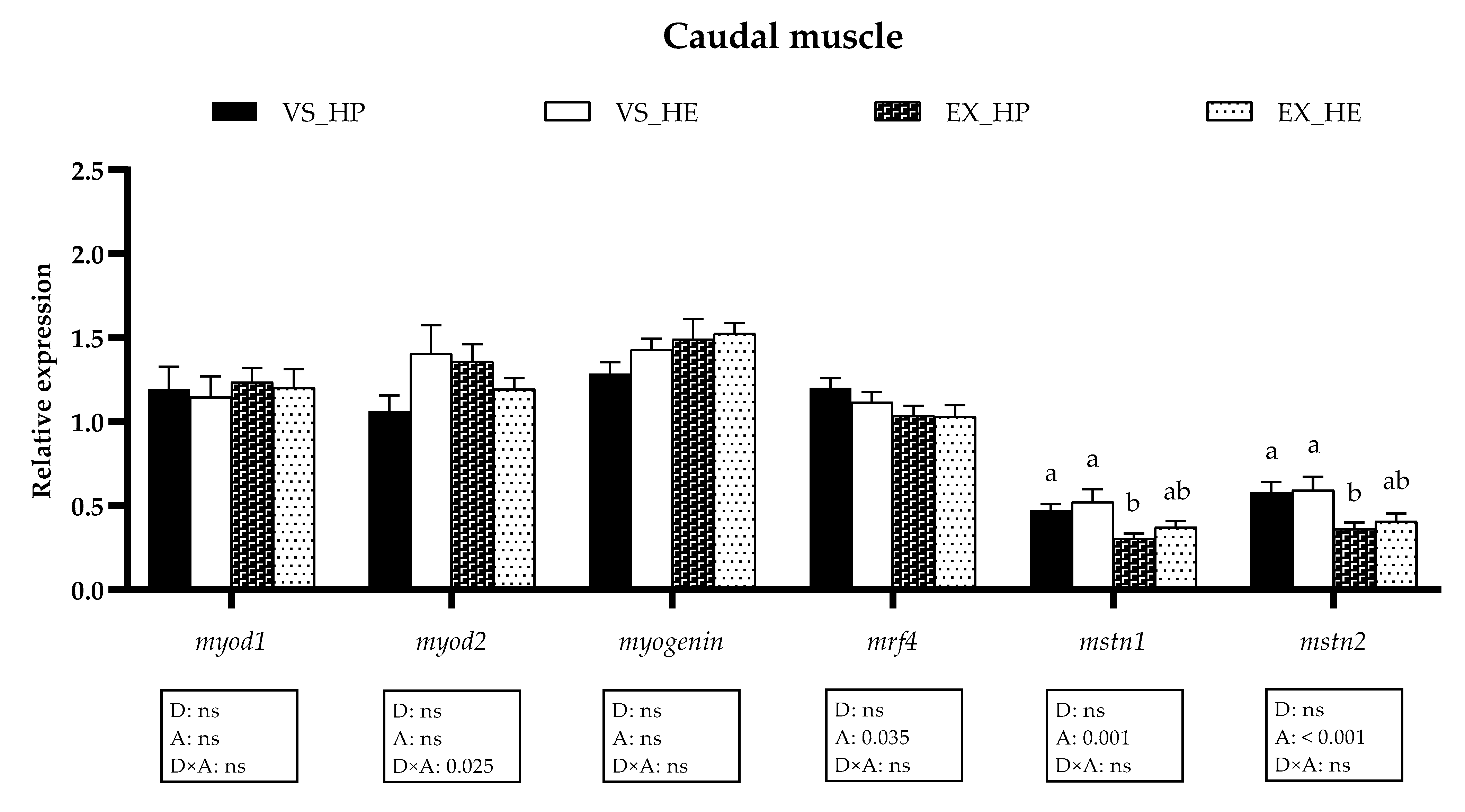
3.5. Myogenic Regulatory Factors and Growth Inhibitors Gene Expression
3.6. Muscle Texture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO and Agriculture Organization of the United Nations. The State of World Fisheries and Aquaculture 2020: Sustainability in Action; FAO: Rome, Italy, 2020; ISBN 978-92-5-132692-3. [Google Scholar]
- Gaylord, T.G.; Barrows, F.T. Multiple amino acid supplementations to reduce dietary protein in plant-based rainbow trout, Oncorhynchus mykiss, feeds. Aquaculture 2009, 287, 180–184. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Hardy, R.W.; Usry, J.L. Plant protein ingredients with lysine supplementation reduce dietary protein level in rainbow trout (Oncorhynchus mykiss) diets, and reduce ammonia nitrogen and soluble phosphorus excretion. Aquaculture 2003, 218, 553–565. [Google Scholar] [CrossRef]
- Martin-Perez, M.; Fernandez-Borras, J.; Ibarz, A.; Millan-Cubillo, A.; Felip, O.; De Oliveira, E.; Blasco, J. New Insights into Fish Swimming: A Proteomic and Isotopic Approach in Gilthead Sea Bream. J. Proteome Res. 2012, 11, 3533–3547. [Google Scholar] [CrossRef]
- Perelló-Amorós, M.; Fernández-Borràs, J.; Sánchez-Moya, A.; Vélez, E.J.; García-Pérez, I.; Gutiérrez, J.; Blasco, J. Mitochondrial Adaptation to Diet and Swimming Activity in Gilthead Seabream: Improved Nutritional Efficiency. Front. Physiol. 2021, 12, 875. [Google Scholar] [CrossRef]
- Ibarz, A.; Felip, O.; Fernández-Borràs, J.; Martín-Pérez, M.; Blasco, J.; Torrella, J.R. Sustained swimming improves muscle growth and cellularity in gilthead sea bream. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2011, 181, 209–217. [Google Scholar] [CrossRef]
- Palstra, A.P.; Roque, A.; Kruijt, L.; Jéhannet, P.; Pérez-Sánchez, J.; Dirks, R.P. Physiological Effects of Water Flow Induced Swimming Exercise in Seabream Sparus aurata. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef]
- Palstra, A.P.; Planas, J.V. Fish under exercise. Fish Physiol. Biochem. 2011, 37, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Felip, O.; Ibarz, A.; Fernández-Borràs, J.; Beltrán, M.; Martin-Perez, M.; Planas, J.V.; Blasco, J. Tracing metabolic routes of dietary carbohydrate and protein in rainbow trout (Oncorhynchus mykiss) using stable isotopes ([13C]starch and [15N]protein): Effects of gelatinisation of starches and sustained swimming. Br. J. Nutr. 2011, 107, 834–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felip, O.; Blasco, J.; Ibarz, A.; Martin-Perez, M.; Fernandez-Borras, J. Beneficial effects of sustained activity on the use of dietary protein and carbohydrate traced with stable isotopes 15N and 13C in gilthead sea bream (Sparus aurata). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2012, 183, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Gurmaches, J.S.; Cruz-Garcia, L.; Ibarz, A.; Fernandez-Borras, J.; Blasco, J.; Gutiérrez, J.; Navarro, I. Insulin, IGF-I, and muscle MAPK pathway responses after sustained exercise and their contribution to growth and lipid metabolism regulation in gilthead sea bream. Domest. Anim. Endocrinol. 2013, 45, 145–153. [Google Scholar] [CrossRef]
- Blasco, J.; Moya, A.; Millán-Cubillo, A.; Vélez, E.J.; Capilla, E.; Pérez-Sánchez, J.; Gutierrez, J.; Fernandez-Borras, J. Growth-promoting effects of sustained swimming in fingerlings of gilthead sea bream (Sparus aurata L.). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2015, 185, 859–868. [Google Scholar] [CrossRef] [Green Version]
- Vélez, E.J.; Azizi, S.; Lutfi, E.; Capilla, E.; Moya, A.; Navarro, I.; Fernandez-Borras, J.; Blasco, J.; Gutiérrez, J. Moderate and sustained exercise modulates muscle proteolytic and myogenic markers in gilthead sea bream (Sparus aurata). Am. J. Physiol. Integr. Comp. Physiol. 2017, 312, R643–R653. [Google Scholar] [CrossRef] [Green Version]
- Vélez, E.J.; Azizi, S.; Millán-Cubillo, A.; Fernandez-Borras, J.; Blasco, J.; Chan, S.J.; Calduch-Giner, J.; Sánchez, J.P.; Navarro, I.; Capilla, E.; et al. Effects of sustained exercise on GH-IGFs axis in gilthead sea bream (Sparus aurata). Am. J. Physiol. Integr. Comp. Physiol. 2016, 310, R313–R322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenzie, D.J.; Palstra, A.P.; Planas, J.; MacKenzie, S.; Bégout, M.; Thorarensen, H.; Vandeputte, M.; Mes, D.; Rey, S.; De Boeck, G.; et al. Aerobic swimming in intensive finfish aquaculture: Applications for production, mitigation and selection. Rev. Aquaculture 2021, 13, 138–155. [Google Scholar] [CrossRef]
- Davison, W. The Effects of Exercise Training on Teleost Fish, a Review of Recent Literature. Comp. Biochem. Physiol. Part A Physiol. 1997, 117, 67–75. [Google Scholar] [CrossRef]
- Reindl, K.M.; Sheridan, M.A. Peripheral regulation of the growth hormone-insulin-like growth factor system in fish and other vertebrates. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2012, 163, 231–245. [Google Scholar] [CrossRef]
- Reinecke, M.; Bjornsson, B.T.; Dickhoff, W.W.; McCormick, S.D.; Navarro, I.; Power, D.M.; Gutiérrez, J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen. Comp. Endocrinol. 2005, 142, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.W.; Duan, C.; Bern, H.A. Insulin-Like Growth Factor Signaling in Fish. Int Rev Cytol. 2005, 243, 215–285. [Google Scholar] [CrossRef]
- Fuentes, E.N.; Valdés, J.A.; Molina, A.; Bjornsson, B.T. Regulation of skeletal muscle growth in fish by the growth hormone—Insulin-like growth factor system. Gen. Comp. Endocrinol. 2013, 192, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández-Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Pérez-Sánchez, J.; Marti-Palanca, H.; Le Bail, P.-Y. Seasonal changes in circulating growth hormone (GH), hepatic GH-binding and plasma insulin-like growth factor-I immunoreactivity in a marine fish, gilthead sea bream, Sparus aurata. Fish Physiol. Biochem. 1994, 13, 199–208. [Google Scholar] [CrossRef]
- Company, R.; Astola, A.; Pendon, C.; Valdivia, M.; Pérez-Sánchez, J. Somatotropic regulation of fish growth and adiposity: Growth hormone (GH) and somatolactin (SL) relationship. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2001, 130, 435–445. [Google Scholar] [CrossRef]
- Benedito-Palos, L.; Saera-Vila, A.; Calduch-Giner, J.; Kaushik, S.; Pérez-Sánchez, J. Combined replacement of fish meal and oil in practical diets for fast growing juveniles of gilthead sea bream (Sparus aurata L.): Networking of systemic and local components of GH/IGF axis. Aquaculture 2007, 267, 199–212. [Google Scholar] [CrossRef] [Green Version]
- Lavajoo, F.; Amorós, M.P.; Vélez, E.J.; Sánchez-Moya, A.; Balbuena-Pecino, S.; Riera-Heredia, N.; Fernández-Borràs, J.; Blasco, J.; Navarro, I.; Capilla, E.; et al. Regulatory mechanisms involved in muscle and bone remodeling during refeeding in gilthead sea bream. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Saera-Vila, A.; Calduch-Giner, J.A.; Prunet, P.; Pérez-Sánchez, J. Dynamics of liver GH/IGF axis and selected stress markers in juvenile gilthead sea bream (Sparus aurata) exposed to acute confinement: Differential stress response of growth hormone receptors. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 154, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Martos-Sitcha, J.A.; Bermejo-Nogales, A.; Calduch-Giner, J.A.; Pérez-Sánchez, J. Gene expression profiling of whole blood cells supports a more efficient mitochondrial respiration in hypoxia-challenged gilthead sea bream (Sparus aurata). Front. Zool. 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, J.; Rašković, B.; Blust, R.; De Boeck, G. Exercise improves growth, alters physiological performance and gene expression in common carp (Cyprinus carpio). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2018, 226, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, J.; Simó-Mirabet, P.; Naya-Català, F.; Martos-Sitcha, J.A.; Perera, E.; Bermejo-Nogales, A.; Benedito-Palos, L.; Calduch-Giner, J.A. Somatotropic Axis Regulation Unravels the Differential Effects of Nutritional and Environmental Factors in Growth Performance of Marine Farmed Fishes. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef]
- Tiago, D.; Laizé, V.; Cancela, M.L. Alternatively spliced transcripts of Sparus aurata insulin-like growth factor 1 are differentially expressed in adult tissues and during early development. Gen. Comp. Endocrinol. 2008, 157, 107–115. [Google Scholar] [CrossRef]
- Nakashima, K.; Ishida, A.; Katsumata, M. Changes in Expression of Proteolytic-Related Genes in Chick Myoblasts during Myogenesis. J. Poult. Sci. 2011, 48, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Bell, R.A.V.; Al-Khalaf, M.; Megeney, L.A. The beneficial role of proteolysis in skeletal muscle growth and stress adaptation. Skelet. Muscle 2016, 6, 16. [Google Scholar] [CrossRef] [Green Version]
- Seiliez, I.; Gutierrez, J.; Salmerón, C.; Skiba-Cassy, S.; Chauvin, C.; Dias, K.; Kaushik, S.; Tesseraud, S.; Panserat, S. An In Vivo and In Vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010, 157, 258–266. [Google Scholar] [CrossRef]
- Seiliez, I.; Dias, K.; Cleveland, B.M. Contribution of the autophagy-lysosomal and ubiquitin-proteasomal proteolytic systems to total proteolysis in rainbow trout (Oncorhynchus mykiss) myotubes. Am. J. Physiol. Integr. Comp. Physiol. 2014, 307, R1330–R1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmerón, C.; Navarro, I.; Johnston, I.A.; Gutiérrez, J.; Capilla, E. Characterisation and expression analysis of cathepsins and ubiquitin-proteasome genes in gilthead sea bream (Sparus aurata) skeletal muscle. BMC Res. Notes 2015, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Serrana, D.G.; Codina, M.; Capilla, E.; Jiménez-Amilburu, V.; Navarro, I.; Du, S.-J.; Johnston, I.; Gutiérrez, J. Characterisation and expression of myogenesis regulatory factors during In Vitro myoblast development and In Vivo fasting in the gilthead sea bream (Sparus aurata). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 167, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Salmerón, C.; De La Serrana, D.G.; Jimenez-Amilburu, V.; Fontanillas, R.; Navarro, I.; Johnston, I.A.; Gutierrez, J.; Capilla, E. Characterisation and Expression of Calpain Family Members in Relation to Nutritional Status, Diet Composition and Flesh Texture in Gilthead Sea Bream (Sparus aurata). PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Mommsen, T.P. Paradigms of growth in fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2001, 129, 207–219. [Google Scholar] [CrossRef]
- Johnston, I.A. Environment and plasticity of myogenesis in teleost fish. J. Exp. Biol. 2006, 209, 2249–2264. [Google Scholar] [CrossRef] [Green Version]
- Seiliez, I.; Sabin, N.; Gabillard, J.-C. Myostatin inhibits proliferation but not differentiation of trout myoblasts. Mol. Cell. Endocrinol. 2012, 351, 220–226. [Google Scholar] [CrossRef]
- Gabillard, J.-C.; Biga, P.; Rescan, P.-Y.; Seiliez, I. Revisiting the paradigm of myostatin in vertebrates: Insights from fishes. Gen. Comp. Endocrinol. 2013, 194, 45–54. [Google Scholar] [CrossRef]
- Martinez-Barbera, J.P.; Pendón, C.; Martí-Palanca, H.; Calduch-Giner, J.A.; Rodríguez, R.B.; Valdivia, M.M.; Pérez-Sánchez, J. The use of recombinant gilthead sea bream (Sparus aurata) growth hormone for radioiodination and standard preparation in radioimmunoassay. Comp. Biochem. Physiol. Part A Physiol. 1995, 110, 335–340. [Google Scholar] [CrossRef]
- Shimizu, M.; Swanson, P.; Fukada, H.; Hara, A.; Dickhoff, W.W. Comparison of Extraction Methods and Assay Validation for Salmon Insulin-like Growth Factor-I Using Commercially Available Components. Gen. Comp. Endocrinol. 2000, 119, 26–36. [Google Scholar] [CrossRef] [PubMed]
- De Celis, S.V.-R.; Rojas, P.; Requeni, P.G.; Albalat, A.; Gutiérrez, J.; Médale, F.; Kaushik, S.; Navarro, I.; Sánchez, J.P. Nutritional assessment of somatolactin function in gilthead sea bream (Sparus aurata): Concurrent changes in somatotropic axis and pancreatic hormones. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2004, 138, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29. [Google Scholar] [CrossRef]
- Vélez, E.J.; Azizi, S.; Verheyden, D.; Salmerón, C.; Lutfi, E.; Sánchez-Moya, A.; Navarro, I.; Gutierrez, J.; Capilla, E. Proteolytic systems’ expression during myogenesis and transcriptional regulation by amino acids in gilthead sea bream cultured muscle cells. PLoS ONE 2017, 12, e0187339. [Google Scholar] [CrossRef]
- Sánchez-Moya, A.; García-Meilán, I.; Heredia, N.R.; Vélez, E.J.; Lutfi, E.; Fontanillas, R.; Gutiérrez, J.; Capilla, E.; Navarro, I. Effects of different dietary vegetable oils on growth and intestinal performance, lipid metabolism and flesh quality in gilthead sea bream. Aquaculture 2020, 519, 734881. [Google Scholar] [CrossRef]
- Huntingford, F.; Kadri, S. Exercise, Stress and Welfare. In Swimming Physiology of Fish: Towards Using Exercise to Farm a Fit Fish in Sustainable Aquaculture; Palstra, A.P., Planas, J.V., Eds.; Springer: Berlin, Germany, 2013; pp. 161–174. ISBN 978-3-642-31049-2. [Google Scholar]
- Pérez-Sánchez, J.; Le Bail, P.-Y. Growth hormone axis as marker of nutritional status and growth performance in fish. Aquaculture 1999, 177, 117–128. [Google Scholar] [CrossRef]
- Martí-Palanca, H.; Martinez-Barbera, J.P.; Pendón, C.; Valdivia, M.M.; Sánchez, J.P.; Kaushik, S. Growth hormone as a function of age and dietary protein: Energy ratio in a marine teleost, the gilthead sea bream (Sparus aurata). Growth Regul. 1996, 6, 253–259. [Google Scholar]
- Cameron, C.; Moccia, R.; Azevedo, P.A.; Leatherland, J.F. Effect of diet and ration on the relationship between plasma GH and IGF-1 concentrations in Arctic charr, Salvelinus alpinus (L.). Aquac. Res. 2007, 38, 877–886. [Google Scholar] [CrossRef]
- Company, R.; Calduch-Giner, J.; Kaushik, S.; Sánchez, J.P. Growth performance and adiposity in gilthead sea bream (Sparus aurata): Risks and benefits of high energy diets. Aquaculture 1999, 171, 279–292. [Google Scholar] [CrossRef]
- Pierce, A.; Shimizu, M.; Beckman, B.; Baker, D.; Dickhoff, W. Time course of the GH/IGF axis response to fasting and increased ration in chinook salmon (Oncorhynchus tshawytscha). Gen. Comp. Endocrinol. 2005, 140, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Gabillard, J.-C.; Kamangar, B.B.; Montserrat, N. Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss). J. Endocrinol. 2006, 191, 15–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saera-Vila, A.; Calduch-Giner, J.A.; Pérez-Sánchez, J. Co-expression of IGFs and GH receptors (GHRs) in gilthead sea bream (Sparus aurata L.): Sequence analysis of the GHR-flanking region. J. Endocrinol. 2007, 194, 361–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gahr, S.A.; Vallejo, R.L.; Weber, G.M.; Shepherd, B.S.; Silverstein, J.T.; Rexroad, C.E. Effects of short-term growth hormone treatment on liver and muscle transcriptomes in rainbow trout (Oncorhynchus mykiss). Physiol. Genom. 2008, 32, 380–392. [Google Scholar] [CrossRef] [Green Version]
- De La Serrana, D.G.; MacQueen, D.J. Insulin-Like Growth Factor-Binding Proteins of Teleost Fishes. Front. Endocrinol. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Vélez, E.J.; Perelló, M.; Azizi, S.; Moya, A.; Lutfi, E.; Sánchez, J.P.; Calduch-Giner, J.; Navarro, I.; Blasco, J.; Fernández-Borràs, J.; et al. Recombinant bovine growth hormone (rBGH) enhances somatic growth by regulating the GH-IGF axis in fingerlings of gilthead sea bream (Sparus aurata). Gen. Comp. Endocrinol. 2018, 257, 192–202. [Google Scholar] [CrossRef]
- Ross, R.J.M.; Esposito, N.; Shen, X.Y.; Von Laue, S.; Chew, S.L.; Dobson, P.R.M.; Postel-Vinay, M.-C.; Finidori, J. A Short Isoform of the Human Growth Hormone Receptor Functions as a Dominant Negative Inhibitor of the Full-Length Receptor and Generates Large Amounts of Binding Protein. Mol. Endocrinol. 1997, 11, 265–273. [Google Scholar] [CrossRef]
- Calduch-Giner, J.; Mingarro, M.; de Celis, S.V.-R.; Boujard, D.; Sánchez, J.P. Molecular cloning and characterization of gilthead sea bream (Sparus aurata) growth hormone receptor (GHR). Assessment of alternative splicing. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 136, 1–13. [Google Scholar] [CrossRef]
- Van Ba, H.; Inho, H. Significant role of μ-calpain (CANP1) in proliferation/survival of bovine skeletal muscle satellite cells. Vitr. Cell. Dev. Biol. Anim. 2013, 49, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Bower, N.I.; Johnston, I.A. Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol. Genom. 2010, 41, 114–130. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, F.; Cardinal, M.; Bugeon, J.; Labbe, L.; Medale, F.; Quillet, E. Selection for muscle fat content and triploidy affect flesh quality in pan-size rainbow trout, Oncorhynchus mykiss. Aquaculture 2015, 448, 569–577. [Google Scholar] [CrossRef] [Green Version]
- Thakur, D.P.; Morioka, K.; Itoh, Y.; Obatake, A. Lipid composition and deposition of cultured yellowtail Seriola quinqueradiata muscle at different anatomical locations in relation to meat texture. Fish Sci. 2003, 69, 487–494. [Google Scholar] [CrossRef] [Green Version]




| Item | HP Diet | HE Diet |
|---|---|---|
| Digestible energy (MJ/kg) | 18 | 19.9 |
| Protein (% dry mass) | 54 | 50 |
| Lipids (% dry mass) | 15 | 20 |
| DHA (% dry mass) | 15 | 20 |
| EPA (% dry mass) | 1 | 1.4 |
| ARA (% dry mass) | 2.5 | 3 |
| DHA/EPA/ARA | 5/12.5/1 | 3.5/7.5/1 |
| Cellulose (% dry mass) | 1.6 | 1.7 |
| Ashes (% dry mass) | 10.5 | 6.4 |
| Total P (% dry mass) | 1.4 | 1.2 |
| Estimated nitrogen-free extract | 20.5 | 23.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perelló-Amorós, M.; García-Pérez, I.; Sánchez-Moya, A.; Innamorati, A.; Vélez, E.J.; Achaerandio, I.; Pujolà, M.; Calduch-Giner, J.; Pérez-Sánchez, J.; Fernández-Borràs, J.; et al. Diet and Exercise Modulate GH-IGFs Axis, Proteolytic Markers and Myogenic Regulatory Factors in Juveniles of Gilthead Sea Bream (Sparus aurata). Animals 2021, 11, 2182. https://doi.org/10.3390/ani11082182
Perelló-Amorós M, García-Pérez I, Sánchez-Moya A, Innamorati A, Vélez EJ, Achaerandio I, Pujolà M, Calduch-Giner J, Pérez-Sánchez J, Fernández-Borràs J, et al. Diet and Exercise Modulate GH-IGFs Axis, Proteolytic Markers and Myogenic Regulatory Factors in Juveniles of Gilthead Sea Bream (Sparus aurata). Animals. 2021; 11(8):2182. https://doi.org/10.3390/ani11082182
Chicago/Turabian StylePerelló-Amorós, Miquel, Isabel García-Pérez, Albert Sánchez-Moya, Arnau Innamorati, Emilio J. Vélez, Isabel Achaerandio, Montserrat Pujolà, Josep Calduch-Giner, Jaume Pérez-Sánchez, Jaume Fernández-Borràs, and et al. 2021. "Diet and Exercise Modulate GH-IGFs Axis, Proteolytic Markers and Myogenic Regulatory Factors in Juveniles of Gilthead Sea Bream (Sparus aurata)" Animals 11, no. 8: 2182. https://doi.org/10.3390/ani11082182
APA StylePerelló-Amorós, M., García-Pérez, I., Sánchez-Moya, A., Innamorati, A., Vélez, E. J., Achaerandio, I., Pujolà, M., Calduch-Giner, J., Pérez-Sánchez, J., Fernández-Borràs, J., Blasco, J., & Gutiérrez, J. (2021). Diet and Exercise Modulate GH-IGFs Axis, Proteolytic Markers and Myogenic Regulatory Factors in Juveniles of Gilthead Sea Bream (Sparus aurata). Animals, 11(8), 2182. https://doi.org/10.3390/ani11082182









