Genomic Epidemiology of Shiga Toxin-Producing Escherichia coli Isolated from the Livestock-Food-Human Interface in South America
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Whole-Genome Sequencing (WGS)
2.2. Publicly Available Genome Sequences
2.3. Virulome and AMR Gene Analysis
2.4. Epidemiological Typing and Phylogenomic Analysis
2.5. Statistical Analysis
3. Results
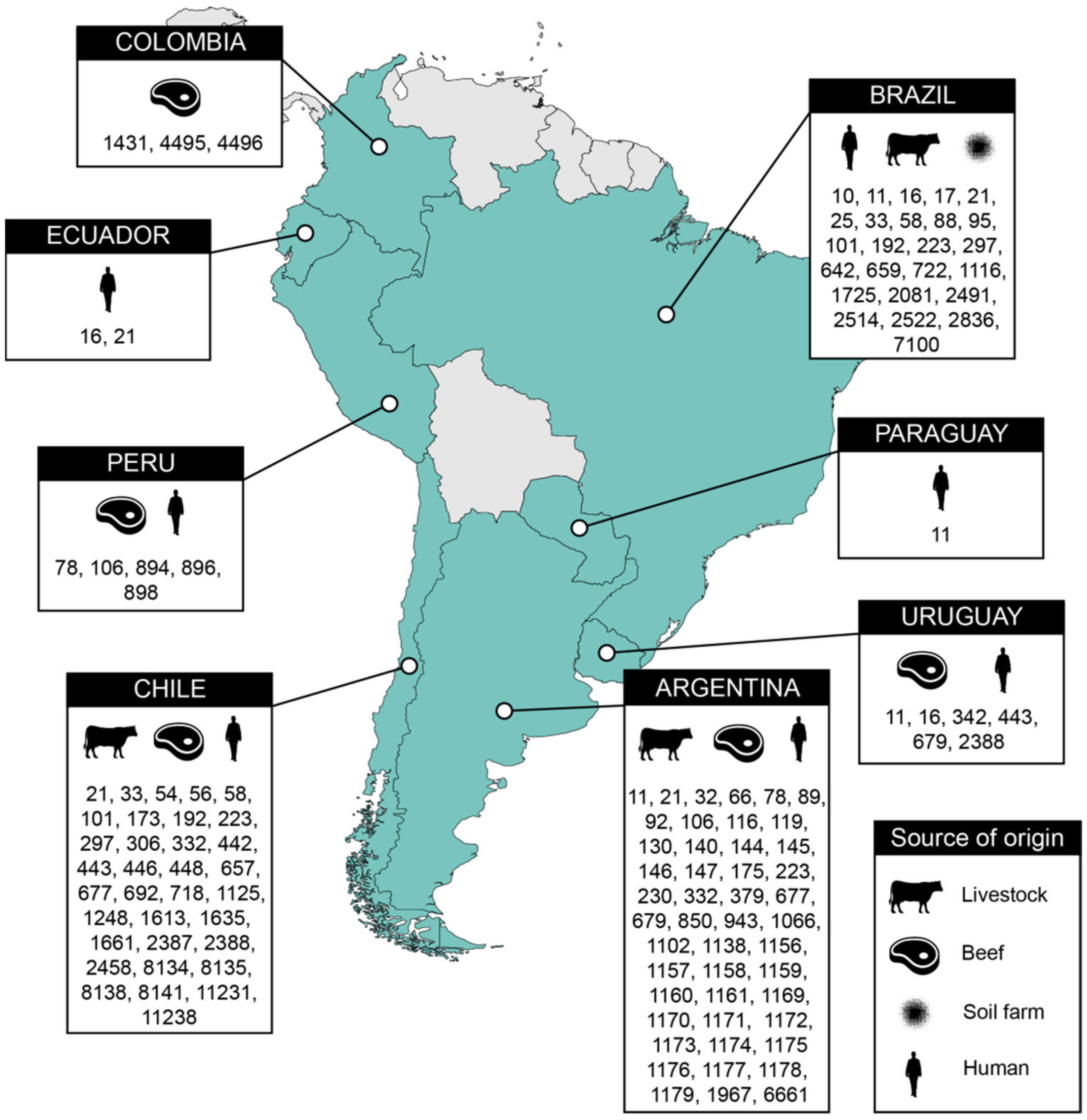
3.1. Source and Origin of the Analyzed STEC Genomes
3.2. Virulome and Antibiotic Resistome of STEC Strains
3.3. Sequence Types, Serotypes and Phylogenomic Analysis of STEC Strains Circulating in South America
3.4. Association Between stx Type and the Geographic Location and Isolation Source of STEC Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, structure, and function. Microbiol. Spectr. 2014, 2, 2–4. [Google Scholar] [CrossRef]
- Gyles, C.L.; Fairbrother, J.M. Escherichia coli. In Pathogenesis of Bacterial Infections in Animals; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 267–308. ISBN 978-0-470-95820-9. [Google Scholar]
- Amézquita-López, B.A.; Soto-Beltrán, M.; Lee, B.G.; Yambao, J.C.; Quiñones, B. Isolation, genotyping and antimicrobial resistance of Shiga toxin-producing Escherichia coli. J. Microbiol. Immunol. Infect. 2018, 51, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Sapountzis, P.; Segura, A.; Desvaux, M.; Forano, E. An overview of the elusive passenger in the gastrointestinal tract of cattle: The Shiga toxin producing Escherichia coli. Microorganisms 2020, 8, 877. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.; Chinen, I.; Guth, B.E.C. Enterohemorrhagic (Shiga toxin-producing) Escherichia coli. In Escherichia coli in the Americas; Torres, A.G., Ed.; Springer International Publishing: Manhattan, NY, USA, 2016; pp. 97–123. ISBN 978-3-319-45092-6. [Google Scholar]
- Fernández, D.; Sanz, M.E.; Parma, A.E.; Padola, N.L. Short Communication: Characterization of Shiga toxin-producing Escherichia coli isolated from newborn, milk-fed, and growing calves in Argentina. J. Dairy Sci. 2012, 95, 5340–5343. [Google Scholar] [CrossRef]
- Rivera, F.P.; Sotelo, E.; Morales, I.; Menacho, F.; Medina, A.M.; Evaristo, R.; Valencia, R.; Carbajal, L.; Ruiz, J.; Ochoa, T.J. Short communication: Detection of Shiga toxin-producing Escherichia coli (STEC) in healthy cattle and pigs in Lima, Peru. J. Dairy Sci. 2012, 95, 1166–1169. [Google Scholar] [CrossRef] [PubMed]
- Galarce, N.; Escobar, B.; Sánchez, F.; Paredes-Osses, E.; Alegría-Morán, R.; Borie, C. Virulence genes, Shiga toxin subtypes, serogroups, and clonal relationship of Shiga toxin-producing Escherichia coli strains isolated from livestock and companion animals. Animals 2019, 9, 733. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.G.M.; Cerqueira, A.M.F. Shiga Toxin-producing Escherichia coli in the animal reservoir and food in Brazil. J. Appl. Microbiol. 2020, 128, 1568–1582. [Google Scholar] [CrossRef]
- Smith, J.L.; Fratamico, P.M. Emerging and re-emerging foodborne pathogens. Foodborne Pathog. Dis. 2018, 15, 737–757. [Google Scholar] [CrossRef]
- Scott, M.E.; Mbandi, E.; Buchanan, S.; Abdelmajid, N.; Gonzalez-Rivera, C.; Hale, K.R.; Jacobsen, L.; Webb, J.; Green, J.; Dolan, P. Salmonella and Shiga toxin–producing Escherichia coli in products sampled in the food safety and inspection service raw pork baseline study. J. Food Protect. 2020, 83, 552–559. [Google Scholar] [CrossRef]
- Franz, E.; Delaquis, P.; Morabito, S.; Beutin, L.; Gobius, K.; Rasko, D.A.; Bono, J.; French, N.; Osek, J.; Lindstedt, B.-A.; et al. Exploiting the explosion of information associated with whole genome sequencing to tackle Shiga toxin-producing Escherichia coli (STEC) in global food production systems. Int. J. Food Microbiol. 2014, 187, 57–72. [Google Scholar] [CrossRef]
- Food Safety and Inspection Service, Department of Agriculture, USA (FSIS). Shiga Toxin-Producing Escherichia coli in Certain Raw Beef Products. Available online: https://www.govinfo.gov/content/pkg/FR-2011-11-23/pdf/2011-30271.pdf (accessed on 31 March 2021).
- Shakerian, A.; Rahimi, E.; Emad, P. Vegetables and restaurant salads as a reservoir for Shiga toxigenic Escherichia coli: Distribution of virulence factors, O-serogroups, and antibiotic resistance properties. J. Food Prot. 2016, 79, 1154–1160. [Google Scholar] [CrossRef]
- Abe, C.M.; Matheus-Guimarães, C.; Garcia, B.G.; Cabilio Guth, B.E. Interactions of Shiga toxin-producing Escherichia coli with leafy green vegetables. Braz. J. Microbiol. 2020, 51, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Cody, E.M.; Dixon, B.P. Hemolytic uremic syndrome. Pediatr. Clin. N. Am. 2019, 66, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M.D. Global Incidence of human Shiga toxin-producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO); World Health Organization (WHO). Attributing Illness Caused by Shiga Toxin-Producing Escherichia coli (STEC) to Specific Foods; Microbiological Risk Assessment Series; FAO-WHO: Rome, Italy, 2019; ISBN 978-92-5-131746-4. [Google Scholar]
- Torres, A.G.; Amaral, M.M.; Bentancor, L.; Galli, L.; Goldstein, J.; Krüger, A.; Rojas-Lopez, M. Recent advances in Shiga toxin-producing Escherichia coli research in Latin America. Microorganisms 2018, 6, 100. [Google Scholar] [CrossRef]
- Castro, V.S.; Teixeira, L.A.C.; Rodrigues, D.D.P.; Dos Santos, L.F.; Conte-Junior, C.A.; de Souza Figueiredo, E.E. Occurrence and antimicrobial resistance of E. coli non-O157 isolated from beef in Mato Grosso, Brazil. Trop. Anim. Health Prod. 2019, 51, 1117–1123. [Google Scholar] [CrossRef]
- Cavalcanti, A.M.F.; Hernandes, R.T.; Takagi, E.H.; Guth, B.E.C.; de Lima Ori, É.; Pinheiro, S.R.S.; de Andrade, T.S.; Oliveira, S.L.; Cergole-Novella, M.C.; Francisco, G.R.; et al. Virulence profiling and molecular typing of Shiga toxin-producing E. coli (STEC) from human sources in Brazil. Microorganisms 2020, 8, 171. [Google Scholar] [CrossRef]
- Organization for Economic Co-Operation and Development (OECD). Food and Agriculture Organization of the United Nations (FAO) Meat. In OECD-FAO Agricultural Outlook 2020–2029; OECD-FAO: Rome, Italy, 2020; Available online: http://www.fao.org/3/ca8861en/Meat.pdf (accessed on 31 March 2021).
- Food and Agriculture Organization of the United Nations Statistics Division (FAOSTAT). Datos Sobre Alimentación y Agricultura. Available online: http://www.fao.org/faostat/es/#home (accessed on 31 March 2021).
- Pianciola, L.; Rivas, M. Genotypic features of clinical and bovine Escherichia coli O157 strains isolated in countries with different associated-disease incidences. Microorganisms 2018, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Pianciola, L.; D’Astek, B.A.; Mazzeo, M.; Chinen, I.; Masana, M.; Rivas, M. Genetic features of human and bovine Escherichia coli O157:H7 strains isolated in Argentina. Int. J. Med. Microbiol. 2016, 306, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Castro, V.S.; Carvalho, R.C.T.; Conte-Junior, C.A.; Figuiredo, E.E.S. Shiga-toxin producing Escherichia coli: Pathogenicity, supershedding, diagnostic methods, occurrence, and foodborne outbreaks. Comp. Rev. Food Sci. Food Saf. 2017, 16, 1269–1280. [Google Scholar] [CrossRef]
- Borie, C.F.; Monreal, Z.; Martinez, J.; Arellano, C.; Prado, V. Detection and characterization of Enterohaemorrhagic Escherichia coli in slaughtered cattle. Zent. Vet. B 1997, 44, 273–279. [Google Scholar] [CrossRef]
- Blanco, M.; Padola, N.L.; Krüger, A.; Sanz, M.E.; Blanco, J.E.; González, E.A.; Dahbi, G.; Mora, A.; Bernárdez, M.I.; Etcheverría, A.I.; et al. Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int. Microbiol. 2004, 7, 269–276. [Google Scholar] [PubMed]
- Pigatto, C.P.; Schocken-Iturrino, R.P.; Souza, E.M.; Pedrosa, F.O.; Comarella, L.; Irino, K.; Kato, M.A.M.F.; Farah, S.M.S.S.; Warth, J.F.; Fadel-Picheth, C.M.T. Virulence properties and antimicrobial susceptibility of Shiga toxin-producing Escherichia coli strains isolated from healthy cattle from Paraná State, Brazil. Can. J. Microbiol. 2008, 54, 588–593. [Google Scholar] [CrossRef]
- Fernández, D.; Irino, K.; Sanz, M.E.; Padola, N.L.; Parma, A.E. Characterization of Shiga toxin-producing Escherichia coli isolated from dairy cows in Argentina. Lett. Appl. Microbiol. 2010, 51, 377–382. [Google Scholar] [CrossRef]
- Iguchi, A.; Shirai, H.; Seto, K.; Ooka, T.; Ogura, Y.; Hayashi, T.; Osawa, K.; Osawa, R. Wide distribution of O157-antigen biosynthesis gene clusters in Escherichia coli. PLoS ONE 2011, 6, e23250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ferdous, M.; Friedrich, A.W.; Grundmann, H.; de Boer, R.F.; Croughs, P.D.; Islam, M.A.; Kluytmans-van den Bergh, M.F.Q.; Kooistra-Smid, A.M.D.; Rossen, J.W.A. Molecular characterization and phylogeny of Shiga toxin-producing Escherichia coli isolates obtained from two Dutch regions using whole genome sequencing. Clin. Microbiol. Infect. 2016, 22, 642.e1. [Google Scholar] [CrossRef][Green Version]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bover-Cid, S.; Chemaly, M.; Davies, R.; Cesare, A.D.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Pathogenicity assessment of Shiga toxin-producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020, 18, e05967. [Google Scholar] [CrossRef]
- Paton, A.W.; Srimanote, P.; Woodrow, M.C.; Paton, J.C. Characterization of Saa, a novel autoagglutinating adhesin produced by Locus of Enterocyte Effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 2001, 69, 6999–7009. [Google Scholar] [CrossRef]
- Montero, D.A.; Velasco, J.; Del Canto, F.; Puente, J.L.; Padola, N.L.; Rasko, D.A.; Farfán, M.; Salazar, J.C.; Vidal, R. Locus of Adhesion and Autoaggregation (LAA), a pathogenicity island present in emerging Shiga toxin–producing Escherichia coli strains. Sci. Rep. 2017, 7, 7011. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Shimamoto, T. Molecular analysis of multidrug resistance in Shiga toxin-producing Escherichia coli O157: H7 isolated from meat and dairy products. Int. J. Food Microbiol. 2015, 193, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-J.; Yoon, J.W.; Heo, E.-J.; Ko, E.-K.; Kim, K.-Y.; Kim, Y.-J.; Yoon, H.-J.; Wee, S.-H.; Park, Y.H.; Moon, J.S. Antibiotic resistance and virulence potentials of Shiga toxin-producing Escherichia coli isolates from raw meats of slaughterhouses and retail markets in Korea. J. Microbiol. Biotechnol. 2015, 25, 1460–1466. [Google Scholar] [CrossRef]
- Colello, R.; Krüger, A.; Conza, J.D.; Rossen, J.W.A.; Friedrich, A.W.; Gutkind, G.; Etcheverría, A.I.; Padola, N.L. Antimicrobial resistance in class 1 integron-positive Shiga toxin-producing Escherichia coli isolated from cattle, pigs, food and farm environment. Microorganisms 2018, 6, 99. [Google Scholar] [CrossRef]
- Furlan, J.P.R.; Gallo, I.F.L.; de Campos, A.C.L.P.; Passaglia, J.; Falcão, J.P.; Navarro, A.; Nakazato, G.; Stehling, E.G. Molecular characterization of multidrug-resistant Shiga toxin-producing Escherichia coli harboring antimicrobial resistance genes obtained from a farmhouse. Pathog. Glob. Health 2019, 113, 268–274. [Google Scholar] [CrossRef]
- Puii, L.H.; Dutta, T.K.; Roychoudhury, P.; Kylla, H.; Chakraborty, S.; Mandakini, R.; Kawlni, L.; Samanta, I.; Bandopaddhay, S.; Singh, S.B. Extended spectrum beta-lactamase producing Shiga-toxin producing-Escherichia coli in piglets, humans and water sources in North East Region of India. Lett. Appl. Microbiol. 2019, 69, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Galarce, N.; Sánchez, F.; Fuenzalida, V.; Ramos, R.; Escobar, B.; Lapierre, L.; Paredes-Osses, E.; Arriagada, G.; Alegría-Morán, R.; Lincopán, N.; et al. Phenotypic and genotypic antimicrobial resistance in non-O157 Shiga toxin-producing Escherichia coli isolated from cattle and swine in Chile. Front. Vet. Sci. 2020, 7, 367. [Google Scholar] [CrossRef] [PubMed]
- Dhanji, H.; Murphy, N.M.; Doumith, M.; Durmus, S.; Lee, S.S.; Hope, R.; Woodford, N.; Livermore, D.M. Cephalosporin resistance mechanisms in Escherichia coli isolated from raw chicken imported into the UK. J. Antimicrob. Chemother. 2010, 65, 2534–2537. [Google Scholar] [CrossRef]
- Alonso, C.A.; Zarazaga, M.; Sallem, R.B.; Jouini, A.; Slama, K.B.; Torres, C. Antibiotic resistance in Escherichia coli in husbandry animals: The African perspective. Lett. Appl. Microbiol. 2017, 64, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Lee, M.-S.; Kim, J.H. Recent updates on outbreaks of Shiga toxin-producing Escherichia coli and its potential reservoirs. Front. Cell Infect. Microbiol. 2020, 10, 273. [Google Scholar] [CrossRef]
- Dallman, T.J.; Byrne, L.; Ashton, P.M.; Cowley, L.A.; Perry, N.T.; Adak, G.; Petrovska, L.; Ellis, R.J.; Elson, R.; Underwood, A.; et al. Whole-genome sequencing for national surveillance of Shiga toxin-producing Escherichia coli O157. Clin. Infect. Dis. 2015, 61, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Baba, H.; Kanamori, H.; Kudo, H.; Kuroki, Y.; Higashi, S.; Oka, K.; Takahashi, M.; Yoshida, M.; Oshima, K.; Aoyagi, T.; et al. Genomic analysis of Shiga toxin-producing Escherichia coli from patients and asymptomatic food handlers in Japan. PLoS ONE 2019, 14, e0225340. [Google Scholar] [CrossRef]
- González-Escalona, N.; Kase, J.A. Virulence gene profiles and phylogeny of Shiga toxin-positive Escherichia coli strains isolated from FDA regulated foods during 2010–2017. PLoS ONE 2019, 14, e0214620. [Google Scholar] [CrossRef]
- Montero, D.A.; Canto, F.D.; Velasco, J.; Colello, R.; Padola, N.L.; Salazar, J.C.; Martin, C.S.; Oñate, A.; Blanco, J.; Rasko, D.A.; et al. Cumulative acquisition of pathogenicity islands has shaped virulence potential and contributed to the emergence of LEE-negative Shiga toxin-producing Escherichia coli strains. Emerg. Microbes. Infect. 2019, 8, 486–502. [Google Scholar] [CrossRef]
- Pavez-Muñoz, E.; González, C.; Fernández-Sanhueza, B.; Sánchez, F.; Escobar, B.; Ramos, R.; Fuenzalida, V.; Galarce, N.; Arriagada, G.; Neira, V.; et al. Antimicrobial usage factors and resistance profiles of Shiga toxin-producing Escherichia coli in backyard production systems from Central Chile. Front. Vet. Sci. 2020, 7, 595149. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Achtman, M.; Agama Study Group. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Contreras, C.A.; Ochoa, T.J.; Ruiz, J.; Lacher, D.W.; Rivera, F.P.; Saenz, Y.; Chea-Woo, E.; Zavaleta, N.; Gil, A.I.; Lanata, C.F.; et al. Phylogenetic relationships of Shiga toxin-producing Escherichia coli isolated from Peruvian Children. J. Med. Microbiol. 2011, 60, 639–646. [Google Scholar] [CrossRef]
- Feng, P.C.H.; Delannoy, S.; Lacher, D.W.; Dos Santos, L.F.; Beutin, L.; Fach, P.; Rivas, M.; Hartland, E.L.; Paton, A.W.; Guth, B.E.C. Genetic diversity and virulence potential of Shiga toxin-producing Escherichia coli O113:H21 strains isolated from clinical, environmental, and food sources. Appl. Environ. Microbiol. 2014, 80, 4757–4763. [Google Scholar] [CrossRef] [PubMed]
- Amézquita-Montes, Z.; Tamborski, M.; Kopsombut, U.G.; Zhang, C.; Arzuza, O.S.; Gómez-Duarte, O.G. Genetic relatedness among Escherichia coli pathotypes isolated from food products for human consumption in Cartagena, Colombia. Foodborne Pathog. Dis. 2015, 12, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Cadona, J.S.; Bustamante, A.V.; González, J.; Sanso, A.M. Genetic relatedness and novel sequence types of non-O157 Shiga toxin-producing Escherichia coli strains isolated in Argentina. Front. Cell Infect. Microbiol. 2016, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Furlan, J.P.R.; da Silva Ferreira, M.E.; Stehling, E.G. Genetic diversity of multidrug-resistant CMY-producing Escherichia coli from feces and soil in a small-scale pig farm. Microb. Drug Resist. 2020, 26, 1365–1371. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Dohoo, I.R.; Martin, S.W.; Stryhn, H. Veterinary Epidemiologic Research; VER: Burbank, CA, USA, 2009; ISBN 978-0-919013-60-5. [Google Scholar]
- Hosmer, D.W.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression, 3rd ed.; Wiley Series in Probability and Statistics; Wiley: Hoboken, NJ, USA, 2013; ISBN 978-0-470-58247-3. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.r-project.org/ (accessed on 31 March 2021).
- Sánchez, F.; Fuenzalida, V.; Ramos, R.; Escobar, B.; Neira, V.; Borie, C.; Lapierre, L.; López, P.; Venegas, L.; Dettleff, P.; et al. Genomic features and antimicrobial resistance patterns of Shiga toxin-producing Escherichia coli strains isolated from food in Chile. Zoonoses Public Health 2021, 68, 226–238. [Google Scholar] [CrossRef]
- Krüger, A.; Lucchesi, P.M.A.; Sanso, A.M.; Etcheverría, A.I.; Bustamante, A.V.; Burgán, J.; Fernández, L.; Fernández, D.; Leotta, G.; Friedrich, A.W.; et al. Genetic characterization of Shiga toxin-producing Escherichia coli O26:H11 strains isolated from animal, food, and clinical samples. Front. Cell Infect. Microbiol. 2015, 5, 74. [Google Scholar] [CrossRef]
- Mota, M.I.; Vázquez, S.; Cornejo, C.; D’Alessandro, B.; Braga, V.; Caetano, A.; Betancor, L.; Varela, G. Does Shiga toxin-producing Escherichia coli and Listeria monocytogenes contribute significantly to the burden of antimicrobial resistance in Uruguay? Front. Vet. Sci. 2020, 7, 583930. [Google Scholar] [CrossRef]
- Serio, A.W.; Magalhães, M.L.; Blanchard, J.S.; Connolly, L.E. Aminoglycosides: Mechanisms of Action and Resistance. In Antimicrobial Drug Resistance: Mechanisms of Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Manhattan, NY, USA, 2017; Volume 1, pp. 213–229. ISBN 978-3-319-46718-4. [Google Scholar]
- Roberts, M.C.; Schwarz, S. Tetracycline and Chloramphenicol Resistance Mechanisms. In Antimicrobial Drug Resistance: Mechanisms of Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Manhattan, NY, USA, 2017; Volume 1, pp. 231–243. ISBN 978-3-319-46718-4. [Google Scholar]
- Bush, K. The Importance of β-Lactamases to the Development of New β-Lactams. In Antimicrobial Drug Resistance: Mechanisms of Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Manhattan, NY, USA, 2017; Volume 1, pp. 165–175. ISBN 978-3-319-46718-4. [Google Scholar]
- Changkaew, K.; Utrarachkij, F.; Siripanichgon, K.; Nakajima, C.; Suthienkul, O.; Suzuki, Y. Characterization of antibiotic resistance in Escherichia coli isolated from shrimps and their environment. J. Food Prot. 2014, 77, 1394–1401. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Walkty, A.J.; Karlowsky, J.A. Fosfomycin: A first-line oral therapy for acute uncomplicated cystitis. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 2082693. [Google Scholar] [CrossRef] [PubMed]
- Schindler, B.D.; Buensalido, J.A.L.; Kaatz, G.W. Fluoroquinolone Resistance in Bacteria. In Antimicrobial Drug Resistance: Mechanisms of Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Manhattan, NY, USA, 2017; Volume 1, pp. 245–263. ISBN 978-3-319-46718-4. [Google Scholar]
- Bonomo, R.A. Mutations as a Basis of Antimicrobial Resistance. In Antimicrobial Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Manhattan, NY, USA, 2017; pp. 77–87. ISBN 978-3-319-46716-0. [Google Scholar]
- Browne, A.S.; Midwinter, A.C.; Withers, H.; Cookson, A.L.; Biggs, P.J.; Marshall, J.C.; Benschop, J.; Hathaway, S.; Haack, N.A.; Akhter, R.N.; et al. Molecular epidemiology of Shiga toxin-producing Escherichia coli (STEC) on New Zealand dairy farms: Application of a culture-independent assay and whole-genome sequencing. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Abdalhamid, B.; Mccutchen, E.; Bouska, A.; Weiwei, Z.; Loeck, B.; Hinrichs, S.; Iwen, P. Whole genome sequencing to characterize Shiga toxin-producing Escherichia coli O26 in a public health setting. J. Infect. Public Health 2019, 12, 884–889. [Google Scholar] [CrossRef]
- Lang, C.; Hiller, M.; Konrad, R.; Fruth, A.; Flieger, A. Whole-genome-based public health surveillance of less common Shiga toxin-producing Escherichia coli serovars and untypeable strains identifies four novel O genotypes. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed]
- Amigo, N.; Mercado, E.; Bentancor, A.; Singh, P.; Vilte, D.; Gerhardt, E.; Zotta, E.; Ibarra, C.; Manning, S.D.; Larzábal, M.; et al. Clade 8 and clade 6 strains of Escherichia coli O157:H7 from cattle in Argentina have hypervirulent-like phenotypes. PLoS ONE 2015, 10, e0127710. [Google Scholar] [CrossRef]
- Manning, S.D.; Motiwala, A.S.; Springman, A.C.; Qi, W.; Lacher, D.W.; Ouellette, L.M.; Mladonicky, J.M.; Somsel, P.; Rudrik, J.T.; Dietrich, S.E.; et al. Variation in virulence among clades of Escherichia coli O157: H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 2008, 105, 4868–4873. [Google Scholar] [CrossRef]
- Neupane, M.; Abu-Ali, G.; Mitra, A.; Lacher, D.; Manning, S.; Riordan, J. Shiga toxin 2 overexpression in Escherichia coli O157:H7 strains associated with severe human disease. Microb. Pathog. 2011, 51, 466–470. [Google Scholar] [CrossRef][Green Version]
- Kawase, J.; Hirai, S.; Yokoyama, E.; Hayashi, F.; Kurosaki, K.; Kawakami, Y.; Fukuma, A.; Sakai, T.; Kotani, M.; Asakura, H. Phylogeny, prevalence, and Shiga Toxin (Stx) production of clinical Escherichia coli O157 clade 2 strains isolated in Shimane Prefecture, Japan. Curr. Microbiol. 2020, 78, 265–273. [Google Scholar] [CrossRef]
- Franz, E.; van Hoek, A.H.A.M.; van der Wal, F.J.; de Boer, A.; Zwartkruis-Nahuis, A.; van der Zwaluw, K.; Aarts, H.J.M.; Heuvelink, A.E. Genetic features differentiating bovine, food, and human isolates of Shiga toxin-producing Escherichia coli O157 in The Netherlands. J. Clin. Microbiol. 2012, 50, 772–780. [Google Scholar] [CrossRef][Green Version]
- Mellor, G.E.; Besser, T.E.; Davis, M.A.; Beavis, B.; Jung, W.; Smith, H.V.; Jennison, A.V.; Doyle, C.J.; Chandry, P.S.; Gobius, K.S.; et al. Multilocus genotype analysis of Escherichia coli O157 isolates from Australia and the United States Provides evidence of geographic divergence. Appl. Environ. Microbiol. 2013, 79, 5050–5058. [Google Scholar] [CrossRef]
- Strachan, N.J.C.; Rotariu, O.; Lopes, B.; MacRae, M.; Fairley, S.; Laing, C.; Gannon, V.; Allison, L.J.; Hanson, M.F.; Dallman, T.; et al. Whole genome sequencing demonstrates that geographic variation of Escherichia coli O157 genotypes dominates host association. Sci. Rep. 2015, 5, 14145. [Google Scholar] [CrossRef] [PubMed]
- Steyert, S.R.; Sahl, J.W.; Fraser, C.M.; Teel, L.D.; Scheutz, F.; Rasko, D.A. Comparative genomics and stx phage characterization of LEE-negative Shiga toxin-producing Escherichia coli. Front. Cell Infect. Microbiol. 2012, 2, 133. [Google Scholar] [CrossRef] [PubMed]
- Bumunang, E.W.; McAllister, T.A.; Zaheer, R.; Ortega Polo, R.; Stanford, K.; King, R.; Niu, Y.D.; Ateba, C.N. Characterization of non-O157 Escherichia coli from cattle faecal samples in the North-West Province of South Africa. Microorganisms 2019, 7, 272. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, A.; Iyoda, S.; Kikuchi, T.; Ogura, Y.; Katsura, K.; Ohnishi, M.; Hayashi, T.; Thomson, N.R. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 2015, 22, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Miko, A.; Delannoy, S.; Fach, P.; Strockbine, N.A.; Lindstedt, B.A.; Mariani-Kurkdjian, P.; Reetz, J.; Beutin, L. Genotypes and virulence characteristics of Shiga toxin-producing Escherichia coli O104 strains from different origins and sources. Int. J. Med. Microbiol. 2013, 303, 410–421. [Google Scholar] [CrossRef]
- Alonso, M.Z.; Krüger, A.; Sanz, M.E.; Padola, N.L.; Lucchesi, P.M.A. Serotypes, virulence profiles and Stx subtypes of Shigatoxigenic Escherichia coli isolated from chicken derived products. Rev. Argent. Microbiol. 2016, 48, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Sanso, A.M.; Bustamante, A.V.; Krüger, A.; Cadona, J.S.; Alfaro, R.; Cáceres, M.E.; Fernández, D.; Lucchesi, P.M.A.; Padola, N.L. Molecular epidemiology of Shiga toxin-producing O113:H21 isolates from cattle and meat. Zoonoses Public Health 2018, 65, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Castillo, D.; Navas-Suárez, P.E.; Gondim, M.F.; Esposito, F.; Sacristán, C.; Fontana, H.; Fuga, B.; Piovani, C.; Kooij, R.; Lincopan, N.; et al. Genomic characterization of multidrug-resistant ESBL-producing Escherichia coli ST58 causing fatal colibacillosis in critically endangered Brazilian Merganser (Mergus octosetaceus). Transbound. Emerg. Dis. 2020, 68, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Chromek, M.; Frykman, A.; Jernberg, C.; Georgieva, V.; Hansson, S.; Zhang, J.; Marits, A.K.; Wan, C.; Matussek, A.; et al. Whole-genome characterization of hemolytic uremic syndrome-causing Shiga toxin-producing Escherichia coli in Sweden. Virulence 2021, 12, 1296–1305. [Google Scholar] [CrossRef]
- Dallman, T.J.; Greig, D.R.; Gharbia, S.E.; Jenkins, C. Phylogenetic context of Shiga toxin-producing Escherichia coli serotype O26:H11 in England. Microb. Genom. 2021, 1–9. [Google Scholar] [CrossRef]
- Jinnerot, T.; Tomaselli, A.T.P.; Johannessen, G.S.; Söderlund, R.; Urdahl, A.M.; Aspán, A.; Sekse, C. The prevalence and genomic context of Shiga toxin 2a genes in E. coli found in cattle. PLoS ONE 2020, 15, e0232305. [Google Scholar] [CrossRef]
- Carter, M.Q.; Quinones, B.; He, X.; Zhong, W.; Louie, J.W.; Lee, B.G.; Yambao, J.C.; Mandrell, R.E.; Cooley, M.B. An environmental Shiga toxin-producing Escherichia coli O145 clonal population exhibits high-level phenotypic variation that includes virulence traits. Appl. Environ. Microbiol. 2016, 82, 1090–1101. [Google Scholar] [CrossRef]
- Koo, H.-J.; Kwak, H.-S.; Yoon, S.-H.; Woo, G.-J. Phylogenetic group distribution and prevalence of virulence genes in Escherichia coli isolates from food samples in South Korea. World J. Microbiol. Biotechnol. 2012, 28, 1813–1816. [Google Scholar] [CrossRef]
- Peng, Z.; Liang, W.; Hu, Z.; Li, X.; Guo, R.; Hua, L.; Tang, X.; Tan, C.; Chen, H.; Wang, X.; et al. O-serogroups, virulence genes, antimicrobial susceptibility, and MLST genotypes of Shiga toxin-producing Escherichia coli from swine and cattle in Central China. BMC Vet. Res. 2019, 15, 427. [Google Scholar] [CrossRef]
- Basu, D.; Tumer, N.E. Do the A subunits contribute to the differences in the toxicity of Shiga toxin 1 and Shiga toxin 2? Toxins 2015, 7, 1467–1485. [Google Scholar] [CrossRef]
- Akiyama, Y.; Futai, H.; Saito, E.; Ogita, K.; Sakae, H.; Fukunaga, M.; Tsuji, H.; Chikahira, M.; Iguchi, A. Shiga toxin subtypes and virulence genes in Escherichia coli isolated from cattle. Jpn. J. Infect. Dis. 2017, 70, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Munns, K.D.; Selinger, L.B.; Stanford, K.; Guan, L.; Callaway, T.R.; McAllister, T.A. Perspectives on super-shedding of Escherichia coli O157:H7 by cattle. Foodborne Pathog. Dis. 2015, 12, 89–103. [Google Scholar] [CrossRef]
- Fitzgerald, S.F.; Beckett, A.E.; Palarea-Albaladejo, J.; McAteer, S.; Shaaban, S.; Morgan, J.; Ahmad, N.I.; Young, R.; Mabbott, N.A.; Morrison, L.; et al. Shiga toxin sub-type 2a increases the efficiency of Escherichia coli O157 transmission between animals and restricts epithelial regeneration in bovine enteroids. PLoS Pathog. 2019, 15, e1008003. [Google Scholar] [CrossRef]
- Sánchez, S.; Llorente, M.T.; Echeita, M.A.; Herrera-León, S. Development of three multiplex PCR assays targeting the 21 most clinically relevant serogroups associated with Shiga toxin-producing E. coli infection in humans. PLoS ONE 2015, 10, e0117660. [Google Scholar] [CrossRef]
- Vélez, M.V.; Colello, R.; Etcheverría, A.I.; Vidal, R.M.; Montero, D.A.; Acuña, P.; Guillén Fretes, R.M.; Toro, M.; Padola, N.L. Distribution of Locus of Adhesion and Autoaggregation and hes Gene in STEC strains from countries of Latin America. Curr. Microbiol. 2020, 77, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- Farfan, M.J.; Cantero, L.; Vidal, R.; Botkin, D.J.; Torres, A.G. Long polar fimbriae of Enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect. Immun. 2011, 79, 3744–3750. [Google Scholar] [CrossRef]
- McWilliams, B.D.; Torres, A.G. Enterohemorrhagic Escherichia coli adhesins. Microbiol. Spectr. 2014, 2, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Prado, J.V.; Cavagnaro, S.M.F. Síndrome hemolítico urémico asociado a infección intestinal por Escherichia coli productora de Shigatoxina (STEC) en pacientes chilenos: Aspectos clínicos y epidemiológicos. Rev. Chil. Infectol. 2008, 25, 435–444. [Google Scholar] [CrossRef][Green Version]
- de Souza, R.L.; Abreu Carvalhaes, J.T.; Sanae Nishimura, L.; de Andrade, M.C.; Cabilio Guth, B.E. Hemolytic uremic syndrome in pediatric intensive care units in São Paulo, Brazil. Open Microbiol. J. 2011, 5, 76–82. [Google Scholar] [CrossRef]
- Pérez, L.; Apezteguía, L.; Piñeyrúa, C.; Dabezies, A.; Bianco, M.N.; Schelotto, F.; Varela, G. Hemolytic uremic syndrome with mild renal involvement due to Shiga toxin-producing Escherichia coli (STEC) O145 strain. Rev. Argent. Microbiol. 2014, 46, 103–106. [Google Scholar] [CrossRef]
- Lorenz, S.C.; Son, I.; Maounounen-Laasri, A.; Lin, A.; Fischer, M.; Kase, J.A. Prevalence of hemolysin genes and comparison of ehxA subtype patterns in Shiga toxin-producing Escherichia coli (STEC) and non-STEC strains from clinical, food, and animal sources. Appl. Environ. Microbiol. 2013, 79, 6301–6311. [Google Scholar] [CrossRef]
- Fu, S.; Bai, X.; Fan, R.; Sun, H.; Xu, Y.; Xiong, Y. Genetic diversity of the enterohaemolysin gene (ehxa) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci. Rep. 2018, 8, 4233. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Sinha, B.; Kuczius, T.; Karch, H. Cytolethal distending toxin from Shiga toxin-producing Escherichia coli O157 causes irreversible G2/M arrest, inhibition of proliferation, and death of human endothelial cells. Infect. Immun. 2005, 73, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Kujawa, M.; McConnell, E.; Paton, A.W.; Paton, J.C.; Wójcik, C. Decreased ER-associated degradation of alpha-TCR induced by Grp78 depletion with the SubAB cytotoxin. Int. J. Biochem. Cell Biol. 2008, 40, 2865–2879. [Google Scholar] [CrossRef] [PubMed]
- Amaral, M.M.; Sacerdoti, F.; Jancic, C.; Repetto, H.A.; Paton, A.W.; Paton, J.C.; Ibarra, C. Action of Shiga toxin type-2 and subtilase cytotoxin on human microvascular endothelial cells. PLoS ONE 2013, 8, e70431. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, C.-A.; Fanning, S.; Karczmarczyk, M.; Byrne, B.; Monaghan, Á.; Bolton, D.; Sweeney, T. Characterizing the multidrug resistance of non-O157 Shiga toxin-producing Escherichia coli isolates from cattle farms and abattoirs. Microb. Drug Resist. 2017, 23, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Valat, C.; Haenni, M.; Saras, E.; Auvray, F.; Forest, K.; Oswald, E.; Madec, J.-Y. CTX-M-15 Extended-spectrum β-lactamase in a Shiga toxin-producing Escherichia coli isolate of serotype O111:H8. Appl. Environ. Microbiol. 2012, 78, 1308–1309. [Google Scholar] [CrossRef]
- Arvand, M.; Bettge-Weller, G.; Fruth, A.; Uphoff, H.; Pfeifer, Y. Extended-spectrum beta-lactamase-producing Shiga toxin gene (stx1)-positive Escherichia coli O91:H14 carrying BlaCTX-M-15 on an IncI1-ST31 plasmid isolated from a human patient in Germany. Int. J. Med. Microbiol. 2015, 305, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Day, M.; Doumith, M.; Jenkins, C.; Dallman, T.J.; Hopkins, K.L.; Elson, R.; Godbole, G.; Woodford, N. Antimicrobial resistance in Shiga toxin-producing Escherichia coli serogroups O157 and O26 Isolated from human cases of diarrhoeal disease in England, 2015. J. Antimicrob. Chemother. 2017, 72, 145–152. [Google Scholar] [CrossRef]
- World Health Organization (WHO) Critically Important Antimicrobials Human Medicine. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf?ua=1 (accessed on 1 April 2021).
- World Organization for Animal Health (OIE) OIE List of Antimicrobial Agents of Veterinary Importance. Available online: https://www.oie.int/fileadmin/Home/esp/Our_scientific_expertise/docs/pdf/AMR/E_OIE_Lista_antimicrobianos_Julio2019.pdf (accessed on 1 April 2021).
- Servicio Agrícola y Ganadero de Chile (SAG) Resolución N°1992. 5 Mayo 2006. Available online: https://www.bcn.cl/leychile (accessed on 1 April 2021).
- Servicio Agrícola y Ganadero de Chile (SAG) Resolución Exenta No:4854/2019, Modifica Resolución Exenta N° 6.801, de 2017. Available online: http://www.sag.cl/sites/default/files/resol_4.854-2019.pdf (accessed on 1 April 2021).
- Normativa y Avisos Legales de Uruguay (IMPO) Decreto N° 98/011 Prohibicion Del Uso de Antibioticos En La Alimentacion Para Animales Ovinos y Bovinos. Available online: https://www.impo.com.uy/bases/decretos/98-2011/1 (accessed on 1 April 2021).


| stx1 | 95% CI | ||||
| Variable | Categories | Odds Ratio | p-Value | Lower | Upper |
| Intercept | 0.238 | 0.059 | 0.054 | 1.057 | |
| Geographical location | Argentina | Reference | |||
| Chile | 0.657 | 0.640 | 0.113 | 3.832 | |
| Other | 3.997 | 0.060 | 0.944 | 16.914 | |
| Isolation source | Beef | Reference | |||
| Cattle | 3.800 | 0.017 | 1.265 | 11.419 | |
| Human | 1.069 | 0.935 | 0.214 | 5.347 | |
| stx1 + stx2 | 95% CI | ||||
| Variable | Categories | Odds Ratio | p-Value | Lower | Upper |
| Intercept | 0.176 | 0.046 | 0.032 | 0.968 | |
| Geographical location | Argentina | Reference | |||
| Chile | 0.688 | 0.714 | 0.094 | 5.061 | |
| Others | 8.541 | 0.037 | 1.142 | 63.859 | |
| Isolation source | Beef | Reference | |||
| Cattle | 4.125 | 0.020 | 1.255 | 13.557 | |
| Human | 0.215 | 0.136 | 0.028 | 1.619 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galarce, N.; Sánchez, F.; Escobar, B.; Lapierre, L.; Cornejo, J.; Alegría-Morán, R.; Neira, V.; Martínez, V.; Johnson, T.; Fuentes-Castillo, D.; et al. Genomic Epidemiology of Shiga Toxin-Producing Escherichia coli Isolated from the Livestock-Food-Human Interface in South America. Animals 2021, 11, 1845. https://doi.org/10.3390/ani11071845
Galarce N, Sánchez F, Escobar B, Lapierre L, Cornejo J, Alegría-Morán R, Neira V, Martínez V, Johnson T, Fuentes-Castillo D, et al. Genomic Epidemiology of Shiga Toxin-Producing Escherichia coli Isolated from the Livestock-Food-Human Interface in South America. Animals. 2021; 11(7):1845. https://doi.org/10.3390/ani11071845
Chicago/Turabian StyleGalarce, Nicolás, Fernando Sánchez, Beatriz Escobar, Lisette Lapierre, Javiera Cornejo, Raúl Alegría-Morán, Víctor Neira, Víctor Martínez, Timothy Johnson, Danny Fuentes-Castillo, and et al. 2021. "Genomic Epidemiology of Shiga Toxin-Producing Escherichia coli Isolated from the Livestock-Food-Human Interface in South America" Animals 11, no. 7: 1845. https://doi.org/10.3390/ani11071845
APA StyleGalarce, N., Sánchez, F., Escobar, B., Lapierre, L., Cornejo, J., Alegría-Morán, R., Neira, V., Martínez, V., Johnson, T., Fuentes-Castillo, D., Sano, E., & Lincopan, N. (2021). Genomic Epidemiology of Shiga Toxin-Producing Escherichia coli Isolated from the Livestock-Food-Human Interface in South America. Animals, 11(7), 1845. https://doi.org/10.3390/ani11071845








