Screening the Presence of Non-Typhoidal Salmonella in Different Animal Systems and the Assessment of Antimicrobial Resistance
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Sample Collection
2.2. Salmonella Isolation
2.3. Serogroup Characterization and Antimicrobial Susceptibility
2.4. Map of the Isolation Sites
2.5. Statistical Analyses
3. Results
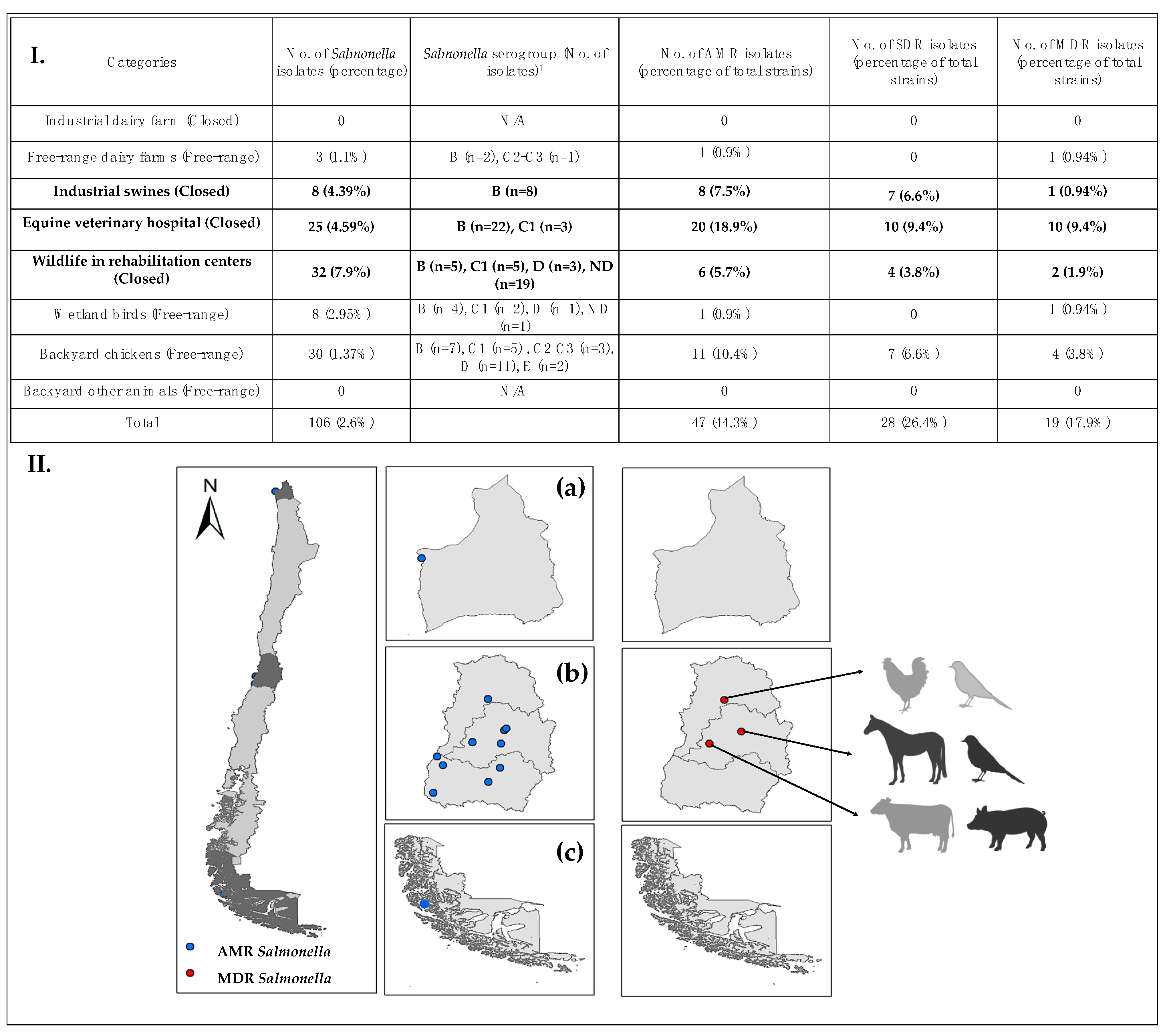
3.1. Salmonella Presence and Serogroups
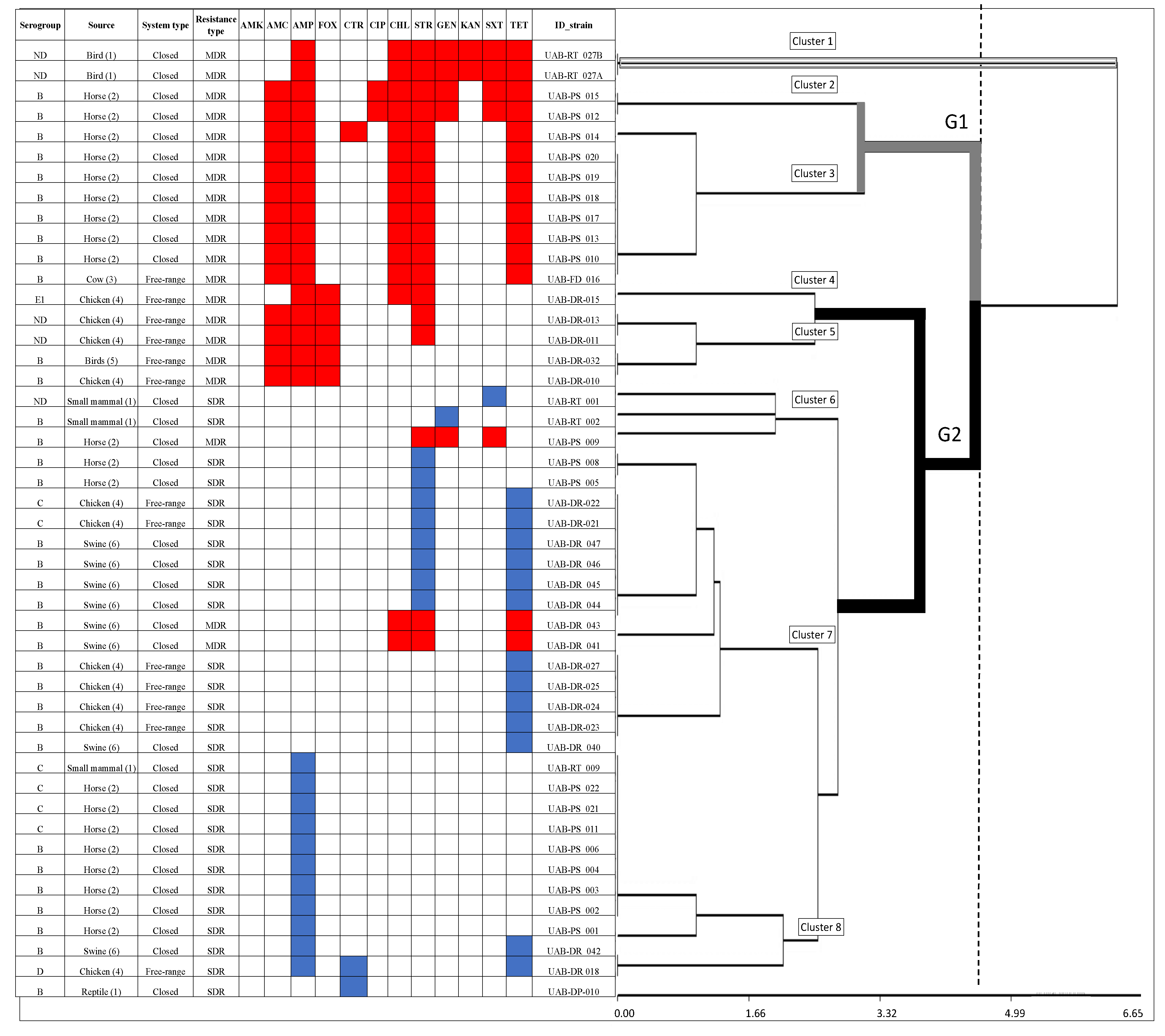
3.2. Antimicrobial Resistance and Susceptibility
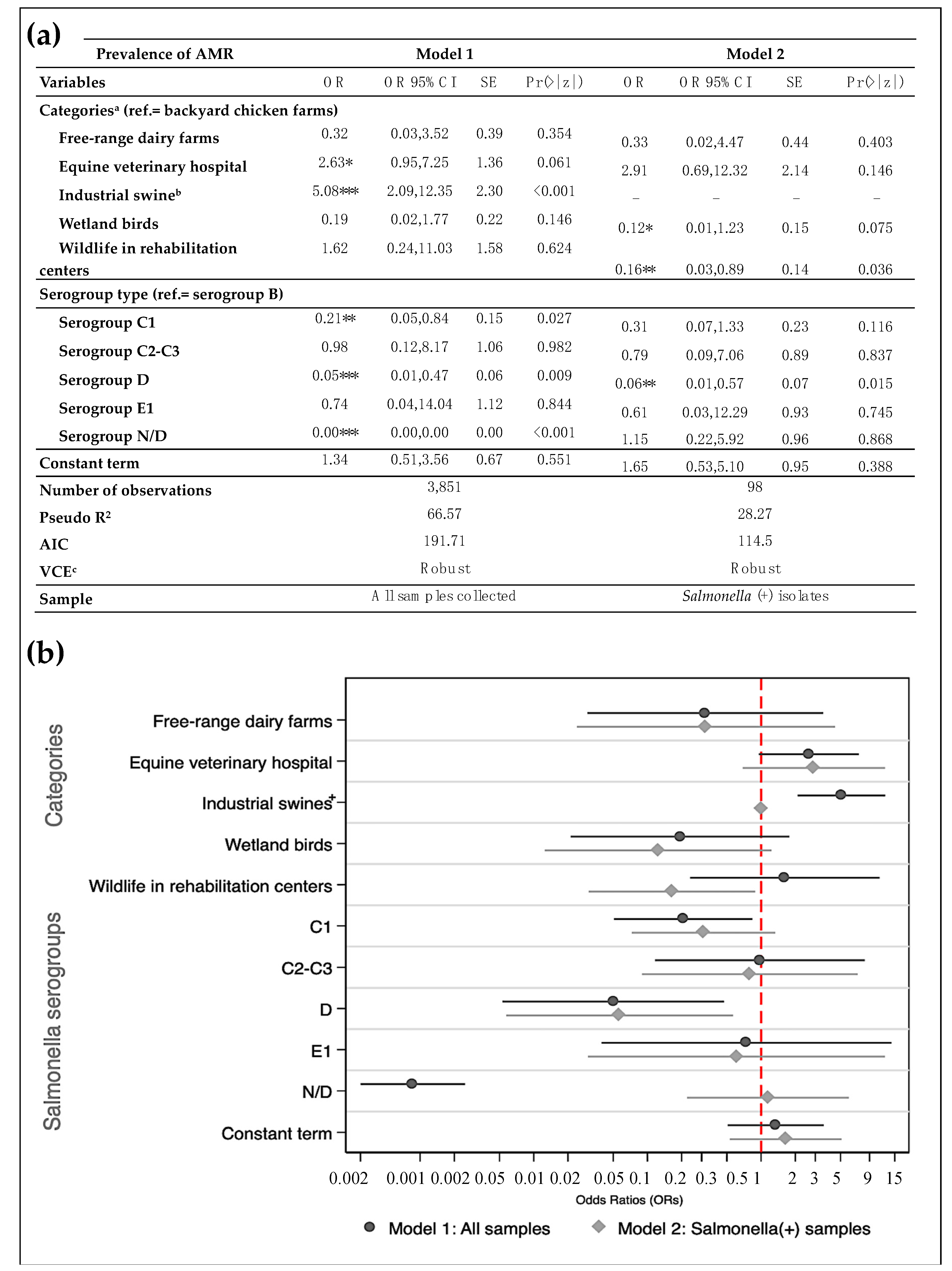
3.3. Likelihood of Antimicrobial-Resistance and Multidrug-Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Bugarel, M.; Tudor, A.; Loneragan, G.; Nightingale, K. Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J. Microbiol. Methods 2017, 134, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Gal-Mor, O. Persistent Infection and Long-Term Carriage of Typhoidal and Nontyphoidal Salmonellae. Clin. Microbiol. Rev. 2018, 32. [Google Scholar] [CrossRef]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.-X. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Agbaje, M.; Begum, R.H.; Oyekunle, M.A.; Ojo, O.E.; Adenubi, O.T. Evolution of Salmonella nomenclature: A critical note. Folia Microbiol. 2011, 56, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Grimont, P.; Weill, F. Antigenic Formulae of the Salmonella Serovars, 9th ed.; World Health Organization (WHO), Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007; Available online: https://www.pasteur.fr/sites/default/files/veng_0.pdf (accessed on 25 March 2019).
- Ranieri, M.L.; Shi, C.; Switt, A.I.M.; Bakker, H.C.D.; Wiedmann, M. Salmonella serovar prediction: Comparison of Typing Methods with a New Procedure Based on Sequence Characterization for Salmonella Serovar Prediction. J. Clin. Microbiol. 2013, 51, 1786–1797. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Global (WHO). Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 25 March 2019).
- Arnott, A.; Wang, Q.; Bachmann, N.; Sadsad, R.; Biswas, C.; Sotomayor, C.; Howard, P.; Rockett, R.; Wiklendt, A.; Iredell, J.R.; et al. Multidrug-Resistant Salmonella enterica 4,[5], 12:i:- Sequence Type 34, New South Wales, Australia, 2016–2017. Emerg. Infect Dis. 2018, 24, 751–753. [Google Scholar] [CrossRef]
- Brown, A.C.; Grass, J.E.; Richardson, L.C.; Nisler, A.L.; Bicknese, A.S.; Gould, L.H. Antimicrobial resistance in Salmonella that caused foodborne disease outbreaks: United States, 2003–2012. Epidemiol. Infect. 2017, 145, 766–774. [Google Scholar] [CrossRef]
- Doyle, M.E. Multidrug-Resistant Pathogens in the Food Supply. Foodborne Pathog. Dis. 2015, 12, 261–279. [Google Scholar] [CrossRef]
- Michael, G.; Schwarz, S. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: An alarming trend? Clin. Microbiol. Infect. 2016, 22, 968–974. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2013. Available online: https://www.cdc.gov/narms/pdf/2013-annual-report-narms-508c.pdf (accessed on 10 March 2020).
- Martínez, M.C.; Retamal, P.; Rojas-Aedo, J.F.; Fernández, J.; Lapierre, L.; Fernández, A. Multidrug-Resistant Outbreak-Associated Salmonella Strains in Irrigation Water from the Metropolitan Region, Chile. Zoonoses Public Health 2016, 64, 299–304. [Google Scholar] [CrossRef]
- Alegria-Moran, R.; Rivera, D.; Toledo, V.; Moreno-Switt, A.I.; Hamilton-West, C. First detection and characterization of Salmonella spp. in poultry and swine raised in backyard production systems in central Chile. Epidemiol. Infect. 2017, 145, 3180–3190. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Castillo, D.; Farfán-López, M.; Esposito, F.; Moura, Q.; Fernandes, M.R.; Lopes, R.; Cardoso, B.; Muñoz, M.E.; Cerdeira, L.; Najle, I.; et al. Wild owls colonized by international clones of extended-spectrum β-lactamase (CTX-M)-producing Escherichia coli and Salmonella Infantis in the Southern Cone of America. Sci. Total Environ. 2019, 674, 554–562. [Google Scholar] [CrossRef]
- Marchant, P.; Hidalgo-Hermoso, E.; Espinoza, K.; Retamal, P. Prevalence of Salmonella enterica and Shiga toxin-producing Escherichia coli in zoo animals from Chile. J. Vet. Sci. 2016, 17, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.; Retamal, P.; Ayers, S.; Barreto, M.; Allard, M.; Brown, E.W.; Gonzalez-Escalona, N. Whole-Genome Sequencing Analysis of Salmonella enterica Serovar Enteritidis Isolates in Chile Provides Insights into Possible Transmission between Gulls, Poultry, and Humans. Appl. Environ. Microbiol. 2016, 82, 6223–6232. [Google Scholar] [CrossRef]
- Tardone, R.; Rivera, D.; Dueñas, F.; Sallaberry-Pincheira, N.; Hamilton-West, C.; Adell, A.D.; Moreno-Switt, A.I. Short Communication: Salmonella in Raptors and Aquatic Wild Birds in Chile. J. Wildl. Dis. 2020, 56, 707. [Google Scholar] [CrossRef]
- Dueñas, F.; Rivera, D.; Toledo, V.; Tardone, R.; Hervé-Claude, L.P.; Hamilton-West, C.; Switt, A.I.M. Short communication: Characterization of Salmonella phages from dairy calves on farms with history of diarrhea. J. Dairy Sci. 2017, 100, 2196–2200. [Google Scholar] [CrossRef] [PubMed]
- Soza-Ossandón, P.; Rivera, D.; Tardone, R.; Riquelme-Neira, R.; García, P.; Hamilton-West, C.; Adell, A.D.; González-Rocha, G.; Moreno-Switt, A.I. Widespread Environmental Presence of Multidrug-Resistant Salmonella in an Equine Veterinary Hospital That Received Local and International Horses. Front. Vet. Sci. 2020, 7, 346. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Vasquez, N.; Baumberger, C.; Jimenez-Bluhm, P.; Di Pillo, F.; Lazo, A.; Sanhueza, J.; Schultz-Cherry, S.; Hamilton-West, C. Risk factors and spatial relative risk assessment for influenza A virus in poultry and swine in backyard production systems of central Chile. Vet. Med. Sci. 2020, 6, 518–526. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, G.G.; Park, J.S.; Jung, Y.H.; Kwak, H.S.; Kim, S.B.; Nam, Y.S.; Kwon, S.T. A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J. Food Prot. 2007, 70, 1656–1662. [Google Scholar] [CrossRef]
- Toro, M.; Rivera, D.; Toledo, V.; Campos-Vargas, R.; Allard, M.W.; Hamilton-West, C.; Moreno-Switt, A.I. Genomics of Salmonella contaminating backyard production systems reveals persistence and transmission of genetically related Salmonella on a farm basis. Zoonoses Public Health 2018, 65, 1008–1014. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. M100, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States 2019; CDC: Atlanta, GA, USA, 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 25 March 2020).
- Instituto de Salud Pública de Chile. Boletín Vigilancia de Laboratorio: Salmonella spp. 2012–2016; Instituto de Salud Pública de Chile: Santiago, Chile, 2017; Available online: http://www.ispch.cl/sites/default/files/BoletinSalmonella-23012017A.pdf (accessed on 4 March 2020).
- Ministerio de Salud de Chile (MINSAL), Departamento de Epidemiología. División de Planificación Sanitaria Ministerio de Salud de Chile. Boletín de Brotes Vol. 1/N° 3. 2018. Available online: http://epi.minsal.cl/wp-content/uploads/2018/03/Bolet%C3%ADn_Brotes_3.pdf (accessed on 4 March 2020).
- Ministerio de Salud de Chile (MINSAL), Departamento de Epidemiología, División de Planificación Sanitaria Ministerio de Salud de Chile. Boletín Epidemiólogico Trimestral. Brote de Enfermedades Transmitidas por Alimentos. 2019. Available online: http://epi.minsal.cl/wp-content/uploads/2020/02/BET_ETA_2019.pdf (accessed on 4 March 2020).
- Lapierre, L.; Cornejo, J.; Zavala, S.; Galarce, N.; Sánchez, F.; Benavides, M.B.; Guzmán, M.; Sáenz, L. Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile. Animals 2020, 10, 1049. [Google Scholar] [CrossRef]
- Colegio Médico Veterinario de Chile. Buenas Prácticas en el Uso de Antimicrobianos en Animales Pequeños. Available online: https://www.colmevet.cl/doc/ManualBuenasPracticasUsoAntimicrobianos.pdf (accessed on 17 May 2021).
- Universidad de Chile. Manual de Buenas Prácticas en el Uso de Antimicrobianos en Salmonicultura Chilena, 4th ed.; Universidad de Chile: Santiago, Chile, 2021; Available online: http://www.sernapesca.cl/sites/default/files/manual_de_buenas_practicas_20210217.pdf (accessed on 17 May 2021).
- Cevidanes, A.; Esperón, F.; Di Cataldo, S.; Neves, E.; Sallaberry-Pincheira, N.; Millán, J. Antimicrobial resistance genes in Andean foxes inhabiting anthropized landscapes in central Chile. Sci. Total Environ. 2020, 724, 138247. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Orellana, D.; Martínez, C. Characterization of Campylobacter jejuni samples coming from poultry meat and feces. Rev. Med. Chile 2017, 145, 1551–1558. [Google Scholar] [CrossRef]
- Lozano-Muñoz, I.; Wacyk, J.; Kretschmer, C.; Vásquez-Martínez, Y.; Martin, M.C.-S. Antimicrobial resistance in Chilean marine-farmed salmon: Improving food safety through One Health. One Health 2021, 12, 100219. [Google Scholar] [CrossRef] [PubMed]
- Vico, J.; Lorenzutti, A.; Zogbi, A.; Aleu, G.; Sánchez, I.; Caffer, M.; Rosmini, M.; Mainar-Jaime, R. Prevalence, associated risk factors, and antimicrobial resistance profiles of non-typhoidal Salmonella in large scale swine production in Córdoba, Argentina. Res. Vet. Sci. 2020, 130, 161–169. [Google Scholar] [CrossRef]
- Ekakoro, J.E.; Okafor, C.C. Antimicrobial use practices of veterinary clinicians at a veterinary teaching hospital in the United States. Vet. Anim. Sci. 2019, 7, 100038. [Google Scholar] [CrossRef] [PubMed]
- Strategy on Antibiotic Resistance (StAR). Newsletter Swiss Strategy on Antibiotic Resistance (StAR). 2021. Available online: https://www.star.admin.ch/star/en/home.html (accessed on 4 March 2021).
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Surveillance for Foodborne Disease Outbreaks, United States, 2016, Annual Report; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2018. Available online: https://www.cdc.gov/fdoss/pdf/2016_FoodBorneOutbreaks_508.pdf (accessed on 4 March 2021).
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A Review of Antibiotic Use in Food Animals: Perspective, Policy, and Potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M.; et al. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef]
- Parmley, E.J.; Pintar, K.; Majowicz, S.; Avery, B.; Cook, A.; Jokinen, C.; Gannon, V.; Lapen, D.R.; Topp, E.; Edge, T.A.; et al. A Canadian Application of One Health: Integration of Salmonella Data from Various Canadian Surveillance Programs (2005–2010). Foodborne Pathog. Dis. 2013, 10, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.; Reginatto, G.A.; Español, A.R.; Colantonio, L.D.; Burrone, M.S. Resistencia antimicrobiana de Salmonella spp aislada de alimentos de origen animal para consumo humano. Rev. Peru. Med. Exp. Salud Pública 2016, 33, 32. [Google Scholar] [CrossRef] [PubMed]
| Categories | Animal Source(s) | Type | Geographical Location | Sample Type | Isolation Years | No. of Farms/Sites | No. of Samples |
|---|---|---|---|---|---|---|---|
| Industrial dairy farms | Cows | Closed | Southern Chile | Environmental | 2015–2017 | 8 | 160 |
| Free-range dairy farms | Cows | Free-range | Central Chile | Environmental | 2016–2017 | 13 | 260 |
| Industrial swine | Pigs | Closed | Central Chile | Environmental | 2015 | 10 | 182 |
| Equine veterinary hospitals | Horses | Closed | Central Chile | Environmental and animal | 2016–2017 | 1 | 545 |
| Wildlife in rehabilitation centers | Small mammals, reptiles and birds | Closed | Central Chile | Animal | 2015–2017 | 3 | 405 |
| Wetland birds | Birds | Free-range | Northern and Southern Chile | Animal | 2016 | 4 | 271 |
| Backyard chickens | Chickens | Free-range | Central Chile | Environmental and animal | 2013–2015 | 329 | 2188 |
| Backyard other animals | Sheep and llamas | Free-range | Northern Chile | Animal | 2016 | 5 | 36 |
| Total | 4047 |
| Variables | Model 1: Sample | Model 2: Sample | ||
|---|---|---|---|---|
| MEAN | 95% CI | MEAN | 95% CI | |
| Salmonella prevalence with AMR (%) | 1.22 | 0.87; 1.56 | 39.8 | 29.93; 49.66 |
| MDR Salmonella prevalence (%) | 0.004 | 0.23; 0.65 | 16.33 | 8.88; 23.77 |
| System | ||||
| Backyard chicken farms (%) | 56.82 | 55.25; 58.38 | 30.61 | 21.32; 39.90 |
| Backyard dairy farms (%) | 6.75 | 5.95; 7.54 | 3.06 | −0.41; 6.53 |
| Equine veterinary hospitals (%) | 14.15 | 13.05; 15.25 | 25.51 | 16.73; 34.29 |
| Swine farms (%) | 4.73 | 4.05; 5.39 | - | - |
| Wetland birds (%) | 7.04 | 6.22; 7.84 | 8.16 | 2.65; 13.68 |
| Wildlife in rehabilitation centers (%) | 10.52 | 6.23; 11.49 | 32.65 | 23.21; 41.10 |
| System type | ||||
| Closed (%) | 29.39 | 27.96; 30.83 | 58.16 | 48.22; 68.10 |
| Open (%) | 70.61 | 69.16; 72.00 | 41.84 | 31.90; 51.78 |
| Sampling type | ||||
| Environment (%) | 31.24 | 29.77; 32.70 | 36.73 | 27.02;46.45 |
| Animal (%) | 68.76 | 67.30; 70.23 | 63.27 | 53.55; 72.98 |
| Serogroups | ||||
| B (%) | 1.25 | 0.90; 1.60 | 40.82 | 30.91; 50.72 |
| C1 (%) | 0.39 | 0.19; 0.59 | 15.31 | 8.05; 22.56 |
| C2–C3 (%) | 0.10 | 0.00; 0.21 | 4.08 | 0.09; 8.07 |
| D (%) | 0.39 | 0.19; 0.59 | 15.31 | 8.05; 22.56 |
| E1 (%) | 0.05 | −0.02; 0.12 | 2.04 | 0.81; 4.89 |
| ND (%) | 97.82 | 97.36; 98.28 | 22.45 | 14.04; 30.86 |
| Number of observations | 3851 | 98 | ||
| Sample | All samples collected | Salmonella (+) strains | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera, D.; Allel, K.; Dueñas, F.; Tardone, R.; Soza, P.; Hamilton-West, C.; Moreno-Switt, A.I. Screening the Presence of Non-Typhoidal Salmonella in Different Animal Systems and the Assessment of Antimicrobial Resistance. Animals 2021, 11, 1532. https://doi.org/10.3390/ani11061532
Rivera D, Allel K, Dueñas F, Tardone R, Soza P, Hamilton-West C, Moreno-Switt AI. Screening the Presence of Non-Typhoidal Salmonella in Different Animal Systems and the Assessment of Antimicrobial Resistance. Animals. 2021; 11(6):1532. https://doi.org/10.3390/ani11061532
Chicago/Turabian StyleRivera, Dácil, Kasim Allel, Fernando Dueñas, Rodolfo Tardone, Paula Soza, Christopher Hamilton-West, and Andrea I. Moreno-Switt. 2021. "Screening the Presence of Non-Typhoidal Salmonella in Different Animal Systems and the Assessment of Antimicrobial Resistance" Animals 11, no. 6: 1532. https://doi.org/10.3390/ani11061532
APA StyleRivera, D., Allel, K., Dueñas, F., Tardone, R., Soza, P., Hamilton-West, C., & Moreno-Switt, A. I. (2021). Screening the Presence of Non-Typhoidal Salmonella in Different Animal Systems and the Assessment of Antimicrobial Resistance. Animals, 11(6), 1532. https://doi.org/10.3390/ani11061532








