Karyological Diversification in the Genus Lyciasalamandra (Urodela: Salamandridae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Cytogenetic Analysis
3. Results
3.1. Chromosome Number and NOR Configuration
3.2. Heterochromatin Distribution and Composition
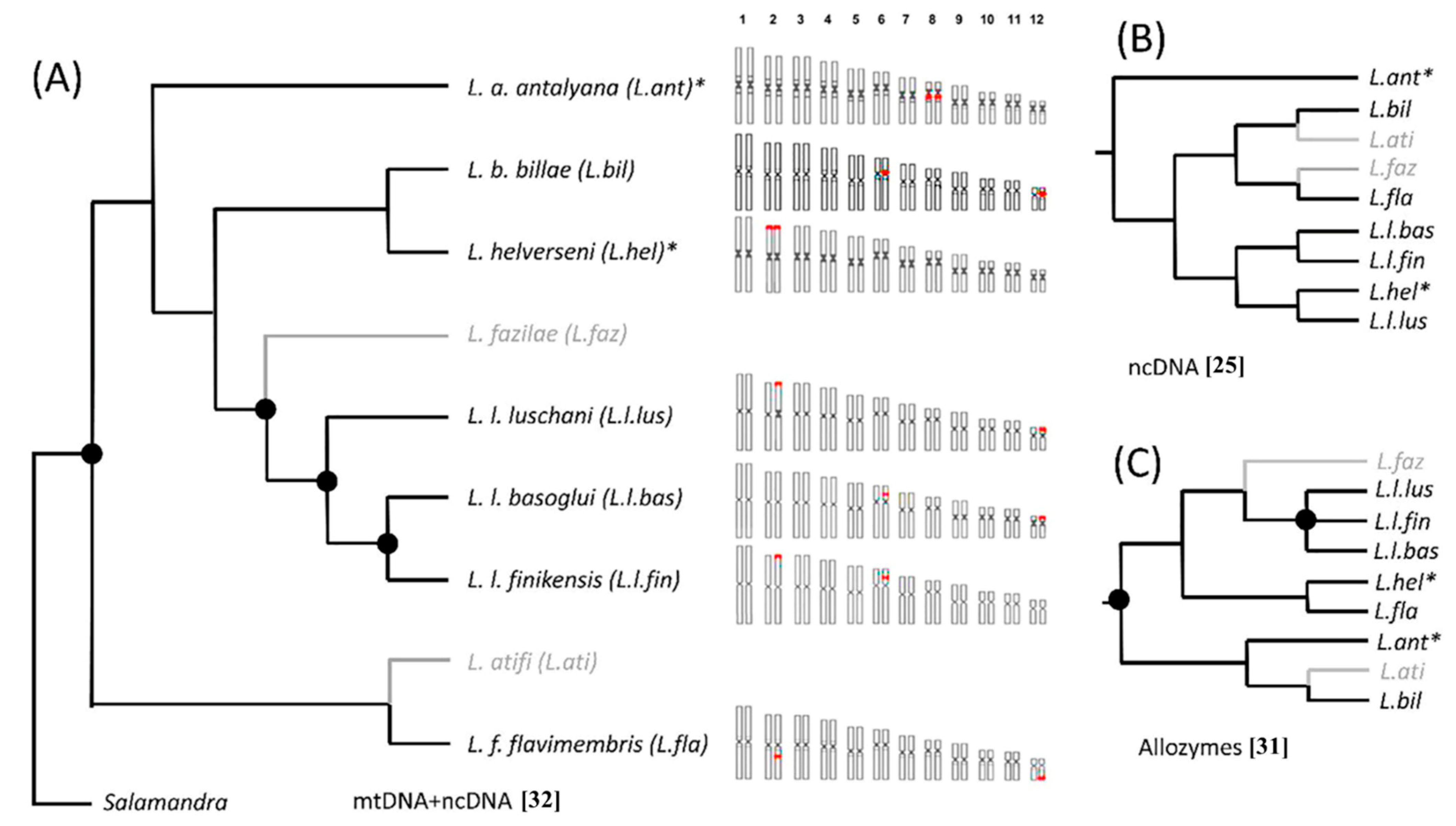
4. Discussion
4.1. Chromosome Number and Morphology
4.2. Heterochromatin Diversity and Distribution
4.3. Variability of NOR Loci
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mezzasalma, M.; Guarino, F.M.; Aprea, G.; Petraccioli, A.; Crottini, A.; Odierna, G. Karyological evidence for diversification of Italian slow worm populations (Squamata, Anguidae). Comp. Cytogenet. 2013, 7, 217–227. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Petraccioli, A.; Odierna, G.; Guarino, F.M. A karyological study of three typhlopid species with some inferences on chromosome evolution in blindsnakes (Scolecophidia). Zool. Anz. 2016, 264, 34–40. [Google Scholar] [CrossRef]
- King, M. Species Evolution: The Role of Chromosome Change; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Mezzasalma, M.; Dall’Asta, A.; Loy, A.; Cheylan, M.; Lymberakis, P.; Zuffi, M.A.L.; Tomović, L.; Odierna, G.; Guarino, F.M. A sisters’ story: Comparative phylogeography and taxonomy of Hierophis viridiflavus and H. gemonensis (Serpentes, Colubridae). Zool. Scr. 2015, 44, 495–508. [Google Scholar] [CrossRef]
- Leaché, A.D.; Banbury, B.L.; Linkem, C.W.; de Oca, A.N. Phylogenomics of a rapid radiation: Is chromosomal evolution linked to increased diversification in north american spiny lizards (Genus Sceloporus)? BMC Evol. Biol. 2016, 16, 63. [Google Scholar] [CrossRef]
- Cuadrado, Á.; de Bustos, A.; Figueroa, R.I. Chromosomal markers in the genus Karenia: Towards an understanding of the evolution of the chromosomes, life cycle patterns and phylogenetic relationships in dinoflagellates. Sci. Rep. 2019, 9, 3072. [Google Scholar] [CrossRef] [PubMed]
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. USA 2005, 102, 6535–6542. [Google Scholar] [CrossRef] [PubMed]
- De Vos, J.M.; Augustijnen, H.; Bätscher, L.; Lucek, K. Speciation through chromosomal fusion and fission in Lepidoptera. Philos. Trans. R. Soc. B Lond. Biol. Sci. 2020, 375, 20190539. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Aprea, G.; Glaw, F.; Odierna, G.; Guarino, F.M. When can chromosomes drive speciation? The peculiar case of the Malagasy tomato frogs (genus Dyscophus). Zool. Anz. 2017, 268, 41–46. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Aprea, G.; Glaw, F.; Odierna, G.; Guarino, F.M. Molecular phylogeny, biogeography and chromosome evolution of Malagasy dwarf geckos of the genus Lygodactylus (Squamata, Gekkonidae). Zool. Scr. 2017, 46, 42–54. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O. Changes in heterochromatin content and ancient chromosome fusion in the endemic Malagasy boid snakes Sanzinia and Acrantophis (Squamata: Serpentes). Salamandra 2019, 55, 140–144. Available online: http://www.salamandra-journal.com (accessed on 8 January 2021).
- Ocalewicz, K.; Dobosz, S.; Kuzminski, H.; Nowosad, J.; Goryczko, K. Chromosome rearrangements and survival of androgenetic rainbow trout (Oncorhynchus mykiss). J. Appl Genet. 2010, 51, 309–317. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Branch, W.R.; Glaw, F.; Guarino, F.M.; Nagy, Z.T.; Odierna, G.; Aprea, G. Chromosome evolution in pseudoxyrhophiine snakes from Madagascar: A wide range of karyotypic variability. Biol. J. Linn. Soc. 2014, 112, 450–460. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Glaw, F.; Odierna, G.; Petraccioli, A.; Guarino, F.M. Karyological analyses of Pseudhymenochirus merlini and Hymenochirus boettgeri provide new insights into the chromosome evolution in the anuran family Pipidae. Zool. Anz. 2015, 258, 47–53. [Google Scholar] [CrossRef]
- AmphibiaWeb. Available online: https://amphibiaweb.org/search/ (accessed on 30 April 2021).
- Sparreboom, M. Salamanders of the Old World: The Salamanders of Europe, Asia and Northern Africa; Brill: Leiden, The Netherlands, 2014. [Google Scholar]
- Steindachner, F. Über einige neue und seltene Reptilien- und Amphibienarten. Sitzungsber. Akad. Wissensch. Wien. Math. Naturwiss. Kl. 1 1891, 100, 289–314. [Google Scholar]
- Wolterstorff, A. Katalog der Amphibien-Sammlung im Museum für Natur- und Heimatkunde. Abh. Ber. Mus. Nat. Heim. Magdeburg 1925, 4, 155–310. [Google Scholar]
- Titus, T.A.; Larson, A. A molecular phylogenetic perspective on the evolutionary radiation of the salamander family Salamandridae. Syst. Biol. 1995, 44, 125–151. [Google Scholar] [CrossRef]
- Veith, M.; Steinfartz, S.; Zardoya, R.; Seitz, A.; Meyer, A. A molecular phylogeny of “true” salamanders (family Salamandridae) and the evolution of terrestriality of reproductive modes. J. Zool. Syst. Evol. Res. 1998, 36, 7–16. [Google Scholar] [CrossRef]
- Weisrock, D.W.; Macey, J.R.; Ugurtas, I.H.; Larson, A.; Papenfuss, T.J. Molecular phylogenetics and historical biogeography among salamandrids of the ‘‘true” salamander clade: Rapid branching of numerous highly divergent lineages in Mertensiella luschani associated with the rise of Anatolia. Mol. Phylogenet. Evol. 2001, 18, 434–448. [Google Scholar] [CrossRef]
- Veith, M.; Steinfartz, S. When non-monophyly results in taxonomic consequences—The case of Mertensiella within the Salamandridae (Amphibia: Urodela). Salamandra 2004, 40, 67–80. [Google Scholar]
- Weisrock, D.W.; Papenfuss, T.J.; Macey, J.R.; Litvinchuk, S.N.; Polymeni, R.; Ugurtas, I.H.; Zhao, E.; Jowkar, H.; Larson, A. A molecular assessment of phylogenetic relationships and lineage accumulation rates within the family Salamandridae (Amphibia, Caudata). Mol. Phylogenet. Evol. 2006, 41, 368–383. [Google Scholar] [CrossRef]
- Zhang, P.; Papenfuss, T.J.; Wake, M.H.; Qu, L.; Wake, D.B. Phylogeny and biogeography of the family Salamandridae (Amphibia Caudata) inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 2008, 49, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Veith, M.; Göçmen, B.; Sotiropoulos, K.; Kieren, S.; Godmann, O.; Steinfartz, S. Seven at one blow: The origin of major lineages of the viviparous Lycian salamanders (Lyciasalamandra) was triggered by a single paleo-historic event. Amphib-Reptilia 2016, 37, 373–387. [Google Scholar] [CrossRef]
- Veith, S.; Bogaerts, F.; Pasmans, S.; Kieren, S. The changing views on the evolutionary relationships of extant Salamandridae (Amphibia: Urodela). PLoS ONE 2018, 13, e0198237. [Google Scholar] [CrossRef]
- Veith, M.; Göçmen, B.; Sotiropoulos, K.; Eleftherakos, K.; Lötters, S.; Godmann, O.; Karış, M.; Oğuz, A.; Ehl, S. Phylogeographic analyses point to long-term survival on the spot in micro-endemic Lycian salamanders. PLoS ONE 2020, 15, e0226326. [Google Scholar] [CrossRef] [PubMed]
- Kieren, S.; Sparreboom, M.; Hochkirch, A.; Veith, M. A biogeographic and ecological perspective to the evolution of reproductive behaviour in the family Salamandridae. Mol. Phylogenet. Evol. 2018, 121, 98–109. [Google Scholar] [CrossRef]
- Göçmen, B.; Arikan, H.; Yalçinkaya, D. A new Lycian Salamander, threatened with extinction, from the Göynük Canyon (Antalya, Anatolia), Lyciasalamandra irfani n. sp. (Urodela: Salamandridae). North West. J. Zool. 2011, 7, 151–160. [Google Scholar]
- Göçmen, B.; Akman, B. Lyciasalamandra arikani n. sp. and L. yehudahi n. sp. (Amphibia: Salamandridae), two new Lycian salamanders from southwestern Anatolia. North West. J. Zool. 2012, 8, 181–194. [Google Scholar]
- Veith, M.; Lipscher, E.; Öz, M.; Kiefer, A.; Baran, I.; Polymeni, R.M.; Steinfartz, S. Cracking the nut: Geographical adjacency of sister taxa supports vicariance in a polytomic salamander clade in the absence of node support. Mol. Phylogenet. Evol. 2008, 47, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Ehl, S.; Vences, M.; Veith, M. Reconstructing evolution at the community level: A case study on Mediterranean amphibians. Mol. Phylogenet. Evol. 2019, 134, 211–225. [Google Scholar] [CrossRef]
- Poulakakis, N.; Pakaki, V.; Mylonas, M.; Lymberakis, P. Molecular phylogeny of the Greek legless skink Ophiomorus punctatissimus (Squamata: Scincidae): The impact of the mid-Aegean trench in its phylogeography. Mol. Phylogenet. Evol. 2008, 4, 396–402. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Di Febbraro, M.; Guarino, F.M.; Odierna, G.; Russo, D. Cold-blooded in the Ice Age: “Refugia within refugia”, inter-and intraspecific biogeographic diversification of European whipsnakes (Squamata, Colubridae, Hierophis). Zoology 2018, 127, 84–94. [Google Scholar] [CrossRef]
- Odierna, G.; Andreone, F.; Aprea, G.; Capriglione, T.; Guarino, F.M. Differenze cromosomiche tra le due sottospecie di salamandra pezzata, Salamandra salamandra salamandra (Linnaeus, 1758) e S. salamandra gigliolii Eiselt and Lanza, 1956, presenti in Italia. Pianura 2001, 13, 73–76. [Google Scholar]
- Sidhom, M.; Said, K.; Chatti, N.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O.; Mezzasalma, M. Karyological characterization of the common chameleon (Chamaeleo chamaeleon) provides insights on the evolution and diversification of sex chromosomes in Chamaeleonidae. Zoology 2020, 141, 125738. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Sahar, E.; Latt, S.A. Energy transfer and binding competition between dyes used to enhance staining differentiation in metaphase chromosomes. Chromosoma 1980, 79, 1–28. [Google Scholar] [CrossRef]
- Howell, W.M.; Black, D.A. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer, A 1-step method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstration of centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Sessions, S.K. Evolutionary cytogenetics in salamanders. Chromosome Res. 2008, 16, 183–201. [Google Scholar] [CrossRef]
- King, M. Amphibia. In Animal Cytogenetics, Chordata 2; John, B., Ed.; Gebrüder Bornträger: Stuttgart, Germany, 1990; Volume 4. [Google Scholar]
- Tan, A.M. Chromosomal variation in the northwestern American newts of the genus Taricha (Caudata: Salamandridae). Chromosome Res. 1994, 2, 281–292. [Google Scholar] [CrossRef]
- Morescalchi, A. Chromosome evolution in Caudata Amphibia. Evol. Biol. 1975, 8, 339–387. [Google Scholar]
- Ragghianti, M.; Bucci-Innocenti, S.; Mancino, G. C-banded karyotype and cytotaxonomy of Mertensiella caucasica (Waga,1876) (Caudata: Salamandridrae). Amphib. Reptil. 1982, 3, 303–307. [Google Scholar] [CrossRef]
- Hutchison, N.; Pardue, M.L. The mitotic chromosomes of Notophthalmus (= Triturus) viridescens: Localization of C-banding regions and DNA sequences complementary to 18S, 28S and 5S ribosomal RNA. Chromosoma 1975, 53, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, H.C.; Sessions, S.K. The biological significance of variation in satellite DNA and heterochromatin in newt of the genus Triturus: An evolutionary prospective. Philos. Trans. R. Soc. B 1986, 312, 243–259. [Google Scholar] [CrossRef]
- Macgregor, H.C. The evolutionary cytogenetics of Triturus (Amphibia, Urodela). An overview. In Symposium on the Evolution of Terrestrial Vertebrates. Selected Symposia and Monographs; Ghiara, G., Angelini, F., Olmo, E., Varano, L., Eds.; Mucchi: Modena, Italy, 1991; Volume 4, pp. 153–169. [Google Scholar]
- Macgregor, H.C. Chromosome heteromorphism in newts (Triturus) and its significance in relation to evolution and development. In Amphibian Cytogenetics and Evolution; Green, D.A., Sessions, S.K., Eds.; Academic Press: San Diego, CA, USA, 1991; pp. 175–196. [Google Scholar]
- John, B. The biology of heterochromatin. In Heterochromatin: Molecular and Structural Aspects; Verma, R.S., Ed.; Cambridge University Press: Cambridge, UK, 1988; pp. 1–128. [Google Scholar]
- Markova, M.; Vyskot, B. New horizons of genomic in situ hybridization. Cytogenet. Genome Res. 2009, 126, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Bellini Bardella, V.; da Rosa, J.A.; Vanzela, A.L.L. Origin and distribution of AT-rich repetitive DNA families in Triatoma infestans (Heteroptera). Infect. Genet. Evol. 2014, 23, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Panzera, F.; Sánchez, A.; Panzera, Y.; Palomeque, T.; Lorite, P. Distribution and evolution of repeated sequences in genomes of triatominae (Hemiptera-Reduviidae) inferred from genomic in situ hybridization. PLoS ONE 2014, 9, e114298. [Google Scholar] [CrossRef] [PubMed]
- Petraccioli, A.; Guarino, F.M.; Kupriyanova, L.; Mezzasalma, M.; Odierna, G.; Picariello, O.; Capriglione, T. Isolation and characterization of interspersed repeated sequences in the common lizard, Zootoca vivipara, and their conservation in Squamata. Cytogenet. Genome Res. 2019, 157, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M. Chromosome banding in Amphibia. VII. Analysis of structure and variability of NORs in Anura. Chromosoma 1982, 87, 327–344. [Google Scholar] [CrossRef]
- Cross, I.; Vega, L.; Rebordinos, L. Nucleolar Organizing Regions in Crassostrea angulata: Chromosomal location and polymorphism. Genetica 2003, 119, 65–74. [Google Scholar] [CrossRef]
- Ocalewicz, K. Cytogenetic analysis of platyfish (Xiphophorus maculatus) shows location of major and minor rDNA on chromosomes. Hereditas 2004, 141, 333–337. [Google Scholar] [CrossRef]
- Kasiroek, W.; Nattawut, L.; Getlekha, N.; Saowakoon, S.; Phinrub, W.; Tanomtong, A. First report on heteromorphic NORs and chromosome analysis of Rolland’s demoiselle, Chrysiptera rollandi (Perciformes, Pomacentrinae) by conventional and Ag-NOR staining techniques. Cytologia 2014, 9, 289–297. [Google Scholar] [CrossRef][Green Version]
- Stock, M.; Lamatsch, D.K.; Steinlein, C.; Epplen, J.T.; Grosse, W.; Hock, R.; Klapperstuck, T.; Lampert, K.P.; Scheer, U.; Schmid, M.; et al. A bisexually reproducing all-triploid vertebrate. Nat. Genet. 2002, 30, 325–328. [Google Scholar] [CrossRef]
- Wallace, H. The balanced lethal system of crested newt. Heredity 1994, 73, 41–46. [Google Scholar] [CrossRef]
- Kekäläinen, J.; Evans, J.P. Gamete-mediated mate choice: Towards a more inclusive view of sexual selection. Proc. R. Soc. B 2018, 285, 20180836. [Google Scholar] [CrossRef] [PubMed]
- Barth, A.; Souza, V.A.; Solé, M.; Costa, M.A. Molecular cytogenetics of nucleolar organizer regions in Phyllomedusa and Phasmahyla species (Hylidae, Phyllomedusinae): A cytotaxonomic contribution. Genet. Mol. Res. 2013, 12, 2400–2408. [Google Scholar] [CrossRef]
- Bruschi, D.P.; Rivera, M.; Pimentel Lima, A.; Zúñiga, A.B.; Recco-Pimentel, S.M. Interstitial Telomeric Sequences (ITS) and major rDNA mapping reveal insights into the karyotypical evolution of Neotropical leaf frogs species (Phyllomedusa, Hylidae, Anura). Mol. Cytogenet. 2014, 7, 22. [Google Scholar] [CrossRef] [PubMed]



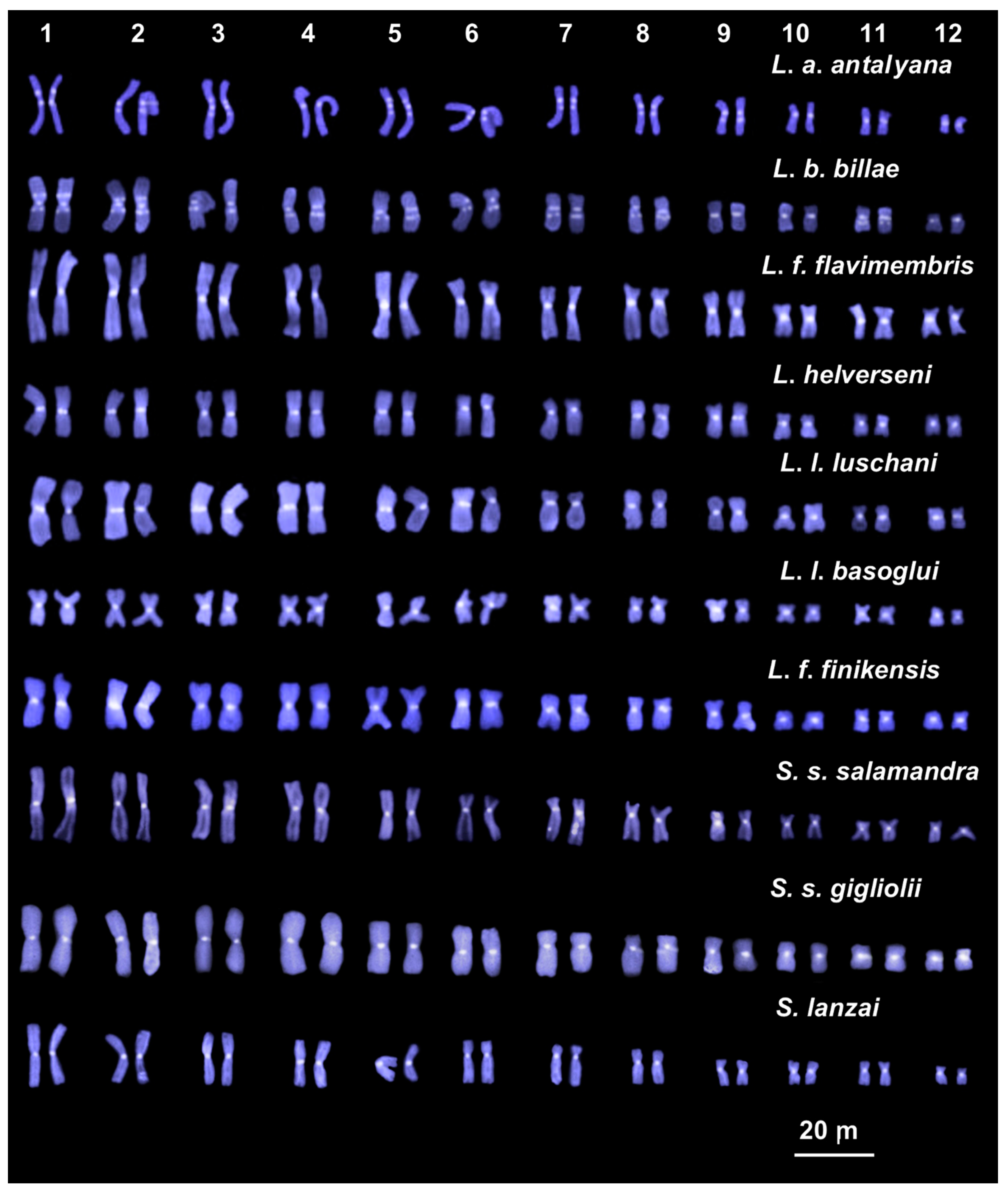
| Genus | Species/Subspecies | Sampling Locality | Number | Sex |
|---|---|---|---|---|
| Lyciasalamandra | ||||
| L. | antalyana antalyana | Hurma (Turkey) | 1 | ♂ |
| L. | billae billae | Kale Tepe (Turkey) | 2 | ♂ |
| L. | flavimembris flavimembris | Marmaris (Turkey) | 1 | ♂ |
| L. | helverseni | Pigadia (Greece) | 1 | ♂ |
| L. | luschani luschani | Letoon (Turkey) | 2 | ♂ |
| L | luschani luschani | Dodurga (Turkey) | 1 | ♂ |
| L. | luschani basoglui | Nadarla (Turkey) | 2 | ♂ |
| L. | luschani finikensis | Finike (Turkey) | 2 | ♂ |
| Salamandra | ||||
| S. | salamandra salamandra | Borgosesia (Italy) | 4 | ♂ |
| S. | salamandra gigliolii | Serino (Italy) | 4 | ♂ |
| S. | salamandra gigliolii | Amalfi (Italy) | 2 | ♂ |
| S. | salamandra gigliolii | Serre (Italy) | 3 | ♂ |
| S. | lanzai | Germanasca (Italy) | 2 | ♂ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezzasalma, M.; Odierna, G.; Petraccioli, A.; Veith, M.; Guarino, F.M. Karyological Diversification in the Genus Lyciasalamandra (Urodela: Salamandridae). Animals 2021, 11, 1709. https://doi.org/10.3390/ani11061709
Mezzasalma M, Odierna G, Petraccioli A, Veith M, Guarino FM. Karyological Diversification in the Genus Lyciasalamandra (Urodela: Salamandridae). Animals. 2021; 11(6):1709. https://doi.org/10.3390/ani11061709
Chicago/Turabian StyleMezzasalma, Marcello, Gaetano Odierna, Agnese Petraccioli, Michael Veith, and Fabio Maria Guarino. 2021. "Karyological Diversification in the Genus Lyciasalamandra (Urodela: Salamandridae)" Animals 11, no. 6: 1709. https://doi.org/10.3390/ani11061709
APA StyleMezzasalma, M., Odierna, G., Petraccioli, A., Veith, M., & Guarino, F. M. (2021). Karyological Diversification in the Genus Lyciasalamandra (Urodela: Salamandridae). Animals, 11(6), 1709. https://doi.org/10.3390/ani11061709









