Beta-Sitosterol Promotes Milk Protein and Fat Syntheses-Related Genes in Bovine Mammary Epithelial Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative Real-Time PCR (qPCR) Analysis
2.3. Western Blot Analysis
2.4. Statistical Analysis
3. Results
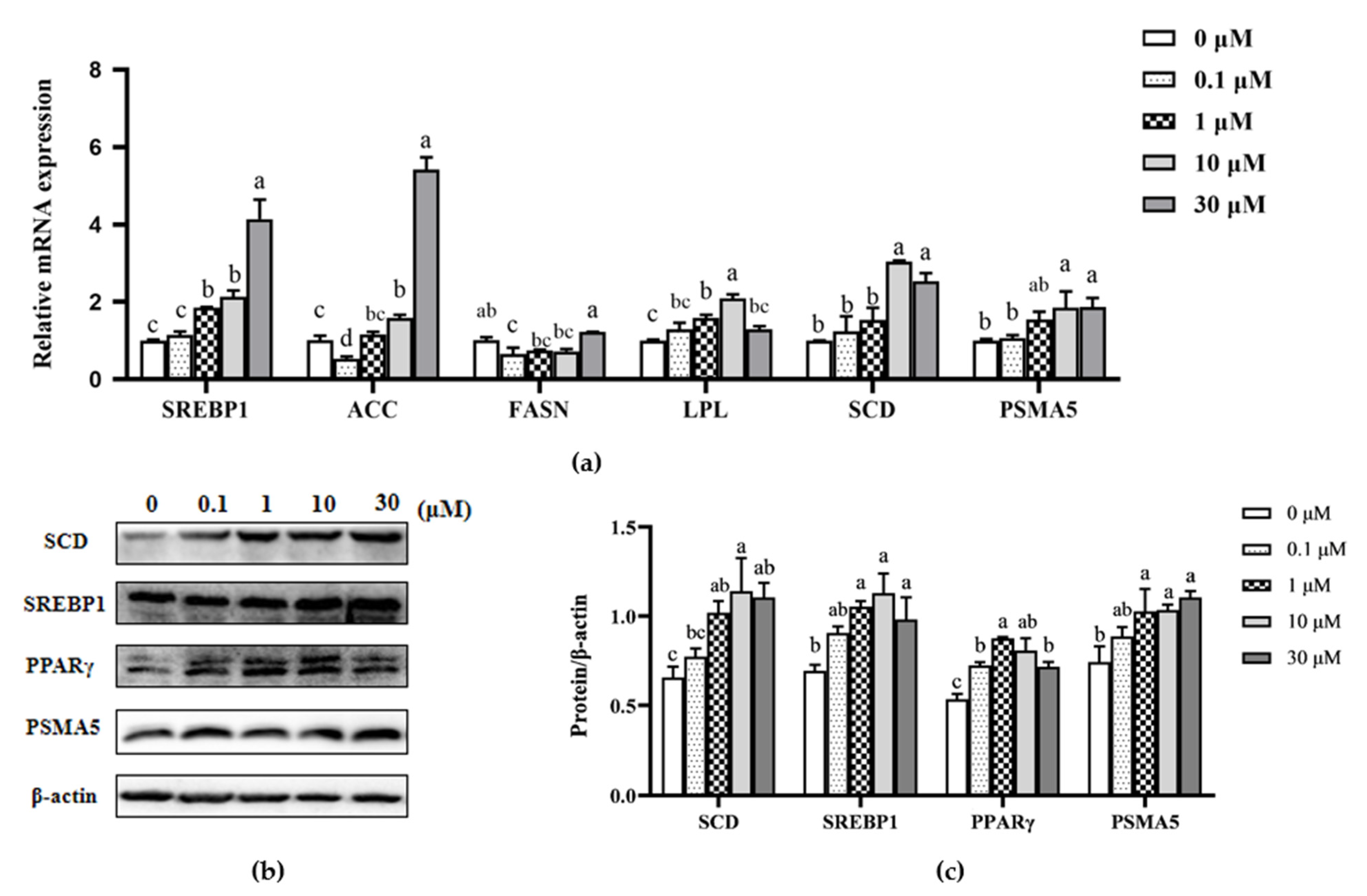
3.1. Effect of β-sitosterol on β-casein mRNA and Protein Expression in MAC-T Cells
3.2. Effect of β-sitosterol on mRNA Expression of Casein Synthesis-Related Genes and Phosphorylation of Casein Synthesis-Related Proteins in MAC-T Cells
3.3. Effect of β-sitosterol on mRNA Expression of Fatty Acid Synthesis-Related Genes in MAC-T Cells
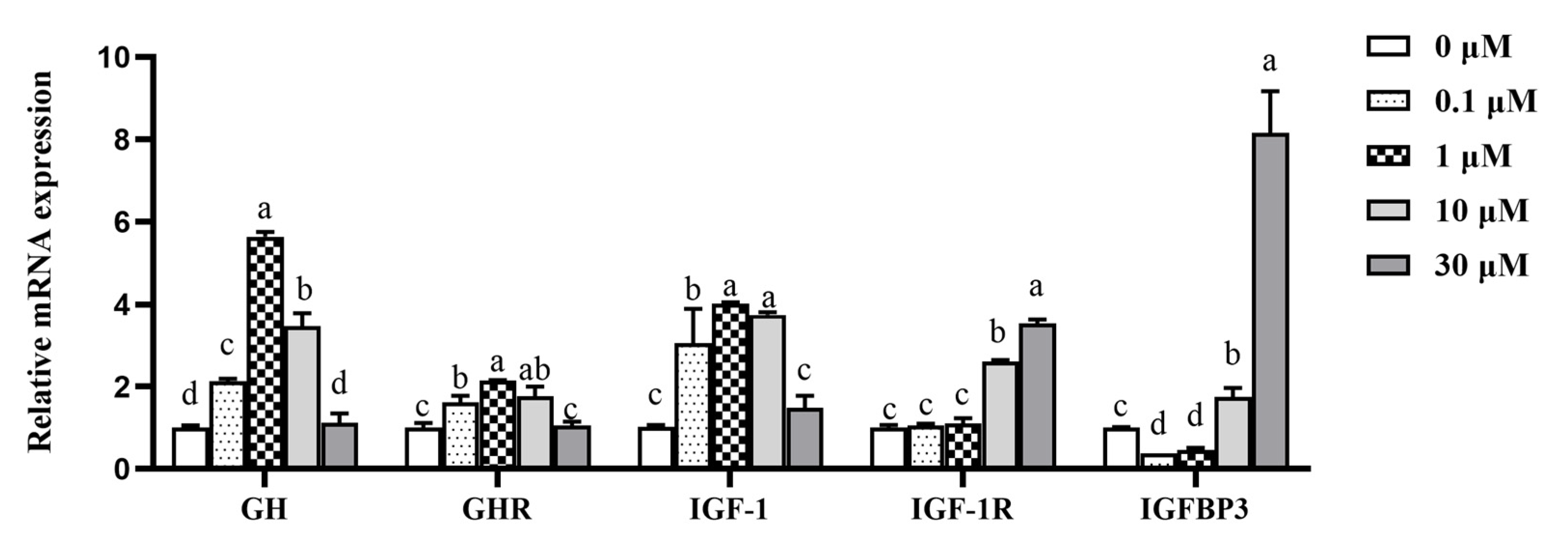
3.4. Effect of β-sitosterol on mRNA Expression of GH-IGF-1 Axis-Related Genes
3.5. Effects of β-sitosterol on the mRNA Expression of HIF-1α and Its Downstream Genes EPO and EPOR
3.6. Effect of β-sitosterol on the mRNA and Protein Expression of SOCS2 and SOCS3
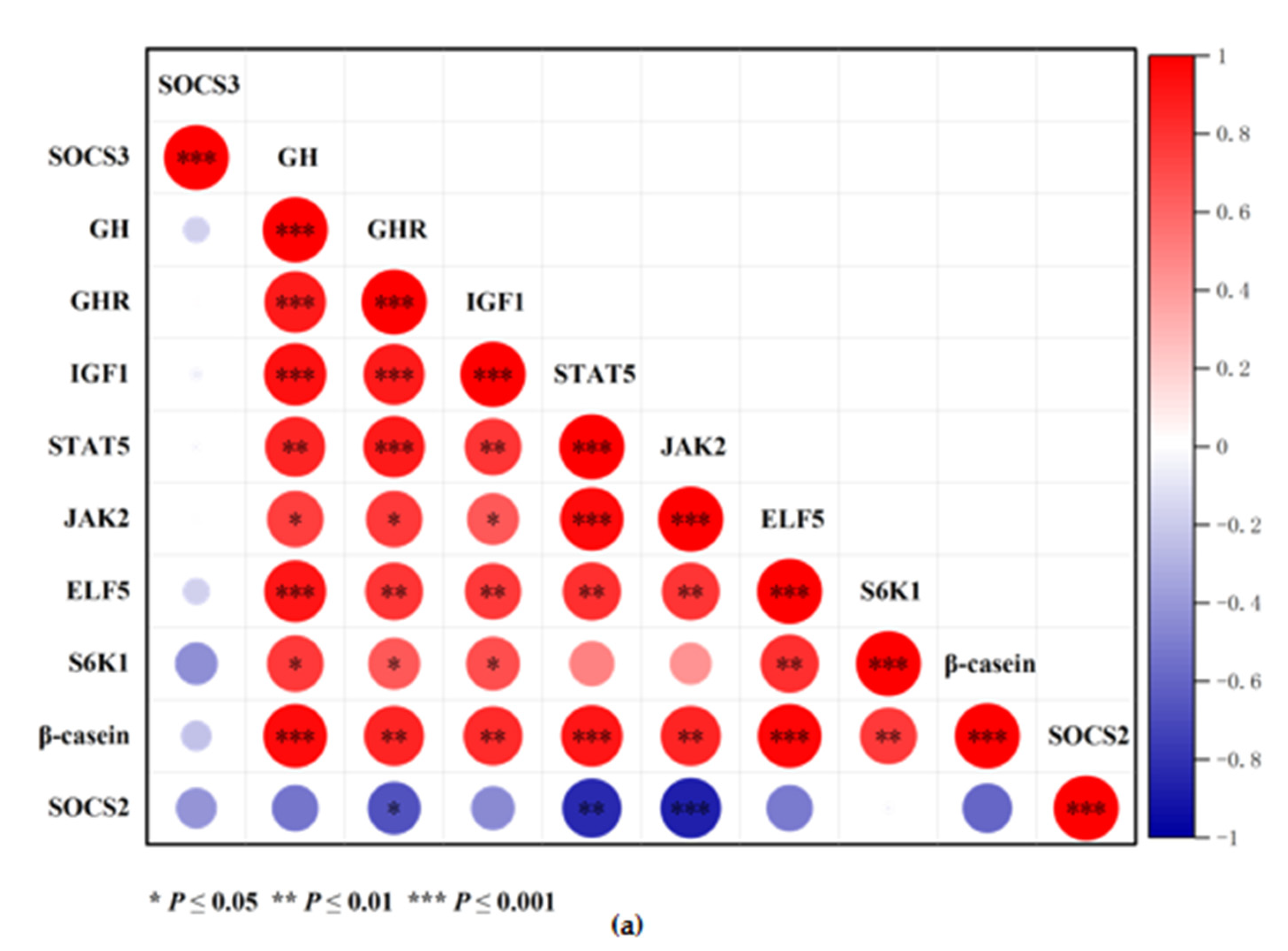
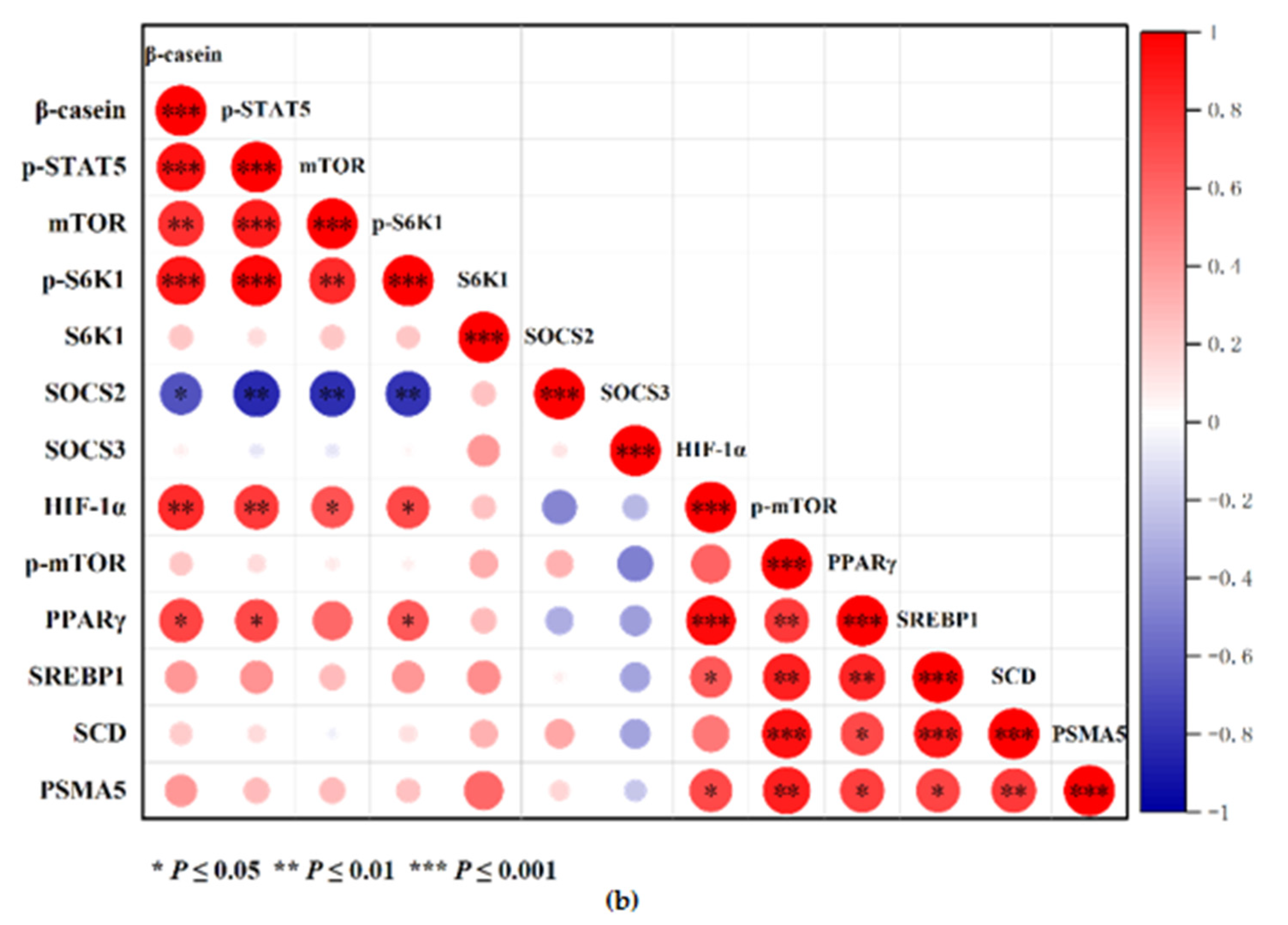
3.7. Correlation Heat Map Analysis
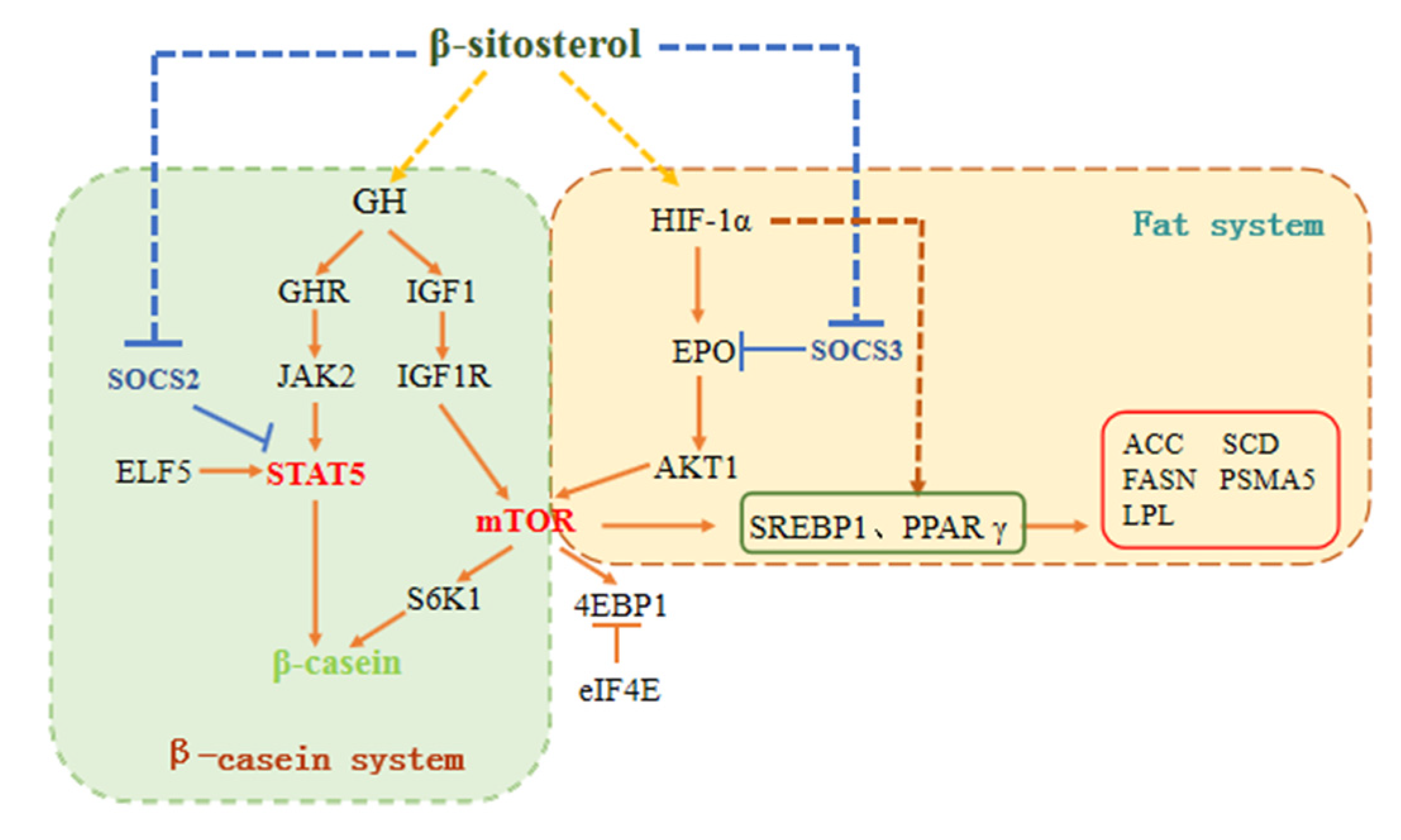
3.8. Schematic Diagram of the Pathways Involved in β-sitosterol-Mediated Regulation of Milk Fat and Protein Synthesis
4. Discussion
4.1. β-sitosterol Affects JAK2/STAT5 and mTOR by Regulating the GH/IGF-1 Axis, Thus Promoting Milk Protein Synthesis
4.2. β-sitosterol Activates HIF-1α/EPO and Promotes Milk Protein and Fat Synthesis
4.3. β-sitosterol Inhibits SOCS Expression and Promotes the Expression of Milk Protein and Fat Synthesis-Related Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, T.L.; Tomanek, L.; Peterson, D.G. A proteomic analysis of the effect of growth hormone on mammary alveolar cell-T (MAC-T) cells in the presence of lactogenic hormones. Domest. Anim. Endocrinol. 2013, 44, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.N.; Zhao, S.G.; Zheng, N.; Zhang, Y.D.; Wang, S.S.; Zhou, X.Q.; Wang, J.Q. Combination of histidine, lysine, methionine, and leucine promotes β-casein synthesis via the mechanistic target of rapamycin signaling pathway in bovine mammary epithelial cells. J. Dairy Sci. 2017, 100, 7696–7709. [Google Scholar] [CrossRef] [PubMed]
- Appuhamy, J.A.; Nayananjalie, W.A.; England, E.M.; Gerrard, D.E.; Akers, R.M.; Hanigan, M.D. Effects of AMP-activated protein kinase (AMPK) signaling and essential amino acids on mammalian target of rapamycin (mTOR) signaling and protein synthesis rates in mammary cells. J. Dairy Sci. 2014, 97, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, X.; Wang, Z.; Lin, X.; Yan, Z.; Cao, Q.; Zhao, M.; Shi, K. MEN1/Menin regulates milk protein synthesis through mTOR signaling in mammary epithelial cells. Sci. Rep. 2017, 7, 5479. [Google Scholar] [CrossRef] [Green Version]
- Miyata, S.; Inoue, J.; Shimizu, M.; Sato, R. Allyl isothiocyanate suppresses the proteolytic activation of sterol regulatory element-binding proteins and de novo fatty acid and cholesterol synthesis. Biosci. Biotechnol. Biochem. 2016, 80, 1006–1011. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Guesthier, M.A.; Burgos, S.A. AMP-activated protein kinase controls lipid and lactose synthesis in bovine mammary epithelial cells. J. Dairy Sci. 2020, 103, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Accornero, P.; Martignani, E.; Miretti, S.; Starvaggi, C.L.; Baratta, M. Epidermal growth factor and hepatocyte growth factor receptors collaborate to induce multiple biological responses in bovine mammary epithelial cells. J. Dairy Sci. 2009, 92, 3667–3675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menzies, K.K.; Lefèvre, C.; Macmillan, K.L.; Nicholas, K.R. Insulin regulates milk protein synthesis at multiple levels in the bovine mammary gland. Funct. Integr. Genomic. 2009, 9, 197–217. [Google Scholar] [CrossRef] [PubMed]
- Sharmin, M.M.; Hayashi, S.; Miyaji, M.; Ishizaki, H.; Matsuyama, H.; Haga, S.; Yonekura, S. IGF-1 induces IRE1-XBP1-dependent endoplasmic reticulum biogenesis in bovine mammary epithelial cells. J. Dairy Sci. 2021, S0022–S0302, 783–789. [Google Scholar] [CrossRef]
- Mullen, M.P.; Lynch, C.O.; Waters, S.M.; Howard, D.J.; O’Boyle, P.; Kenny, D.A.; Buckley, F.; Horan, B.; Diskin, M.G. Single nucleotide polymorphisms in the growth hormone and insulin-like growth factor-1 genes are associated with milk production, body condition score and fertility traits in dairy cows. Genet. Mol. Res. 2011, 10, 1819–1830. [Google Scholar] [CrossRef]
- Sevrin, T.; Boquien, C.Y.; Gandon, A.; Grit, I.; de Coppet, P.; Darmaun, D.; Alexandre-Gouabau, M.C. Fenugreek Stimulates the Expression of Genes Involved in Milk Syntheses and Milk Flow through Modulation of Insulin/GH/IGF-1 Axis and Oxytocin Secretion. Genes 2020, 11, 1208. [Google Scholar] [CrossRef]
- Nawathe, A.R.; Christian, M.; Kim, S.H.; Johnson, M.; Savvidou, M.D.; Terzidou, V. Insulin-like growth factor axis in pregnancies affected by fetal growth disorders. Clin. Epigenetics 2016, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Z.; Lin, J.; Ye, L.; Zhang, Q.; Chen, J.; Yang, Q.; Yu, Q. Modulation of Mammary Gland Development and Milk Production by Growth Hormone Expression in GH Transgenic Goats. Front. Physiol. 2016, 7, 278. [Google Scholar] [CrossRef] [Green Version]
- Dehkhoda, F.; Lee, C.M.M.; Medina, J.; Brooks, A.J. The Growth Hormone Receptor: Mechanism of Receptor Activation, Cell Signaling, and Physiological Aspects. Front. Endocrinol. 2018, 9, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacchetti, T.; Masciangelo, S.; Bicchiega, V.; Bertoli, E.; Ferretti, G. Phytosterols, phytostanols and their esters: From natural to functional foods. Mediterr. J. Nutr. Metab. 2011, 4, 165–172. [Google Scholar] [CrossRef]
- Yuan, C.; Zhang, X.; Long, X.; Jin, J.; Jin, R. Effect of β-sitosterol self-microemulsion and β-sitosterol ester with linoleic acid on lipid-lowering in hyperlipidemic mice. Lipids Health Dis. 2019, 18, 157. [Google Scholar] [CrossRef] [Green Version]
- Liz, R.; Zanatta, L.; dos Reis, G.O.; Horst, H.; Pizzolatti, M.G.; Silva, F.R.; Fröde, T.S. Acute effect of β-sitosterol on calcium uptake mediates anti-inflammatory effect in murine activated neutrophils. J. Pharm. Pharmacol. 2013, 65, 115–122. [Google Scholar] [CrossRef]
- Nieminen, P.; Pölönen, I.; Mustonen, A.M. Increased reproductive success in the white American mink (Neovison vison) with chronic dietary beta-sitosterol supplement. Anim. Reprod. Sci. 2010, 119, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Domené, H.M.; Wit, J.M.; Frank, S.J. Physiology of GH action and associated human disorders. Mol. Cell. Endocrinol. 2021, 15, 111078. [Google Scholar] [CrossRef] [PubMed]
- Chilliard, Y.; Cissé, M.; Lefaivre, R.; Rémond, B. Body composition of dairy cows according to lactation stage, somatotropin treatment, and concentrate supplementation. J. Dairy Sci. 1991, 74, 3103–3116. [Google Scholar] [CrossRef]
- McCoard, S.A.; Hayashi, A.A.; Sciascia, Q.; Rounce, J.; Sinclair, B.; McNabb, W.C.; Roy, N.C. Mammary transcriptome analysis of lactating dairy cows following administration of bovine growth hormone. Animal 2016, 10, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Macias, H.; Hinck, L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 533–557. [Google Scholar] [CrossRef] [Green Version]
- Peel, C.J.; Bauman, D.E.; Gorewit, R.C.; Sniffen, C.J. Effect of exogenous growth hormone on lactational performance in high yielding dairy cows. J. Nutr. 1981, 111, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Shi, L.; Men, J.; Li, Q.; Hou, X.; Wang, C.; Zhao, F. Controlled synchronization of prolactin/STAT5 and AKT1/mTOR in bovine mammary epithelial cells. Vitr. Cell. Dev. Biol. Anim. 2020, 56, 243–252. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, J.Q.; Lin, J.; Yu, Q.H.; Yu, H.Q.; Xu, X.J.; Liu, G.H.; Yang, Q. Production GH transgenic goat improving mammogenesis by somatic cell nuclear transfer. Mol. Biol. Rep. 2014, 41, 4759–4768. [Google Scholar] [CrossRef]
- Flint, D.J.; Gardner, M. Evidence that growth hormone stimulates milk synthesis by direct action on the mammary gland and that prolactin exerts effects on milk secretion by maintenance of mammary deoxyribonucleic acid content and tight junction status. Endocrinology 1994, 135, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Herrington, J.; Carter-Su, C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol. Metab. 2001, 12, 252–257. [Google Scholar] [CrossRef]
- González, L.; Curto, L.M.; Miquet, J.G.; Bartke, A.; Turyn, D.; Sotelo, A.I. Differential regulation of membrane associated-growth hormone binding protein (MA-GHBP) and growth hormone receptor (GHR) expression by growth hormone (GH) in mouse liver. Growth Horm. IGF Res. 2007, 17, 104–112. [Google Scholar] [CrossRef]
- Brown, R.J.; Adams, J.J.; Pelekanos, R.A.; Wan, Y.; McKinstry, W.J.; Palethorpe, K.; Seeber, R.M.; Monks, T.A.; Eidne, K.A.; Parker, M.W.; et al. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat. Struct. Mol. Biol. 2005, 12, 814–821. [Google Scholar] [CrossRef]
- Waters, M.J.; Hoang, H.N.; Fairlie, D.P.; Pelekanos, R.A.; Brown, R.J. New insights into growth hormone action. J. Mol. Endocrinol. 2006, 36, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jiang, M.; Shi, Y.; Song, S.; Hou, X.; Lin, Y. Prolactin regulates LAT1 expression via STAT5 (signal transducer and activator of transcription 5) signaling in mammary epithelial cells of dairy cows. J. Dairy Sci. 2020, 103, 6627–6634. [Google Scholar] [CrossRef]
- Buitenhuis, M.; Coffer, P.J.; Koenderman, L. Signal transducer and activator of transcription 5 (STAT5). Int. J. Biochem. Cell Biol. 2004, 36, 2120–2124. [Google Scholar] [CrossRef]
- Bauman, D.E. Bovine somatotropin and lactation: From basic science to commercial application. Domest. Anim. Endocrinol. 1999, 17, 101–116. [Google Scholar] [CrossRef]
- Velez, J.C.; Donkin, S.S. Bovine somatotropin increases hepatic phosphoenolpyruvate carboxykinase mRNA in lactating dairy cows. J. Dairy Sci. 2004, 87, 1325–1335. [Google Scholar] [CrossRef] [Green Version]
- Rose, M.T.; Weekes, T.E.; Rowlinson, P. Correlation of blood and milk components with the milk yield response to bovine somatotropin in dairy cows. Domest. Anim. Endocrinol. 2005, 28, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Chicharro, J.L.; López-Calderon, A.; Hoyos, J.; Martín-Velasco, A.I.; Villa, G.; Villanúa, M.A.; Lucía, A. Effects of an endurance cycling competition on resting serum insulin-like growth factor I (IGF-I) and its binding proteins IGFBP-1 and IGFBP-3. Br. J. Sports Med. 2001, 35, 303–307. [Google Scholar] [CrossRef] [Green Version]
- Ling, W.H.; Jones, P.J. Dietary phytosterols: A review of metabolism, benefits and side effects. Life Sci. 1995, 57, 195–206. [Google Scholar] [CrossRef]
- Muldoon, T.G. Interplay between estradiol and prolactin in the regulation of steroid hormone receptor levels, nature, and functionality in normal mouse mammary tissue. Endocrinology 1981, 109, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, O.; Marino, R.; De Luca, F.; Phillip, M.; Baron, J. Endocrine regulation of the growth plate. Horm. Res. 2005, 64, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Babu, S.; Krishnan, M.; Rajagopal, P.; Periyasamy, V.; Veeraraghavan, V.; Govindan, R.; Jayaraman, S. Beta-sitosterol attenuates insulin resistance in adipose tissue via IRS-1/Akt mediated insulin signaling in high fat diet and sucrose induced type-2 diabetic rats. Eur. J. Pharmacol. 2020, 873, 173004. [Google Scholar] [CrossRef]
- Abryelska, A.; Karuga, F.F.; Szmyd, B.; Białasiewicz, P. HIF-1α as a Mediator of Insulin Resistance, T2DM, and Its Complications: Potential Links With Obstructive Sleep Apnea. Front. Physiol. 2020, 11, 1035. [Google Scholar] [CrossRef] [PubMed]
- Kimba, Y.; Abe, T.; Wu, J.L.; Inoue, R.; Fukiki, M.; Kohno, K.; Kobayashi, H. Mutant IkappaBalpha suppresses hypoxia-induced VEGF expression through downregulation of HIF-1alpha and COX-2 in human glioma cells. Oncol. Res. 2005, 15, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Obara, N.; Imagawa, S.; Nakano, Y.; Suzuki, N.; Yamamoto, M.; Nagasawa, T. Suppression of erythropoietin gene expression by cadmium depends on inhibition of HIF-1, not stimulation of GATA-2. Arch. Toxicol. 2003, 77, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Jarero-Basulto, J.J.; Rivera-Cervantes, M.C.; Gasca-Martínez, D.; García-Sierra, F.; Gasca-Martínez, Y.; Beas-Zárate, C. Current Evidence on the Protective Effects of Recombinant Human Erythropoietin and Its Molecular Variants against Pathological Hallmarks of Alzheimer’s Disease. Pharmaceuticals 2020, 13, 424. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shang, Y.; Sun, S.; Liang, H.; Liu, R. Erythropoietin prevents PC12 cells from 1-methyl-4-phenylpyridinium ion-induced apoptosis via the Akt/GSK-3beta/caspase-3 mediated signaling pathway. Apoptosis 2007, 12, 1365–1375. [Google Scholar] [CrossRef]
- Sola, A.; Wen, T.C.; Hamrick, S.E.; Ferriero, D.M. Potential for protection and repair following injury to the developing brain: A role for erythropoietin? Pediatr. Res. 2005, 57, 110R–117R. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiese, K. Erythropoietin and mTOR: A “One-Two Punch” for Aging-Related Disorders Accompanied by Enhanced Life Expectancy. Curr. Neurovasc. Res. 2016, 13, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Richard, D.; Laplante, M. The Roles of mTOR Complexes in Lipid Metabolism. Annu. Rev. Nutr. 2015, 35, 321–348. [Google Scholar] [CrossRef]
- Carreño, D.; Hervás, G.; Toral, P.G.; Castro-Carrera, T.; Frutos, P. Fish oil-induced milk fat depression and associated downregulation of mammary lipogenic genes in dairy ewes. J. Dairy Sci. 2016, 99, 7971–7981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Lin, Y.; Liu, L.; Wang, L.; Bian, Y.; Gao, X.; Li, Q. Regulation of peroxisome proliferator-activated receptor gamma on milk fat synthesis in dairy cow mammary epithelial cells. Vitr. Cell. Dev. Biol. Anim. 2016, 52, 1044–1059. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, K.; Liu, J. Effects of glucose availability on expression of the key genes involved in synthesis of milk fat, lactose and glucose metabolism in bovine mammary epithelial cells. PLoS ONE 2013, 8, e66092. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhang, Q. Identification and functional analysis of candidate gene VPS28 for milk fat in bovine mammary epithelial cells. Biochem. Biophys. Res. Commun. 2019, 510, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Sharma, V.; Verma, V.; Pandey, D.; Yadav, S.K.; Maikhuri, J.P.; Gupta, G. Apigenin manipulates the ubiquitin-proteasome system to rescue estrogen receptor-β from degradation and induce apoptosis in prostate cancer cells. Eur. J. Nutr. 2015, 54, 1255–1267. [Google Scholar] [CrossRef]
- Jin, Y.C.; Li, Z.H.; Hong, Z.S.; Xu, C.X.; Han, J.A.; Choi, S.H.; Yin, J.L.; Zhang, Q.K.; Lee, K.B.; Kang, S.K.; et al. Conjugated linoleic acid synthesis-related proteasome 20s subunit α5 (PSMA5) is increased by vaccenic acid treatment in goat mammary tissue. J. Dairy Sci. 2012, 95, 4286–4297. [Google Scholar] [CrossRef]
- García-Fuentes, E.; Santiago-Fernández, C.; Gutiérrez-Repiso, C.; Mayas, M.D.; Oliva-Olivera, W.; Coín-Aragüez, L.; Alcaide, J.; Ocaña-Wilhelmi, L.; Vendrell, J.; Tinahones, F.J.; et al. Hypoxia is associated with a lower expression of genes involved in lipogenesis in visceral adipose tissue. J. Transl. Med. 2015, 13, 373. [Google Scholar] [CrossRef] [Green Version]
- Ezzeddini, R.; Taghikhani, M.; Somi, M.H.; Samadi, N.; Rasaee, M.J. Clinical importance of FASN in relation to HIF-1α and SREBP-1c in gastric adenocarcinoma. Life Sci. 2019, 224, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhao, F.Q. Emerging evidence of the physiological role of hypoxia in mammary development and lactation. J. Anim. Sci. Biotechnol. 2014, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, S.A.; Baganizi, D.R.; Sahu, R.; Singh, S.R.; Dennis, V.A. SOCS Proteins as Regulators of Inflammatory Responses Induced by Bacterial Infections: A Review. Front. Microbiol. 2017, 8, 2431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Morales, A.; Greenhalgh, C.J.; Norstedt, G.; Rico-Bautista, E. Negative regulation of growth hormone receptor signaling. Mol. Endocrinol. 2006, 20, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Vesterlund, M.; Zadjali, F.; Persson, T.; Nielsen, M.L.; Kessler, B.M.; Norstedt, G.; Flores-Morales, A. The SOCS2 ubiquitin ligase complex regulates growth hormone receptor levels. PLoS ONE 2011, 6, e25358. [Google Scholar] [CrossRef]
- Vočanec, D.; Prijatelj, T.; Debeljak, N.; Kunej, T. Genetic variants of erythropoietin (EPO) and EPO receptor genes in familial erythrocytosis. Int. J. Lab. Hematol. 2019, 41, 162–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arun, S.J.; Thomson, P.C.; Sheehy, P.A.; Khatkar, M.S.; Raadsma, H.W.; Williamson, P. Targeted Analysis Reveals an Important Role of JAK-STAT-SOCS Genes for Milk Production Traits in Australian Dairy Cattle. Front. Genet. 2015, 15, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.Z.; Khan, A.; Xiao, J.; Ma, Y.; Ma, J.; Gao, J.; Cao, Z. Role of the JAK-STAT Pathway in Bovine Mastitis and Milk Production. Animals 2020, 10, 2107. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Shen, J.; Zong, J.; Liu, J.; Jin, Y. Beta-Sitosterol Promotes Milk Protein and Fat Syntheses-Related Genes in Bovine Mammary Epithelial Cells. Animals 2021, 11, 3238. https://doi.org/10.3390/ani11113238
Liu X, Shen J, Zong J, Liu J, Jin Y. Beta-Sitosterol Promotes Milk Protein and Fat Syntheses-Related Genes in Bovine Mammary Epithelial Cells. Animals. 2021; 11(11):3238. https://doi.org/10.3390/ani11113238
Chicago/Turabian StyleLiu, Xinlu, Jinglin Shen, Jinxin Zong, Jiayi Liu, and Yongcheng Jin. 2021. "Beta-Sitosterol Promotes Milk Protein and Fat Syntheses-Related Genes in Bovine Mammary Epithelial Cells" Animals 11, no. 11: 3238. https://doi.org/10.3390/ani11113238
APA StyleLiu, X., Shen, J., Zong, J., Liu, J., & Jin, Y. (2021). Beta-Sitosterol Promotes Milk Protein and Fat Syntheses-Related Genes in Bovine Mammary Epithelial Cells. Animals, 11(11), 3238. https://doi.org/10.3390/ani11113238





