Molecular Detection of Infectious Laryngotracheitis Virus in Chickens with a Microfluidic Chip
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Chemicals and Materials
2.3. Virus and Clinical Samples
2.4. DNA Extraction
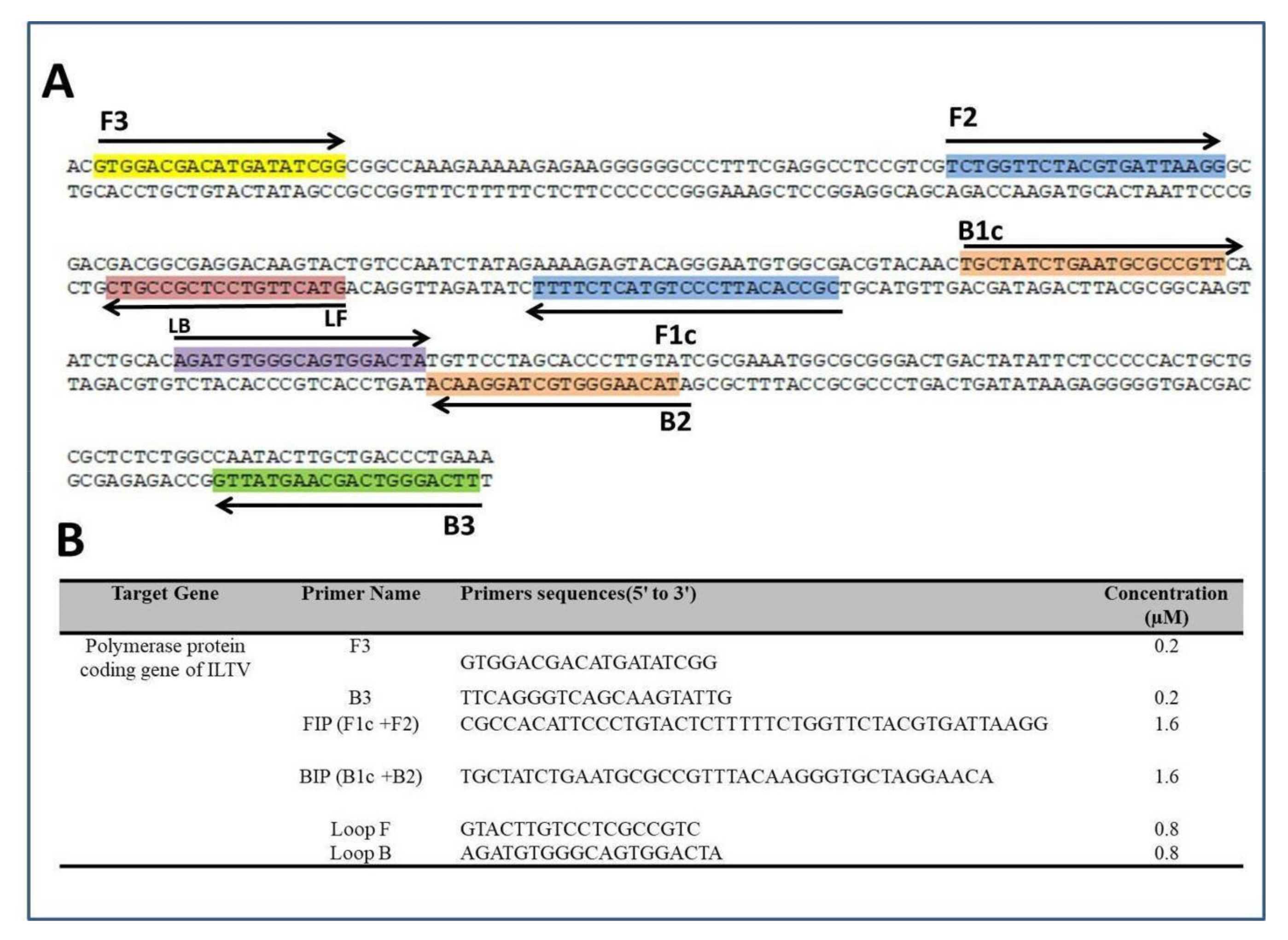
2.5. LAMP Primers
2.6. Benchtop LAMP Amplification
2.7. Benchtop qPCR Amplification
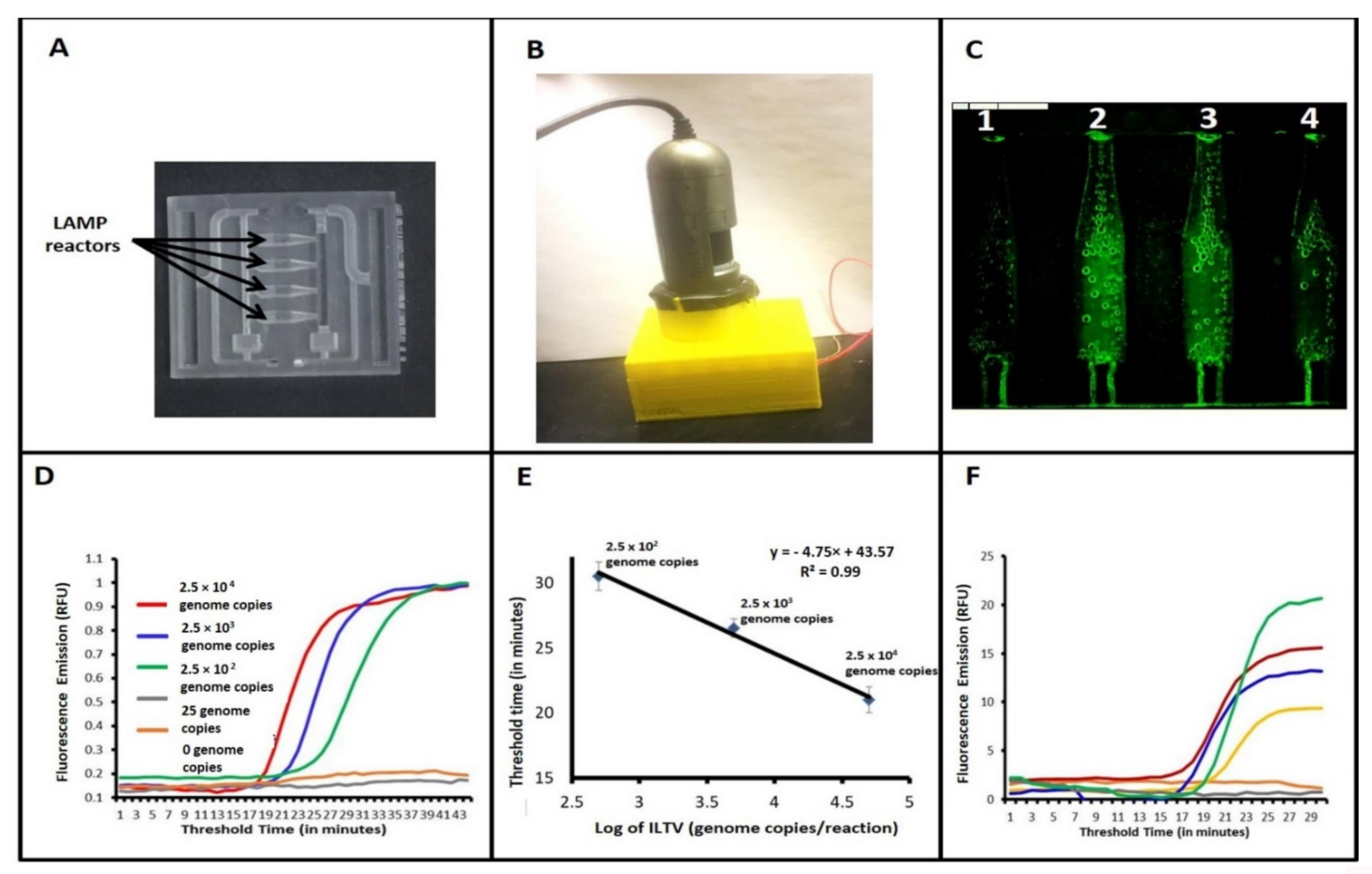
2.8. Microfluidic Chip for Real-Time-LAMP
2.9. The Analytical Sensitivity
2.10. Detection of Nucleic Acids from Clinical Samples
3. Results
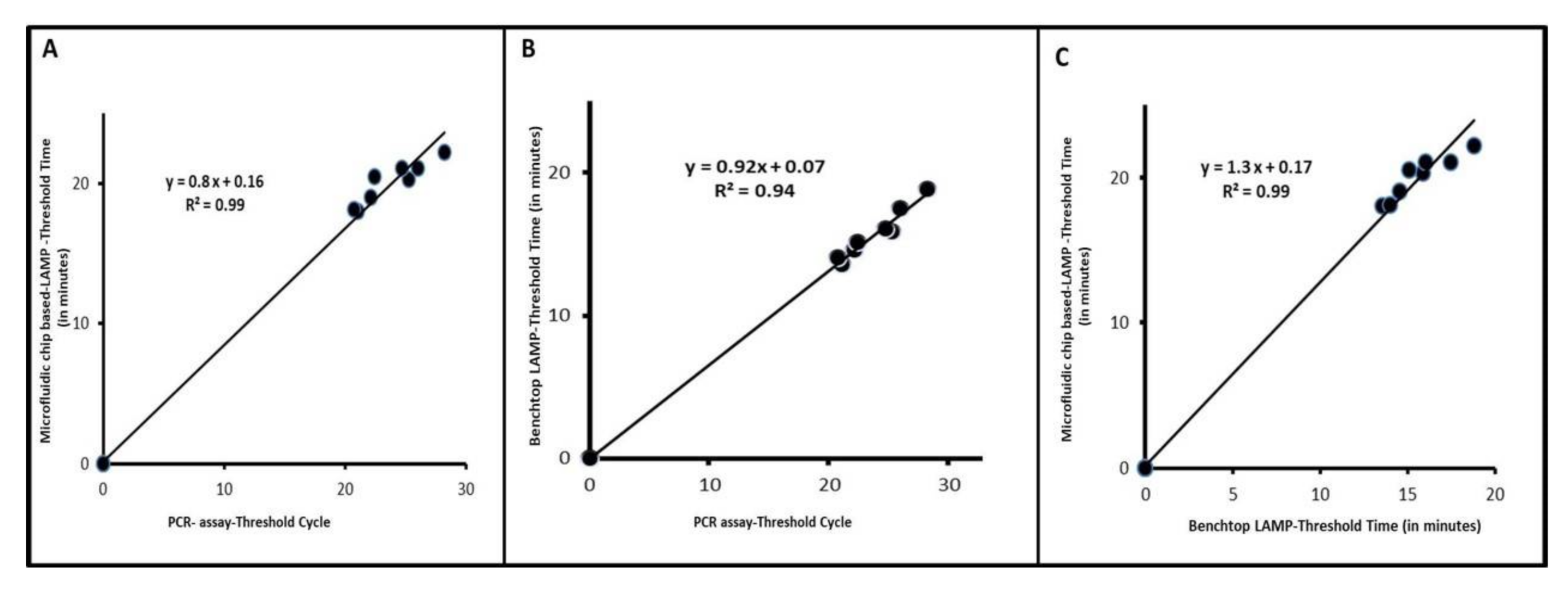
3.1. ILTV LAMP Performs on Par with qPCR
3.2. Detection of ILTV with Our Microfluidic Chip and Real-Time LAMP
3.3. Clinical Performance of Our Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davison, A.J.; Eberle, R.; Hayward, G.S.; Mcgeoch, D.J.; Minson, A.C.; Pellet, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Spatz, S.J.; Guy, J.S. Infectious laryngotracheitis. In Diseases of Poultry, 13th ed.; Swayne, D.E., Glisson, J.R., McDougald, L.R., Nolan, L.K., Suarez, D.L., Nair, V., Eds.; Blackwell Publishing: Ames, IA, USA, 2013; pp. 161–179. [Google Scholar]
- Menendez, K.R.; García, M.; Spatz, S.; Tablante, N.L. Molecular epidemiology of infectious laryngotracheitis: A review. Avian Pathol. 2014, 43, 108–117. [Google Scholar] [CrossRef] [Green Version]
- García, M.; Volkening, J.; Riblet, S.; Spatz, S. Genomic sequence analysis of the United States infectious laryngotracheitis vaccine strains chicken embryo origin (CEO) and tissue culture origin (TCO). Virology 2013, 440, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Ide, P.R. Sensitivity and specificity of the fluorescent antibody technique for detection of infectious laryngotracheitis virus. Can. J. Comp. Med. 1978, 42, 54–62. [Google Scholar] [PubMed]
- Kotiw, M.; Wilks, C.R.; May, J.T. Differentiation of infectious laryngotracheitis virus strains using restriction endonucleases. Avian Dis. 1982, 26, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.G.; Turner, A.J. Characterization of infectious laryngotracheitis viruses, antigenic comparison by kinetics of neutralization and immunization studies. Can. J. Comp. Med. 1983, 47, 163–171. [Google Scholar] [PubMed]
- York, J.J.; Young, J.G.; Fahey, K.J. The appearance of viral antigen and antibody in the trachea of naïve and vaccinated chickens infected with infectious laryngotracheitis virus. Avian Pathol. 1989, 18, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Shirley, M.W.; Kemp, D.J.; Sheppard, M.; Fahey, K.J. Detection of DNA from infectious laryngotracheitis virus by colourimetric analyses of polymerase chain reactions. J. Virol. Methods 1990, 30, 251–259. [Google Scholar] [CrossRef]
- Vogtlin, A.; Bruckner, L.; Ottiger, H.P. Use of polymerase chain reaction (PCR) for the detection of vaccine contamination by infectious laryngotracheitis virus. Vaccine 1999, 17, 2501–2506. [Google Scholar] [CrossRef]
- Humberd, J.; Garcia, M.; Riblet, S.M.; Resurreccion, R.S.; Brown, T.P. Detection of infectious laryngotracheitis virus in formalin-fixed, paraffinembedded tissues by nested polymerase chain reaction. Avian Dis. 2002, 46, 64–74. [Google Scholar] [CrossRef]
- Mahmoudian, A.; Kirkpatrick, N.C.; Coppo, M.; Lee, S.W.; Devlin, J.M.; Markham, P.F.; Browning, G.F.; Noormohammadi, A.H. Development of a SYBR Green quantitative polymerase chain reaction assay for rapid detection and quantification of infectious laryngotracheitis virus. Avian Pathol. 2011, 40, 237–242. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, C.; Cui, X.; Cui, H.; Shi, X.; Zhang, X.; Hu, S.; Hao, L.; Wang, Y. Detection of Infectious Laryngotracheitis Virus by RealTime PCR in Naturally and Experimentally Infected Chickens. PLoS ONE 2013, 8, e67598. [Google Scholar]
- Laamiri, N.; Aouini, R.; Marnissi, B.; Ghram, A.; Hmila, I. A multiplex real-time RT PCR for simultaneous detection of four most common avian respiratory viruses. Virology 2018, 515, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.M.; Nakajima, C.; Ohashi, K.; Onuma, M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J. Clin. Microbiol. 2005, 43, 1646–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.T.; Zhang, J.; Sun, D.H.; Ma, L.N.; Liu, X.T.; Cai, X.P.; Liu, Y.S. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods 2008, 151, 200–203. [Google Scholar] [CrossRef]
- Li, Q.; Xue, C.; Qin, J.; Zhou, Q.; Chen, F.; Bi, Y.; Cao, Y. An improved reverse transcription loop-mediated isothermal amplification assay for sensitive and specific detection of Newcastle disease virus. Arch. Virol. 2009, 154, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.T.; Zhang, J.; Ma, Y.P.; Ma, L.N.; Ding, Y.Z.; Liu, X.T.; Cai, X.P.; Ma, L.Q.; Zhang, Y.G.; Liu, Y.S. Reverse transcription loop-mediated isothermal amplification for the rapid detection of infectious bronchitis virus in infected chicken tissues. Mol. Cell. Probes 2010, 24, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.C.; Giambrone, J.J.; Macklin, K.S. Comparison of a TaqMan real-time polymerase chain reaction assay with a loop-mediated isothermal amplification assay for detection of Gallid herpesvirus 1. J. Vet. Diagn. Investig. 2012, 24, 138–141. [Google Scholar] [CrossRef] [Green Version]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef]
- Gandelman, O.A.; Church, V.L.; Moore, C.A.; Kiddle, G.; Carne, C.A.; Parmar, S.; Jalal, H.; Tisi, L.C.; Murray, J.A. Novel bioluminescent quantitative detection of nucleic acid amplification in real-time. PLoS ONE 2010, 5, e14155. [Google Scholar] [CrossRef]
- Kiddle, G.; Hardinge, P.; Buttigieg, N.; Gandelman, O.; Pereira, C.; McElgunn, C.J.; Rizzoli, M.; Jackson, R.; Appleton, N.; Moore, C.; et al. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) sui for field use. BMC Biotechnol. 2012, 12, 15. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Domesle, K.J.; Wang, F.; Ge, B. Rapid detection of Salmonella in food and feed by coupling loop-mediated isothermal amplification with bioluminescent assay in real-time. BMC Microbiol. 2016, 16, 112. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Liu, C.; Mauk, M.G.; Peng, J.; Schoenfeld, T.; Bau, H.H. A Multifunctional Reactor with Dry-Stored Reagents for Enzymatic Amplification of Nucleic Acids. Anal. Chem. 2018, 90, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Pandian, V.; Mauk, M.G.; Bau, H.H.; Cherry, S.; Tisi, L.C.; Liu, C. Smartphone-Based Mobile Detection Platform for Molecular Diagnostics and Spatiotemporal Disease Mapping. Anal. Chem. 2018, 90, 4823–4831. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Wang, J.; Liu, L.; Zhang, R.; Shi, R.; Han, Q.; Sun, J.; Yuan, W. A real-time loop-mediated isothermal amplification method for rapid detection of Lawsonia intracellularis in porcine fecal samples. J. Microbiol. Methods 2018, 151, 62–65. [Google Scholar] [CrossRef]
- Dou, M.; Dominguez, D.C.; Li, X.; Sanchez, J.; Scott, G. A versatile PDMS/paper hybrid microfluidic platform for sensitive infectious disease diagnosis. Anal. Chem. 2014, 86, 7978–7986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, M.; Sanchez, J.; Tavakoli, H.; Gonzalez, J.E.; Sun, J.; Dien Bard, J.; Li, X. A low-cost microfluidic platform for rapid and instrument-free detection of whooping cough. Anal. Chim. Acta 2019, 1065, 71–78. [Google Scholar] [CrossRef]
- Sharma, S.; Zapatero-Rodriguez, J.; Estrela, P.; O’Kennedy, R. Point-of-care diagnostics in low resource settings: Present status and future role of microfluidics. Biosensors 2015, 5, 577–601. [Google Scholar] [CrossRef] [Green Version]
- Pandey, C.M.; Augustine, S.; Kumar, S.; Kumar, S.; Nara, S.; Srivastava, S.; Malhotra, B.D. Microfluidics Based Point-of-Care Diagnostics. Biotechnol. J. 2018, 13, 1700047. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, Z.; Han, Q.; Qiu, J.; Chen, J.; Zhang, G.; Li, Z.; Lou, S.; Li, N. Development of one-step SYBR Green real-time RT-PCR for quantifying bovine viral diarrhea virus type-1 and its comparison with conventional RT-PCR. Virol. J. 2011, 8, 374. [Google Scholar] [CrossRef] [Green Version]
- El-Tholoth, M.; Bai, H.; Mauk, M.G.; Saif, L.; Bau, H.H. A portable, 3D printed, microfluidic device for multiplexed, real time, molecular detection of the porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus at the point of need. Lab Chip 2021, 21, 1118–1130. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, F.; Prinyawiwatkul, W.; Ge, B. Robustness of Salmonella loop-mediated isothermal amplification assays for food applications. J. Appl. Microbiol. 2014, 116, 81–88. [Google Scholar] [CrossRef]
- Dou, M.; Sanjay, S.T.; Dominguez, D.C.; Zhan, S.; Li, X. A paper/polymer hybrid CD-like microfluidic SpinChip integrated with DNA-functionalized graphene oxide nanosensors for multiplex qLAMP detection. Chem. Commun. 2017, 53, 10886–10889. [Google Scholar] [CrossRef]
- Li, R.J.; Mauk, M.G.; Seok, Y.; Bau, H.H. Electricity-free chemical heater for isothermal nucleic acid amplification with applications in COVID-19 home testing. Analyst 2021, 146, 4212–4218. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.M.; Bitew, M.; Fokam, J.; Lelo, E.A.; Ahidjo, A.; Asmamaw, K.; Beloumou, G.A.; Bulimo, W.D.; Buratti, E.; Chenwi, C.; et al. Diagnostic performance of a colorimetric RT -LAMP for the identification of SARS-CoV-2: A multicenter prospective clinical evaluation in sub-Saharan Africa. EClinicalMedicine 2021, 40, 101101. [Google Scholar] [CrossRef]
- Dou, M.; Sanjay, S.T.; Dominguez, D.C.; Liu, P.; Xu, F.; Li, X. Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip. Biosens. Bioelectron. 2017, 87, 865–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Macias, N.; Shen, F.; Bard, J.D.; Domínguez, D.C.; Li, X. Rapid and Accurate Diagnosis of the Respiratory Disease Pertussis on a Point-of-Care Biochip. EClinicalMedicine 2019, 8, 72–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokkinos, P.A.; Ziros, P.G.; Bellou, M.; Vantarakis, A. Loop-mediated isothermal amplification (LAMP) for the detection of Salmonella in food. Food Anal. Methods 2014, 7, 512–526. [Google Scholar] [CrossRef]
- Liao, S.-C.; Peng, J.; Mauk, M.G.; Awasthi, S.; Song, J.; Friedman, H.; Bau, H.H.; Liu, C. Smart Cup: A Minimally Instrumented, Smartphone-Based Point-of-Care Molecular Diagnostic Device. Sens. Actuators B Chem. 2016, 229, 232–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Macdonald, J.; Von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Tholoth, M.; Bai, H.; Mauk, M.G.; Anis, E.; Bau, H.H. Molecular Detection of Infectious Laryngotracheitis Virus in Chickens with a Microfluidic Chip. Animals 2021, 11, 3203. https://doi.org/10.3390/ani11113203
El-Tholoth M, Bai H, Mauk MG, Anis E, Bau HH. Molecular Detection of Infectious Laryngotracheitis Virus in Chickens with a Microfluidic Chip. Animals. 2021; 11(11):3203. https://doi.org/10.3390/ani11113203
Chicago/Turabian StyleEl-Tholoth, Mohamed, Huiwen Bai, Michael G. Mauk, Eman Anis, and Haim H. Bau. 2021. "Molecular Detection of Infectious Laryngotracheitis Virus in Chickens with a Microfluidic Chip" Animals 11, no. 11: 3203. https://doi.org/10.3390/ani11113203
APA StyleEl-Tholoth, M., Bai, H., Mauk, M. G., Anis, E., & Bau, H. H. (2021). Molecular Detection of Infectious Laryngotracheitis Virus in Chickens with a Microfluidic Chip. Animals, 11(11), 3203. https://doi.org/10.3390/ani11113203





