Impact of Soil Microbes and Oxygen Availability on Bacterial Community Structure of Decomposing Poultry Carcasses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
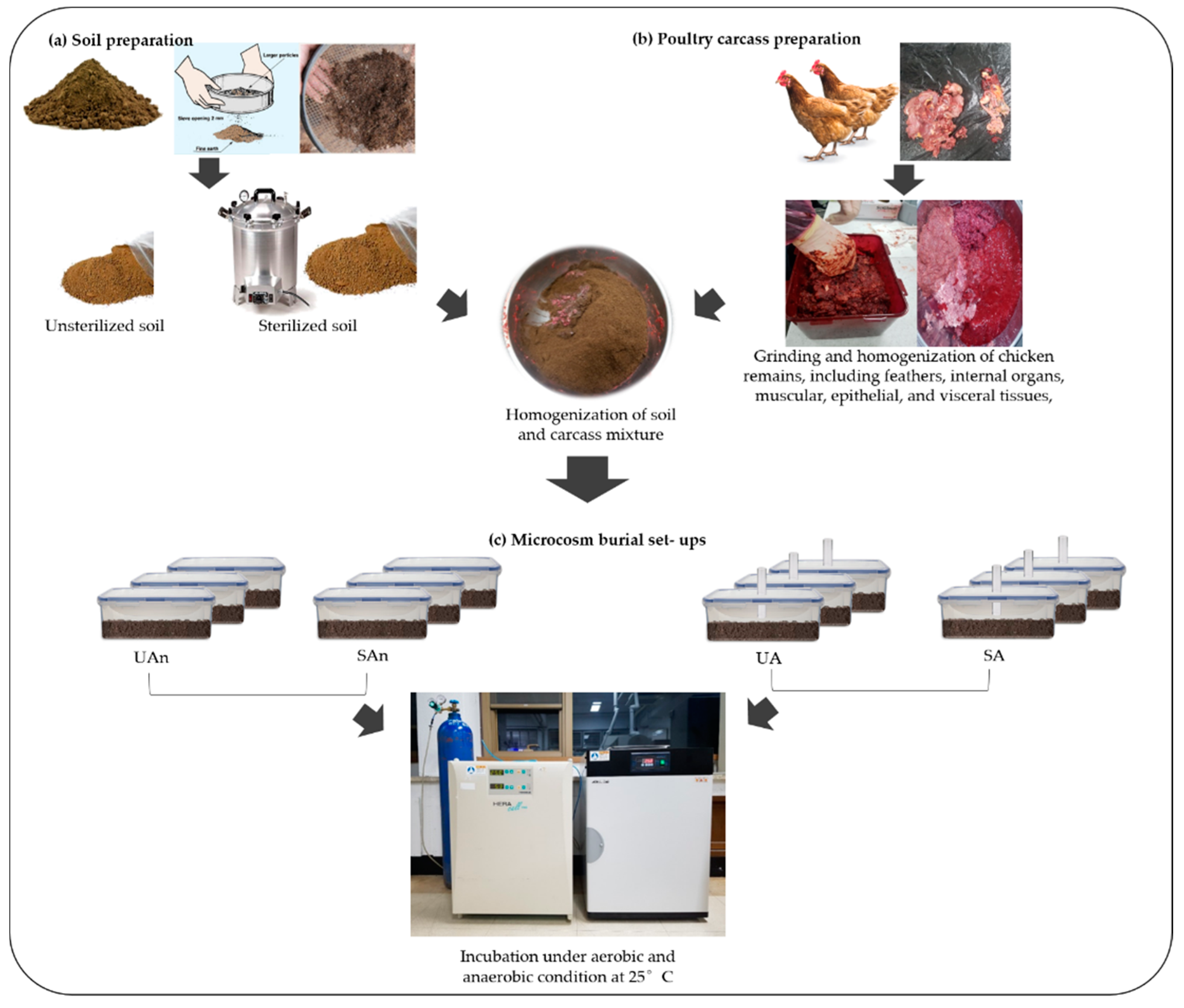
2.2. Carcass and Soil Preparation
2.3. In Vitro Set-Up for Carcass Decomposition
2.4. Sample Collection
2.5. Moisture Content and pH during Carcass Decomposition
2.6. DNA Extraction, PCR Amplification and 16S rRNA Amplicon Sequencing
2.7. Sequence Data Processing and Metataxonomic Analysis
2.8. Statistical Analysis
3. Results
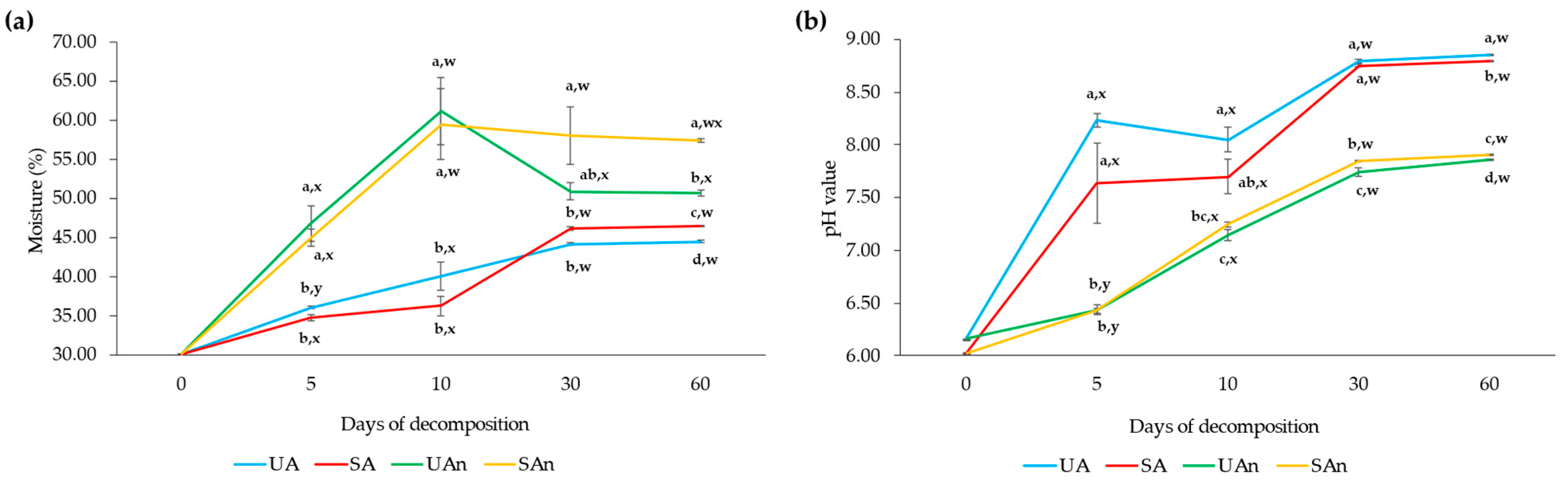
3.1. Effect of Oxygen Availability and Sterilized Soil in Moisture Content and pH during Carcass Decomposition
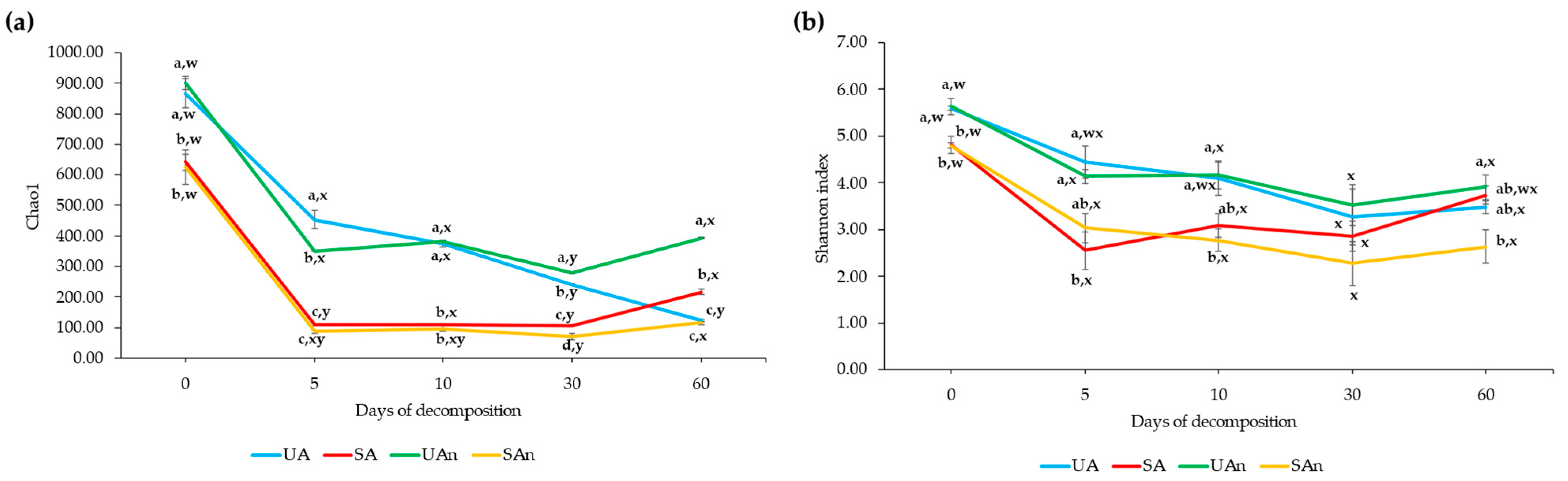
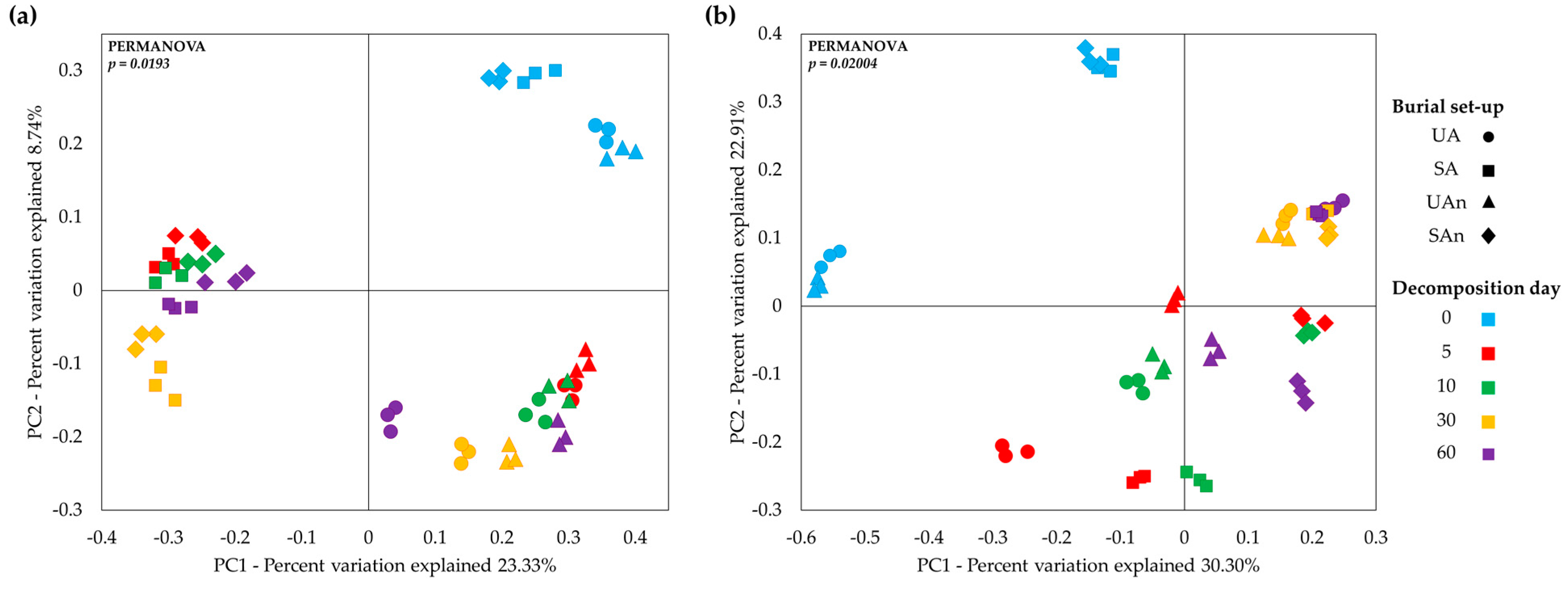
3.2. Species Richness and Diversity of Bacterial Community in Different Burial Set-Ups
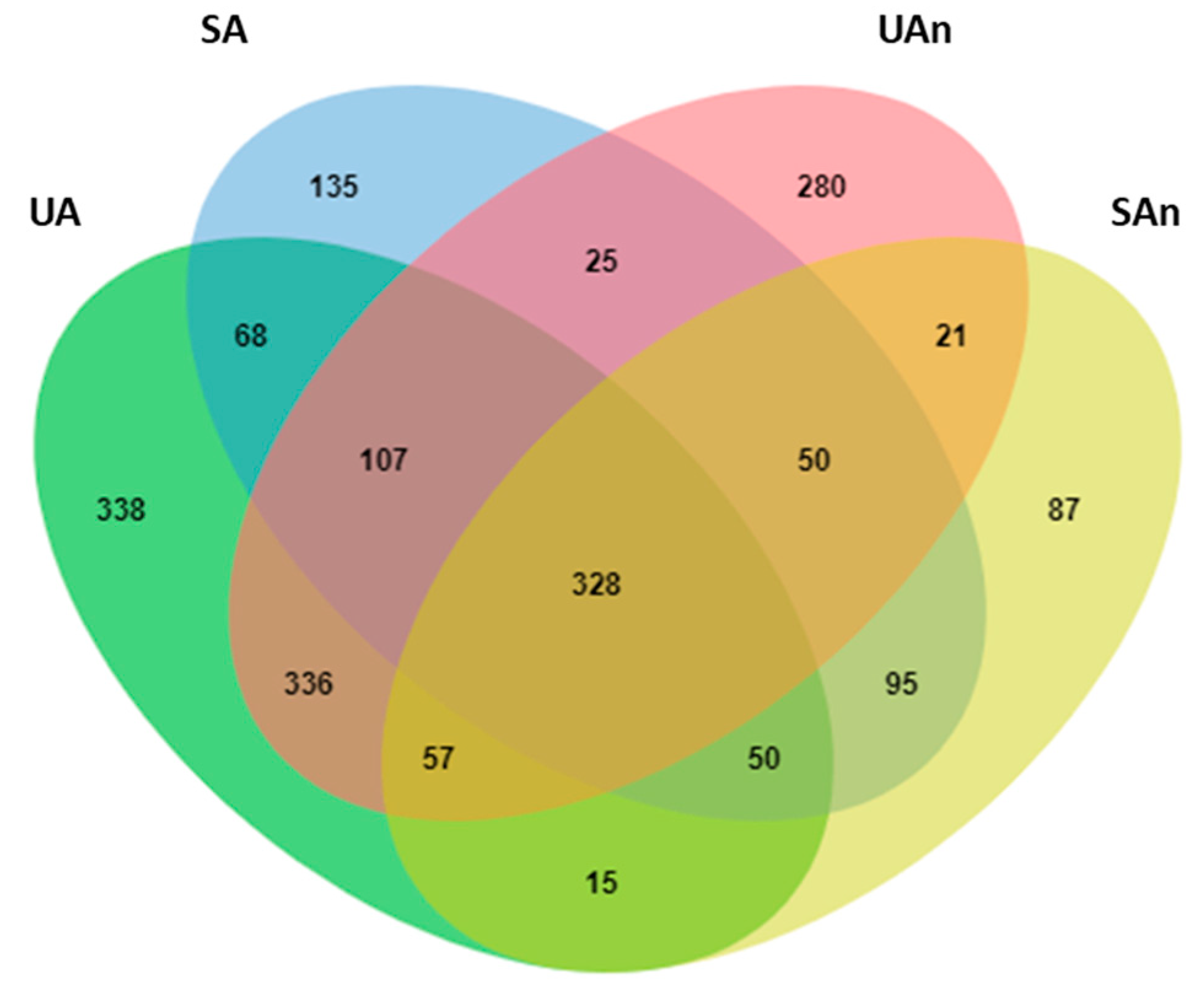
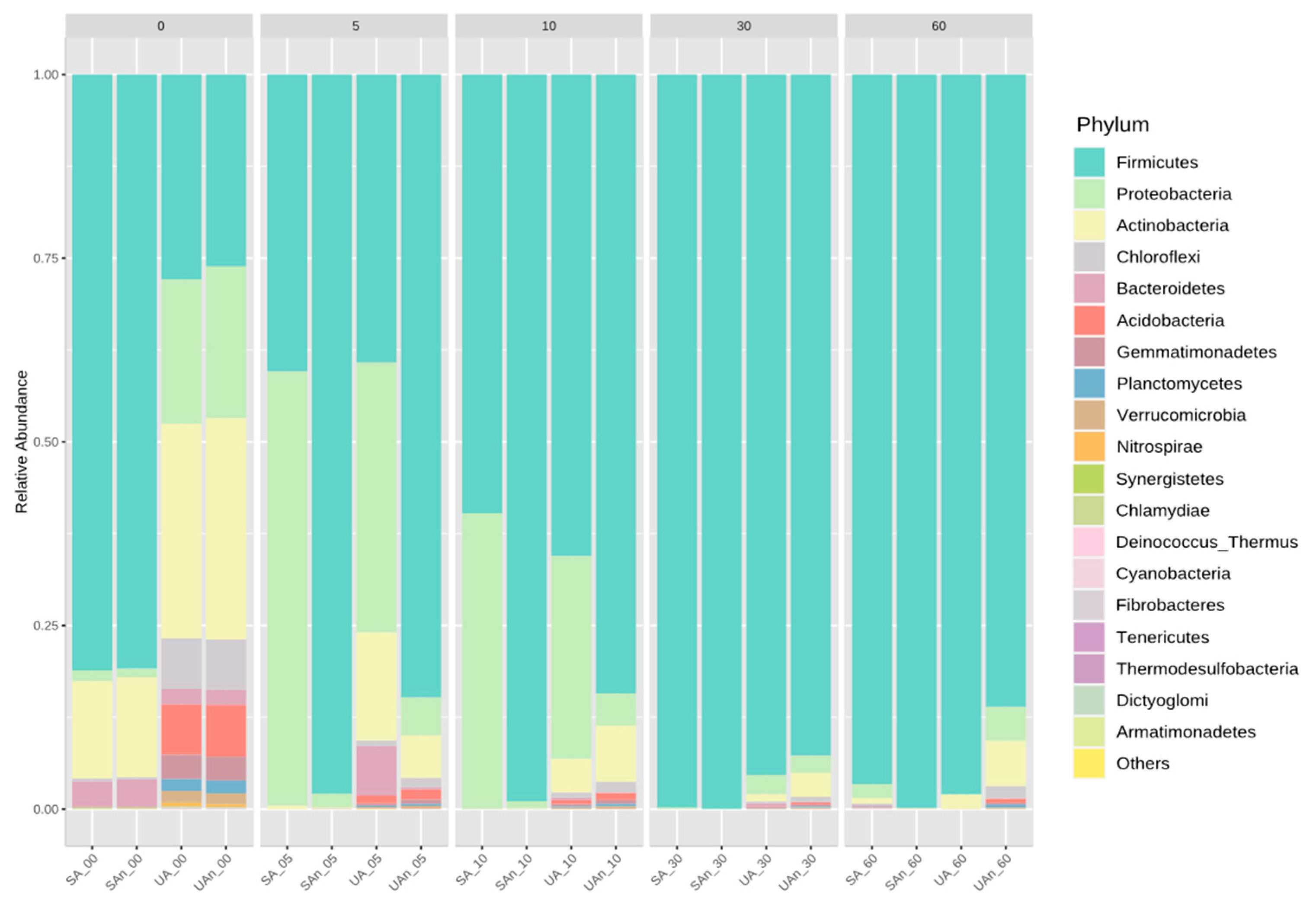
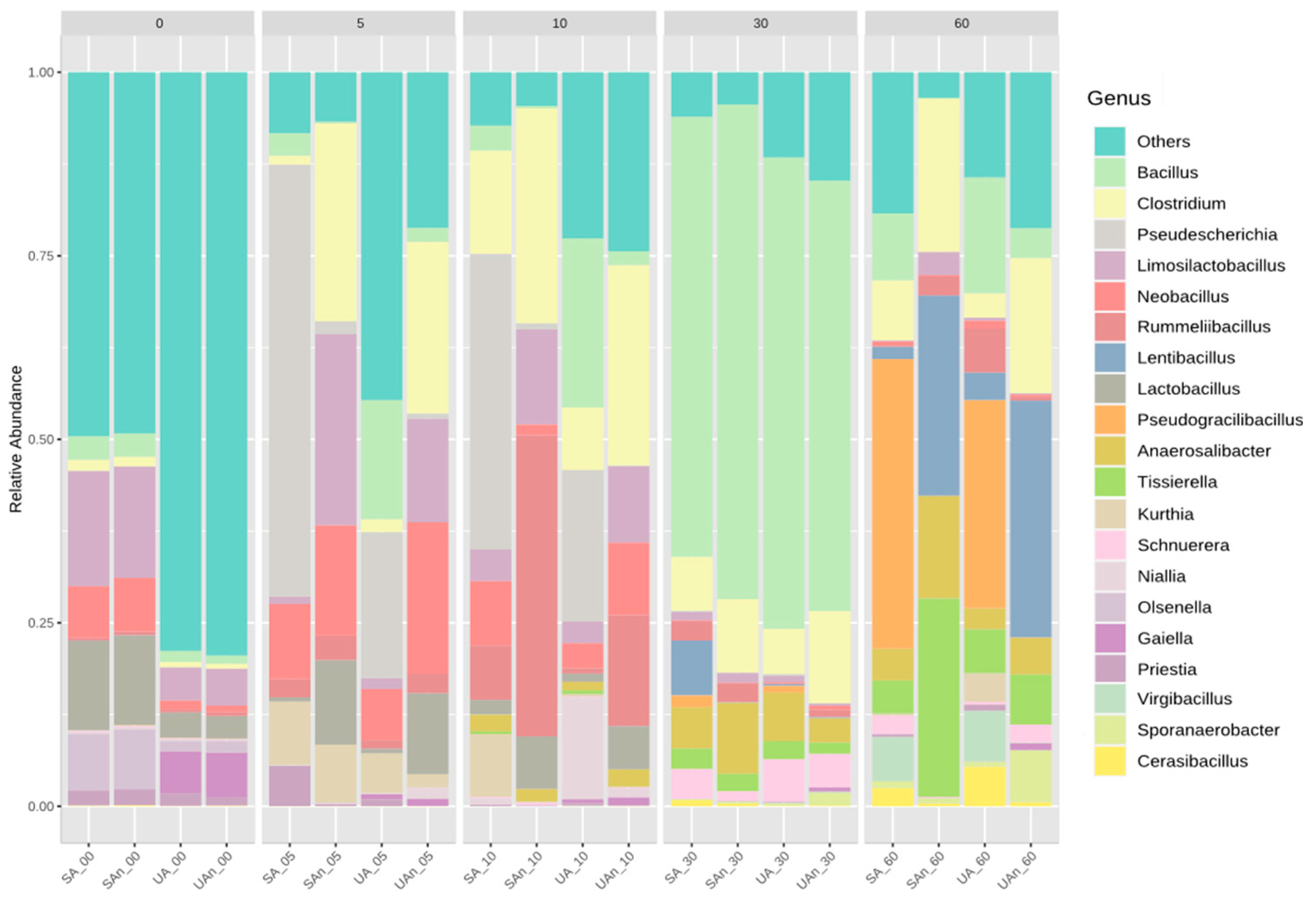
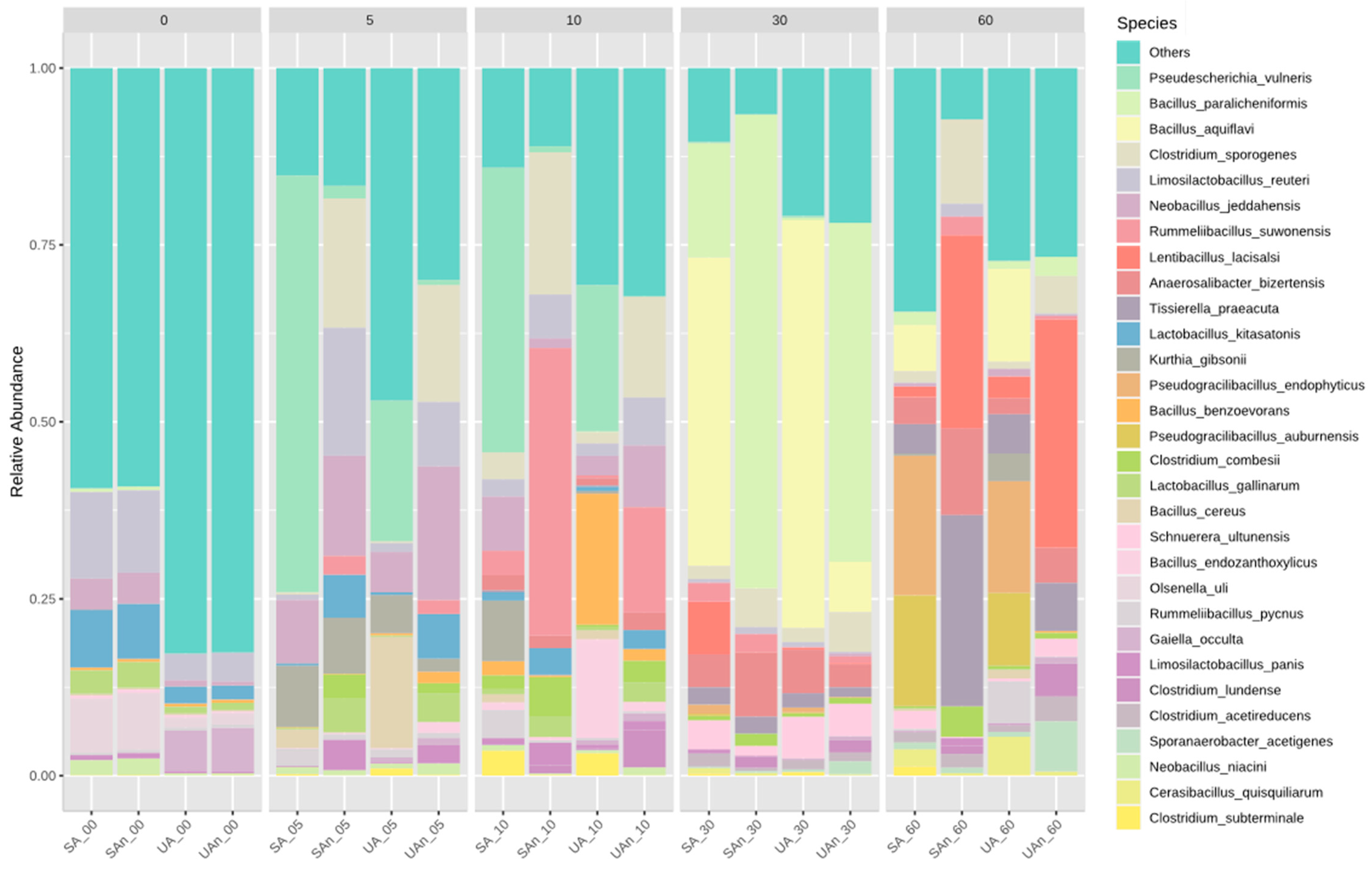
3.3. Bacterial Community Composition during Poultry Carcass Decomposition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Biggs, P.M.; Blaxter, K.L.; Fowden, L. Infectious animal disease and its control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1985, 310, 259–274. [Google Scholar] [CrossRef]
- Costa, T.; Akdeniz, N. A Review of the animal disease outbreaks and biosecure animal mortality composting systems. Waste Manag. 2019, 90, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Whiting, T.L. Foreign animal disease outbreaks, the animal welfare implications for Canada: Risks apparent from international experience. Can. Vet. J. 2003, 44, 805–815. [Google Scholar]
- Pfeiffer, D.U.; Otte, M.J.; Roland-Holst, D.; Zilberman, D. A one health perspective on HPAI H5N1 in the Greater Mekong sub-region. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 309–319. [Google Scholar] [CrossRef]
- Proença-Módena, J.L.; Macedo, I.S.; Arruda, E. H5N1 avian influenza virus: An overview. Braz. J. Infect. Dis. 2007, 11, 125–133. [Google Scholar] [CrossRef][Green Version]
- Miller, L.P.; Miknis, R.A.; Flory, G.A. Carcass Management Guidelines: Effective Disposal of Animal Carcasses and Contaminated Materials on Small to Medium-Sized Farms; FAO Animal Production and Health Guidelines: Rome, Italy, 2020; ISBN 978-92-5-133743-1. [Google Scholar]
- Baba, I.A.; Banday, M.; Khan, A.; Nighat, N. Traditional methods of carcass disposal: A review. J. Dairy Vet. Anim. Res. 2017, 5, 00128. [Google Scholar] [CrossRef]
- Gwyther, C.L.; Williams, A.P.; Golyshin, P.N.; Edwards-Jones, G.; Jones, D.L. The Environmental and Biosecurity Characteristics of Livestock Carcass Disposal Methods: A Review; Pergamon: Oxford, UK, 2011; Volume 3. [Google Scholar]
- Ki, B.M.; Kim, Y.M.; Jeon, J.M.; Ryu, H.W.; Cho, K.S. Characterization of odor emissions and microbial community structure during degradation of pig carcasses using the soil burial-composting method. Waste Manag. 2018, 77, 30–42. [Google Scholar] [CrossRef]
- Dangerfield, C.R.; Frehner, E.H.; Buechley, E.R.; Şekercioğlu, Ç.H.; Brazelton, W.J. Succession of bacterial communities on carrion is independent of vertebrate scavengers. PeerJ 2020, 8, e9307. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Metcalf, J.L.; Keepers, K.; Ackermann, G.; Carter, D.O.; Knight, R. Vertebrate decomposition is accelerated by soil microbes. Appl. Environ. Microbiol. 2014, 80, 4920–4929. [Google Scholar] [CrossRef] [PubMed]
- Iancu, L.; Dean, D.E.; Purcarea, C. Temperature influence on prevailing necrophagous diptera and bacterial taxa with forensic implications for postmortem interval estimation: A review. J. Med. Entomol. 2018, 55, 1369–1379. [Google Scholar] [CrossRef]
- Swift, M.J.; Heal, O.W.; Anderson, J.M.; Anderson, J.M. Decomposition in Terrestrial Ecosystems; University of California Press: Oakland, CA, USA, 1979. [Google Scholar]
- Carter, D.O.; Yellowlees, D.; Tibbett, M. Moisture Can be the dominant environmental parameter governing cadaver decomposition in soil. Forensic Sci. Int. 2010, 200, 60–66. [Google Scholar] [CrossRef]
- Vass, A. Beyond the grave—Understanding human decomposition. Microbiol. Today 2001, 28, 190–192. [Google Scholar]
- Howard, G.T.; Duos, B.; Watson-Horzelski, E.J. Characterization of the soil microbial community associated with the decomposition of a swine carcass. Int. Biodeterior. Biodegrad. 2010, 64, 300–304. [Google Scholar] [CrossRef]
- Harrison, L.; Kooienga, E.; Speights, C.; Tomberlin, J.; Lashley, M.; Barton, B.; Jordan, H. Microbial succession from a subsequent secondary death event following mass mortality. BMC Microbiol. 2020, 20, 309. [Google Scholar] [CrossRef]
- Finley, S.J.; Benbow, M.E.; Javan, G.T. Microbial communities associated with human decomposition and their potential use as postmortem clocks. Int. J. Legal Med. 2015, 129, 623–632. [Google Scholar] [CrossRef]
- Metcalf, J.L.; Carter, D.O.; Knight, R. Microbiology of death. Curr. Biol. 2016, 26, R561–R563. [Google Scholar] [CrossRef] [PubMed]
- Rapp, D.; Potier, P.; Jocteur-Monrozier, L.; Richaume, A. Prion degradation in soil: Possible role of microbial enzymes stimulated by the decomposition of buried carcasses. Environ. Sci. Technol. 2006, 40, 6324–6329. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.S.; Pontual, E.V.; Coelho, L.C.B.B.; Paiva, P.M.G. Saprophytic, symbiotic and parasitic bacteria: Importance to environment, biotechnological applications and biocontrol. Adv. Res. 2014, 2, 250–265. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Acosta-Mercado, D.; Sotomayor-Ramírez, D.; Cruz-Rodríguez, L. Microbial communities and enzymatic activities under different management in semiarid soils. Appl. Soil Ecol. 2008, 38, 249–260. [Google Scholar] [CrossRef]
- Yang, S.H.; Ahn, H.K.; Kim, B.S.; Chang, S.S.; Chung, K.Y.; Lee, E.M.; Ki, K.S.; Kwon, E.G. Comparison of bacterial communities in leachate from decomposing bovine carcasses. Asian-Australas. J. Anim. Sci. 2017, 30, 1660–1666. [Google Scholar] [CrossRef]
- Parmenter, R.R.; MacMahon, J.A. Carrion decomposition and nutrient cycling in a semiarid shrub–steppe ecosystem. Ecol. Monogr. 2009, 79, 637–661. [Google Scholar] [CrossRef]
- Stahl, D.; Tiedje, J. Microbial Ecology and Genomics: A Crossroads of Opportunity; American Academy of Microbiology: Washington, DC, USA, 2002. [Google Scholar] [CrossRef]
- Carter, D.O.; Yellowlees, D.; Tibbett, M. Autoclaving kills soil microbes yet soil enzymes remain active. Pedobiologia 2007, 51, 295–299. [Google Scholar] [CrossRef]
- Tiwari, S.C.; Tiwari, B.K.; Mishra, R.R. Enzyme activities in soils: Effects of leaching, ignition, autoclaving and fumigation. Soil Biol. Biochem. 1988, 20, 583–585. [Google Scholar] [CrossRef]
- Mant, A.K. Knowledge acquired from post-war exhumations. In Death, Decay and Reconstruction: Approaches to Archaeology and Forensic Science; Boddington, A., Garland, A.N., Janaway, R.C., Eds.; Manchester University Press: Manchester, UK, 1987; pp. 65–80. [Google Scholar]
- Illumina Inc. 16S Metagenomic Sequencing Library Preparation–Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. 2013. Available online: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 18 May 2019).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Zhu, Y.-L.; Zheng, G.-D.; Gao, D.; Chen, T.-B.; Wu, F.-K.; Niu, M.-J.; Zou, K.-H. Odor composition analysis and odor indicator selection during sewage sludge composting. J. Air Waste Manag. Assoc. 2016, 66, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Hassen, A.; Belguith, K.; Jedidi, N.; Cherif, A.; Cherif, M.; Boudabous, A. Microbial characterization during composting of municipal solid waste. Bioresour. Technol. 2001, 80, 217–225. [Google Scholar] [CrossRef]
- Wong, J.W.C.; Mak, K.F.; Chan, N.W.; Lam, A.; Fang, M.; Zhou, L.X.; Wu, Q.T.; Liao, X.D. Co-composting of soybean residues and leaves in Hong Kong. Bioresour. Technol. 2001, 76, 99–106. [Google Scholar] [CrossRef]
- Guo, R.; Li, G.; Jiang, T.; Schuchardt, F.; Chen, T.; Zhao, Y.; Shen, Y. Effect of aeration rate, C/N ratio and moisture content on the stability and maturity of compost. Bioresour. Technol. 2012, 112, 171–178. [Google Scholar] [CrossRef]
- König, S.; Vogel, H.-J.; Harms, H.; Worrich, A. Physical, chemical and biological effects on soil bacterial dynamics in microscale models. Front. Ecol. Evol. 2020, 8, 53. [Google Scholar] [CrossRef]
- Prosser, J.I.; Bohannan, B.J.M.; Curtis, T.P.; Ellis, R.J.; Firestone, M.K.; Freckleton, R.P.; Green, J.L.; Green, L.E.; Killham, K.; Lennon, J.J.; et al. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 2007, 5, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Minick, K.J.; Strickland, M.S.; Wickings, K.G.; Crippen, T.L.; Tarone, A.M.; Benbow, M.E.; Sufrin, N.; Tomberlin, J.K.; Pechal, J.L. Temporal and spatial impact of human cadaver decomposition on soil bacterial and arthropod community structure and function. Front. Microbiol. 2018, 8, 2616. [Google Scholar] [CrossRef]
- Metcalf, J.L.; Parfrey, L.W.; Gonzalez, A.; Lauber, C.L.; Knights, D.; Ackermann, G.; Humphrey, G.C.; Gebert, M.J.; Van Treuren, W.; Berg-Lyons, D.; et al. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. eLife 2013, 2013, e01104. [Google Scholar] [CrossRef]
- Cobaugh, K.L.; Schaeffer, S.M.; DeBruyn, J.M. Functional and structural succession of soil microbial communities below decomposing human cadavers. PLoS ONE 2015, 10, e0130201. [Google Scholar] [CrossRef]
- Pascual, J.; von Hoermann, C.; Rottler-Hoermann, A.-M.; Nevo, O.; Geppert, A.; Sikorski, J.; Huber, K.J.; Steiger, S.; Ayasse, M.; Overmann, J. Function of bacterial community dynamics in the formation of cadaveric semiochemicals during in situ carcass decomposition. Environ. Microbiol. 2017, 19, 3310–3322. [Google Scholar] [CrossRef] [PubMed]
- Ki, B.-M.; Kim, Y.M.; Jeon, J.M.; Ryu, H.W.; Cho, K.-S. Characterization of bacterial community dynamics during the decomposition of pig carcasses in simulated soil burial and composting systems. J. Microbiol. Biotechnol. 2017, 27, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Pechal, J.L.; Crippen, T.L.; Tarone, A.M.; Lewis, A.J.; Tomberlin, J.K.; Benbow, M.E. Microbial community functional change during vertebrate carrion decomposition. PLoS ONE 2013, 8, 79035. [Google Scholar] [CrossRef]
- Löki, V.; Deák, B.; Lukács, A.B.; Molnár, V.A. Biodiversity potential of burial places—A review on the flora and fauna of cemeteries and churchyards. Glob. Ecol. Conserv. 2019, 18, e00614. [Google Scholar] [CrossRef]
- Keenan, S.W.; DeBruyn, J.M. Changes to vertebrate tissue stable isotope (Δ15N) composition during decomposition. Sci. Rep. 2019, 9, 9929. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting soil PH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef]
- Cho, S.-J.; Kim, M.-H.; Lee, Y.-O. Effect of PH on soil bacterial diversity. J. Ecol. Environ. 2016, 40, 10. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil PH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Bartram, A.K.; Jiang, X.; Lynch, M.D.J.; Masella, A.P.; Nicol, G.W.; Dushoff, J.; Neufeld, J.D. Exploring links between PH and bacterial community composition in soils from the craibstone experimental farm. FEMS Microbiol. Ecol. 2014, 87, 403–415. [Google Scholar] [CrossRef]
- Janaway, R. The decomposition of materials associated with buried cadavers. In Soil Analysis in Forensic Taphonomy; CRC Press: Boca Raton, FL, USA, 2008; pp. 153–201. [Google Scholar]
- Hyde, E.R.; Haarmann, D.P.; Lynne, A.M.; Bucheli, S.R.; Petrosino, J.F. The living dead: Bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLoS ONE 2013, 8, e77733. [Google Scholar] [CrossRef]
- Chiba, A.; Uchida, Y.; Kublik, S.; Vestergaard, G.; Buegger, F.; Schloter, M.; Schulz, S. Soil bacterial diversity is positively correlated with decomposition rates during early phases of maize litter decomposition. Microorganisms 2021, 9, 357. [Google Scholar] [CrossRef]
- Sanapareddy, N.; Hamp, T.J.; Gonzalez, L.C.; Hilger, H.A.; Fodor, A.A.; Clinton, S.M. Molecular diversity of a north carolina wastewater treatment plant as revealed by pyrosequencing. Appl. Environ. Microbiol. 2009, 75, 1688–1696. [Google Scholar] [CrossRef]
- Li, A.; Chu, Y.; Wang, X.; Ren, L.; Yu, J.; Liu, X.; Yan, J.; Zhang, L.; Wu, S.; Li, S. A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol. Biofuels 2013, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Carter, D.O.; Metcalf, J.L.; Knight, R. Carcass mass has little influence on the structure of gravesoil microbial communities. Int. J. Legal Med. 2016, 130, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Fu, X.; Liao, H.; Hu, Z.; Long, L.; Yan, W.; Ding, Y.; Zha, L.; Guo, Y.; Yan, J.; et al. potential use of bacterial community succession for estimating post-mortem interval as revealed by high-throughput sequencing. Sci. Rep. 2016, 6, 24197. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Du, X.; Zhang, Y.; Li, T.; Liao, X. Effect of substrate on identification of microbial communities in poultry carcass composting and microorganisms associated with poultry carcass decomposition. J. Agric. Food Chem. 2016, 64, 6838–6847. [Google Scholar] [CrossRef]
- Benbow, M.E.; Pechal, J.L.; Lang, J.M.; Erb, R.; Wallace, J.R. The potential of high-throughput metagenomic sequencing of aquatic bacterial communities to estimate the postmortem submersion interval. J. Forensic Sci. 2015, 60, 1500–1510. [Google Scholar] [CrossRef]
- Yang, S.H.; Lim, J.S.; Khan, M.A.; Kim, B.S.; Choi, D.Y.; Lee, E.Y.; Ahn, H.K. High-throughput nucleotide sequence analysis of diverse bacterial communities in leachates of decomposing pig carcasses. Genet. Mol. Biol. 2015, 38, 373–380. [Google Scholar] [CrossRef]
- Tracy, B.P.; Jones, S.W.; Fast, A.G.; Indurthi, D.C.; Papoutsakis, E.T. Clostridia: The importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotechnol. 2012, 23, 364–381. [Google Scholar] [CrossRef]
- Berge, A.C.B.; Glanville, T.D.; Millner, P.D.; Klingborg, D.J. Methods and microbial risks associated with composting of animal carcasses in the United States. J. Am. Vet. Med. Assoc. 2009, 234, 47–56. [Google Scholar] [CrossRef]
- Alou, M.T.; Ndongo, S.; Frégère, L.; Labas, N.; Andrieu, C.; Richez, M.; Couderc, C.; Baudoin, J.-P.; Abrahão, J.; Brah, S.; et al. Taxonogenomic description of four new clostridium species isolated from human gut: ‘clostridium amazonitimonense’, ‘clostridium merdae’, ‘clostridium massilidielmoense’ and ‘clostridium nigeriense’. New Microbes New Infect. 2018, 21, 128–139. [Google Scholar] [CrossRef]
- Semenov, M.V.; Krasnov, G.S.; Rybka, K.Y.; Kharitonov, S.L.; Zavgorodnyaya, Y.A.; Yudina, A.V.; Shchegolkova, N.M. Spatial changes in microbial communities along different functional zones of a free-water surface wetland. Microorganisms 2020, 8, 1604. [Google Scholar] [CrossRef]
- Longa, C.M.O.; Nicola, L.; Antonielli, L.; Mescalchin, E.; Zanzotti, R.; Turco, E.; Pertot, I. Soil microbiota respond to green manure in organic vineyards. J. Appl. Microbiol. 2017, 123, 1547–1560. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, T.; Zhang, F.; Wang, E.T.; He, J.F.; Ding, H.; Zhang, B.T.; Liu, J.; Ran, X.B.; Zang, J.Y. Diversity and structure of soil bacterial communities in the fildes region (Maritime Antarctica) as revealed by 454 pyrosequencing. Front. Microbiol. 2015, 6, 1188. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.A.; Riedel, T.; Oren, A.; Overmann, J.; Lawley, T.D.; Clavel, T. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun. 2021, 1, 1–16. [Google Scholar] [CrossRef]
- Oakley, B.B.; Morales, C.A.; Line, J.; Berrang, M.E.; Meinersmann, R.J.; Tillman, G.E.; Wise, M.G.; Siragusa, G.R.; Hiett, K.L.; Seal, B.S. The poultry-associated microbiome: Network analysis and farm-to-fork characterizations. PLoS ONE 2013, 8, e57190. [Google Scholar] [CrossRef]
- Scupham, A.J.; Jones, J.A.; Rettedal, E.A.; Weber, T.E. Antibiotic manipulation of intestinal microbiota to identify microbes associated with campylobacter jejuni exclusion in poultry. Appl. Environ. Microbiol. 2010, 76, 8026–8032. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kim, G.B.; Cha, C.J. Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult. Sci. 2014, 93, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Kilani, B.; Ammari, L.; Benaïssa, H.T.; Fendri, C.; Chaabane, T.B. Escherichia vulneris as a cause of bacteremia in a patient with chronic lymphocytic leukemia. Int. J. Infect. Dis. 2008, 12, 110–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brenner, D.J.; McWhorter, A.C.; Knutson, J.K.; Steigerwalt, A.G. Escherichia vulneris: A new species of enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 1982, 15, 1133–1140. [Google Scholar] [CrossRef]
- Jiménez, S.M.; Tiburzi, M.C.; Salsi, M.S.; Pirovani, M.E.; Moguilevsky, M.A. The role of visible faecal material as a vehicle for generic escherichia coli, coliform, and other enterobacteria contaminating poultry carcasses during slaughtering. J. Appl. Microbiol. 2003, 95, 451–456. [Google Scholar] [CrossRef]
- Her, J.; Kim, J. Rummeliibacillus suwonensis sp. nov., isolated from soil collected in a mountain area of South Korea. J. Microbiol. 2013, 51, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Bittar, F.; Bibi, F.; Ramasamy, D.; Lagier, J.-C.; Azhar, E.I.; Jiman-Fatani, A.A.; Al-Ghamdi, A.K.; Nguyen, T.T.; Yasir, M.; Fournier, P.-E.; et al. Non contiguous-finished genome sequence and description of bacillus jeddahensis sp. nov. Stand. Genom. Sci. 2015, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-H.; Hong, S.H.; Cho, S.B.; Lim, J.S.; Bae, S.E.; Ahn, H.; Lee, E.Y. Characterization of microbial community in the leachate associated with the decomposition of entombed pigs. J. Microbiol. Biotechnol. 2012, 22, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Rezgui, R.; Maaroufi, A.; Fardeau, M.-L.; Ben Ali Gam, Z.; Cayol, J.-L.; Ben Hamed, S.; Labat, M. 2012 Anaerosalibacter bizertensis Gen. Nov., Sp. Nov., a halotolerant bacterium isolated from sludge. Int. J. Syst. Evol. Microbiol. 2012, 62, 2469–2474. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.-K.; Yun, B.-R.; Han, J.-H.; Kim, S.B. 2018 Pseudogracilibacillus endophyticus Sp. Nov., a moderately thermophilic and halophilic species isolated from plant root. Int. J. Syst. Evol. Microbiol. 2018, 68, 165–169. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Kang, K.H.; Park, Y.-H. Lentibacillus salicampi Gen. Nov., Sp. Nov., a moderately halophilic bacterium isolated from a salt field in Korea. Int. J. Syst. Evol. Microbiol. 2002, 52, 2043–2048. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miguel, M.A.; Kim, S.-H.; Lee, S.-S.; Cho, Y.-I. Impact of Soil Microbes and Oxygen Availability on Bacterial Community Structure of Decomposing Poultry Carcasses. Animals 2021, 11, 2937. https://doi.org/10.3390/ani11102937
Miguel MA, Kim S-H, Lee S-S, Cho Y-I. Impact of Soil Microbes and Oxygen Availability on Bacterial Community Structure of Decomposing Poultry Carcasses. Animals. 2021; 11(10):2937. https://doi.org/10.3390/ani11102937
Chicago/Turabian StyleMiguel, Michelle A., Seon-Ho Kim, Sang-Suk Lee, and Yong-Il Cho. 2021. "Impact of Soil Microbes and Oxygen Availability on Bacterial Community Structure of Decomposing Poultry Carcasses" Animals 11, no. 10: 2937. https://doi.org/10.3390/ani11102937
APA StyleMiguel, M. A., Kim, S.-H., Lee, S.-S., & Cho, Y.-I. (2021). Impact of Soil Microbes and Oxygen Availability on Bacterial Community Structure of Decomposing Poultry Carcasses. Animals, 11(10), 2937. https://doi.org/10.3390/ani11102937




