Fast and Slow-Growing Management Systems: Characterisation of Broiler Caecal Microbiota Development throughout the Growing Period
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiment Design
2.2. Sample Collection
2.3. DNA Extraction
2.4. 16S rRNA Sequencing Analysis
2.5. Data Availability
3. Results
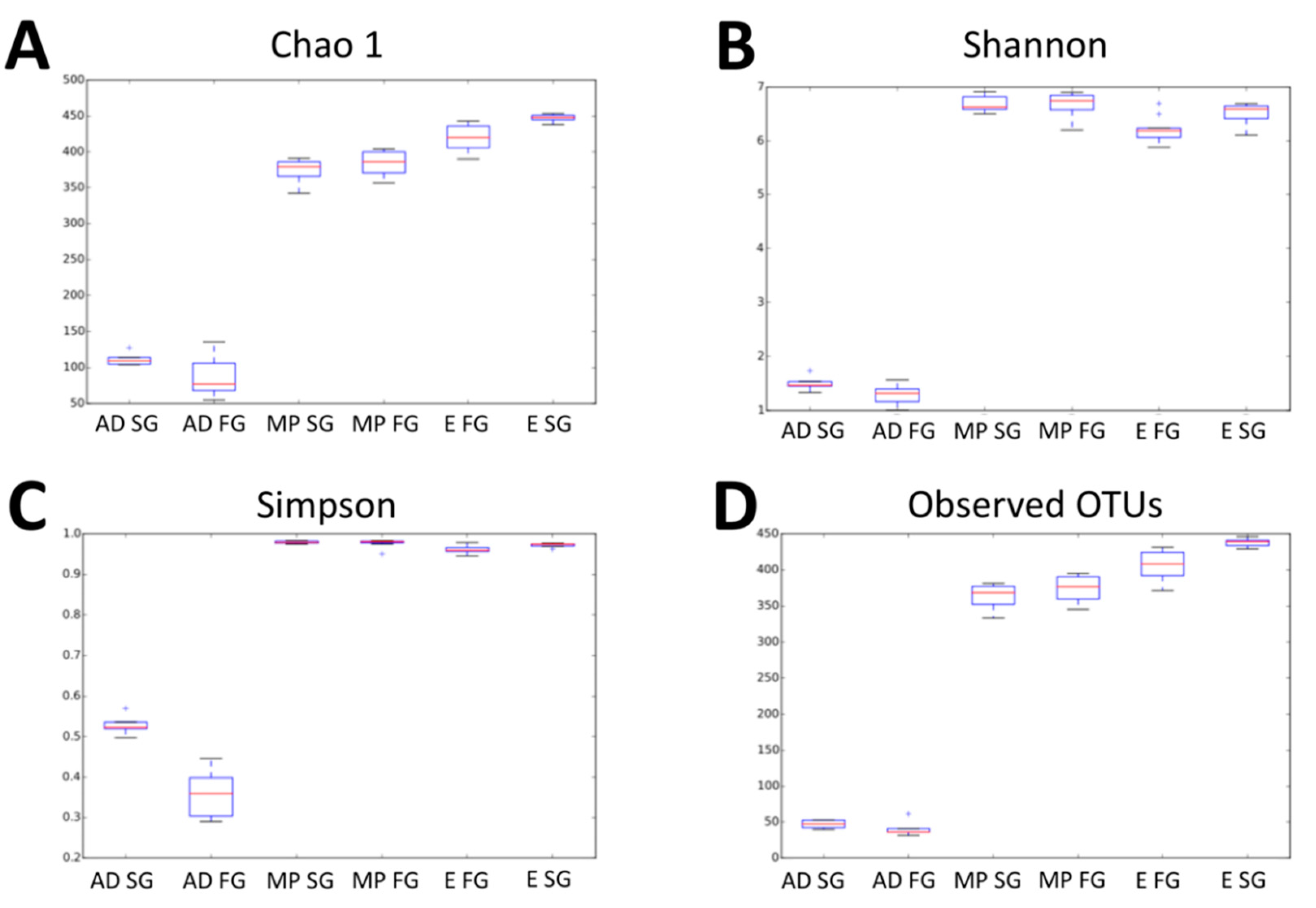
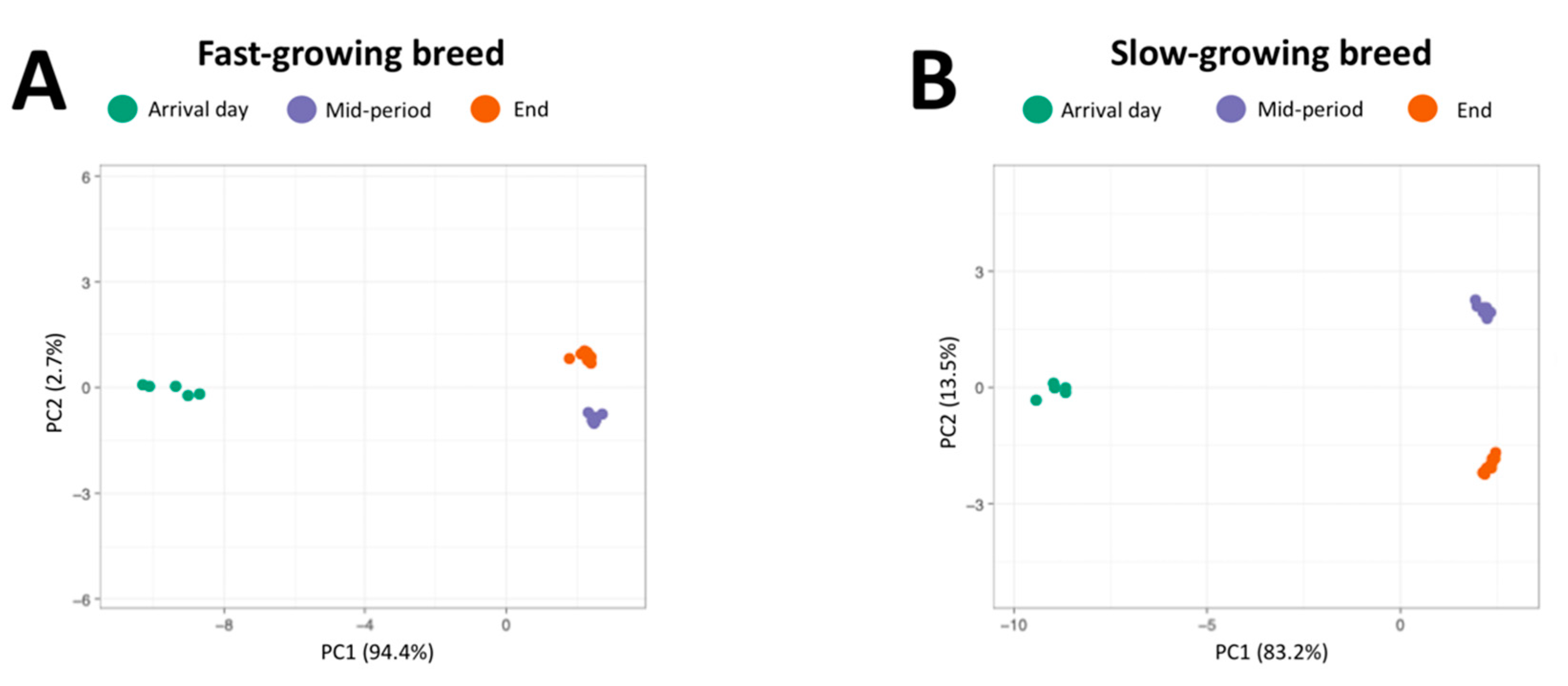
3.1. 16 rRNA Profiling of Fast and Slow-Growing Management Systems
3.2. Differential Gut Microbiota Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Pourabedin, M.; Zhao, X. Prebiotics and gut microbiota in chickens. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14. [Google Scholar] [CrossRef]
- Banerjee, S.; Sar, A.; Misra, A.; Pal, S.; Chakraborty, A.; Dam, B. Increased productivity in poultry birds by sub-lethal dose of antibiotics is arbitrated by selective enrichment of gut microbiota, particularly short-chain fatty acid producers. Microbiology (United Kingdom) 2018, 164, 142–153. [Google Scholar] [CrossRef]
- Clavijo, V.; Flórez, M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poult. Sci. 2018, 97, 1006–1021. [Google Scholar] [CrossRef]
- Pandit, R.J.; Hinsu, A.T.; Patel, N.V.; Koringa, P.G.; Jakhesara, S.J.; Thakkar, J.R.; Shah, T.M.; Limon, G.; Psifidi, A.; Guitian, J.; et al. Microbial diversity and community composition of caecal microbiota in commercial and indigenous Indian chickens determined using 16s rDNA amplicon sequencing. Microbiome 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Carrasco, J.M.D.; Casanova, N.A.; Miyakawa, M.E.F. Microbiota, gut health and chicken productivity: What is the connection? Microorganisms 2019, 7, 374. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Caetano, M.; Antunes, L.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Stegeman, J.A.; Smidt, H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- WHO|Antimicrobial Resistance: Global Report on Surveillance 2014; World Health Organization: Geneva, Switzerland, 2014.
- Alós, J. Resistencia bacteriana a los antibióticos: Una crisis global. Enferm. Infecc. Microbiol. Clin. 2015, 33, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Gadde, U.; Kim, W.H.; Oh, S.T.; Lillehoj, H.S. Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: A review. Anim. Health Res. Rev. 2017, 18, 26–45. [Google Scholar] [CrossRef]
- Montoro-Dasi, L.; Villagra, A.; Sevilla-Navarro, S.; Pérez-Gracia, M.T.; Vega, S.; Marin, C. The dynamic of antibiotic resistance in commensal Escherichia coli throughout the growing period in broiler chickens: Fast-growing vs. slow-growing breeds. Poult. Sci. 2020, 99, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Nijnik, A.; Philpott, D.J. Modulating immunity as a therapy for bacterial infections for bacterial infections. Nat. Rev. Microbiol. 2012, 10, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Hao, H.; Xie, S.; Wang, X.; Dai, M.; Huang, L.; Yuan, Z. Antibiotic alternatives: The substitution of antibiotics in animal husbandry? Front. Microbiol. 2014, 5, 1–15. [Google Scholar] [CrossRef]
- Castellini, C.; Bosco, A.D. Animal Welfare and Poultry Meat in Alternative Production Systems (and Ethics of Poultry Meat Production); Elsevier Ltd.: Amsterdam, The Netherlands, 2017; ISBN 9780081007631. [Google Scholar]
- Polycarpo, G.V.; Andretta, I.; Kipper, M.; Dadalt, J.C.; Rodrigues, P.H.M.; Albuquerque, R. Meta-analytic study of organic acids as an alternative performance-enhancing feed additive to antibiotics for broiler chickens. Poult. Sci. 2017, 96, 3645–3653. [Google Scholar] [CrossRef]
- Suresh, G.; Das, R.K.; Brar, S.K.; Rouissi, T.; Ramirez, A.; Chorfi, Y.; Godbout, S. Critical Reviews in Microbiology Alternatives to antibiotics in poultry feed: Molecular perspectives. Crit. Rev. Microbiol. 2017, 1–18. [Google Scholar] [CrossRef]
- Alagawany, M.; Abd El-Hack, M.E.; Farag, M.R.; Sachan, S.; Karthik, K.; Dhama, K. The use of probiotics as eco-friendly alternatives for antibiotics in poultry nutrition. Environ. Sci. Pollut. Res. 2018, 25, 10611–10618. [Google Scholar] [CrossRef]
- Sevilla-Navarro, S.; Marín, C.; Cortés, V.; García, C.; Vega, S.; Catalá-Gregori, P. Autophage as a control measure for Salmonella in laying hens. Poult. Sci. 2018, 97, 4367–4373. [Google Scholar] [CrossRef]
- Kogut, M.H.; Yin, X.; Yuan, J.; Broom, L. Gut health in poultry. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2017, 12. [Google Scholar] [CrossRef]
- Pedroso, A.A.; Menten, J.F.M.; Lambais, M.R. The Structure of Bacterial Community in the Intestines of Newly Hatched Chicks. J. Appl. Poult. Res. 2005, 14, 232–237. [Google Scholar] [CrossRef]
- Oakley, B.B.; Morales, C.A.; Line, J.; Berrang, M.E.; Meinersmann, R.J.; Tillman, G.E.; Wise, M.G.; Siragusa, G.R.; Hiett, K.L.; Seal, B.S. The Poultry-Associated Microbiome: Network Analysis and Farm-to-Fork Characterizations. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Richards, P.; Fothergill, J.; Bernardeau, M.; Wigley, P. Development of the caecal microbiota in three broiler breeds. Front. Vet. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Xi, Y.; Shuling, N.; Kunyuan, T.; Qiuyang, Z.; Hewen, D.; ChenCheng, G.; Tianhe, Y.; Liancheng, L.; Xin, F. Characteristics of the intestinal flora of specific pathogen free chickens with age. Microb. Pathog. 2019, 132, 325–334. [Google Scholar] [CrossRef]
- Brisbin, J.T.; Gong, J.; Sharif, S. Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim. Health Res. Rev. 2008, 9, 101–110. [Google Scholar] [CrossRef]
- Rasschaert, G.; Houf, K.; Godard, C.; Wildemauwe, C.; Pastuszczak-Frąk, M.; De Zutter, L. Contamination of carcasses with Salmonella during poultry slaughter. J. Food Prot. 2008, 71, 146–152. [Google Scholar] [CrossRef]
- Ellerbroek, L.I.; Lienau, J.A.; Klein, G. Campylobacter spp. in broiler flocks at farm level and the potential for cross-contamination during slaughter. Zoonoses Public Health 2010, 57. [Google Scholar] [CrossRef]
- Sevilla-Navarro, S.; Marin, C.; Cortés, V.; García, C.; Catalá-Gregori, P. Campylobacter prevalence and risk factors associated with exceeding allowable limits in poultry slaughterhouses in Spain. Vet. Rec. 2020, 186. [Google Scholar] [CrossRef]
- Kogut, M.H. The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Technol. 2019, 250, 32–40. [Google Scholar] [CrossRef]
- Spain 2013. Royal Degree 53/2013, 1st of Febrary, por el que se Establecen las Normas Básicas Aplicables Para la Protección de los Animales Utilizados en Experimentación y Otros Fines Científicos, Incluyendo la Docencia; Boletín Oficial del Estado: Madrid, Spain, 2013; Volume 34, pp. 11370–11421. [Google Scholar]
- Ross. Ross 308/Ross 308 FF broiler: Performance Objectives. Available online: http://es.aviagen.com/assets/Tech_Center/Ross_Broiler/Ross308-308FF-BroilerPO2019-EN.pdf (accessed on 17 June 2020).
- Valls, M. Reproductoras y Pollos de Crecimiento Lento Como Producto Diferenciado. Available online: https://avicultura.info/reproductoras-y-pollos-de-crecimiento-lento/ (accessed on 17 June 2020).
- Santomá, G.; Mateos, G.G. Necesidades Nutricionales para Avicultura: Normas FEDNA, 2nd ed.; FEDNA (Fundación Española Desarrollo Nutrición Animal): Madrid, Spain; ISBN 9788409065295.
- Illumina Support. 16S Metagenomic Sequencing Library Preparation. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 17 June 2020).
- Babraham Bioinformatics. FastQC A Quality Control tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 June 2020).
- Babraham Bioinformatics. Trim Galore! Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 17 June 2020).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Qiime (Quantitative Insights Into Microbial Ecology). Compare_alpha_diversity.py–This Script Compares Alpha Diversities Based on a Two-Sample t-test Using Either Parametric or Non-Parametric (Monte Carlo) Methods. Available online: http://qiime.org/scripts/compare_alpha_diversity.html (accessed on 18 June 2020).
- Paulson, J.N.; Colin Stine, O.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10. [Google Scholar] [CrossRef]
- Mancabelli, L.; Ferrario, C.; Milani, C.; Mangifesta, M.; Turroni, F.; Duranti, S.; Lugli, G.A.; Viappiani, A.; Ossiprandi, M.C.; van Sinderen, D.; et al. Insights into the biodiversity of the gut microbiota of broiler chickens. Environ. Microbiol. 2016, 18, 4727–4738. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 8, 1–31. [Google Scholar] [CrossRef]
- Wang, J.; Nesengani, L.T.; Gong, Y.; Yang, Y.; Lu, W. 16S rRNA gene sequencing reveals effects of photoperiod on cecal microbiota of broiler roosters. PeerJ 2018, 2. [Google Scholar] [CrossRef]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and Succession of the Intestinal Bacterial Community of the Maturing Broiler Chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef]
- Mohd Shaufi, M.A.; Sieo, C.C.; Chong, C.W.; Gan, H.M.; Ho, Y.W. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 2015, 7, 1–12. [Google Scholar] [CrossRef]
- Ocejo, M.; Oporto, B.; Hurtado, A. 16S rRNA amplicon sequencing characterization of caecal microbiome composition of broilers and free-range slow-growing chickens throughout their productive lifespan. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Stanley, D.; Denman, S.E.; Hughes, R.J.; Geier, M.S.; Crowley, T.M.; Chen, H.; Haring, V.R.; Moore, R.J. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl. Microbiol. Biotechnol. 2012, 96, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Schokker, D.; Veninga, G.; Vastenhouw, S.A.; Bossers, A.; de Bree, F.M.; Kaal-Lansbergen, L.M.T.E.; Rebel, J.M.J.; Smits, M.A. Early life microbial colonization of the gut and intestinal development differ between genetically divergent broiler lines. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Qu, A.; Brulc, J.M.; Wilson, M.K.; Law, B.F.; Theoret, J.R.; Joens, L.A.; Konkel, M.E.; Angly, F.; Dinsdale, E.A.; Edwards, R.A.; et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE 2008, 3. [Google Scholar] [CrossRef]
- Wei, S.; Morrison, M.; Yu, Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013, 92, 671–683. [Google Scholar] [CrossRef]
- Rychlik, I. Composition and function of chicken gut microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef]
- Ballou, A.L.; Ali, R.A.; Mendoza, M.A.; Ellis, J.C.; Hassan, H.M.; Croom, W.J.; Koci, M.D. Development of the chick microbiome: How early exposure influences future microbial diversity. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef]
- Ducatelle, R.; Goossens, E.; De Meyer, F.; Eeckhaut, V.; Antonissen, G.; Haesebrouck, F.; Van Immerseel, F. Biomarkers for monitoring intestinal health in poultry: Present status and future perspectives. Vet. Res. 2018, 49, 1–9. [Google Scholar] [CrossRef]
- Yacoubi, N.; Saulnier, L.; Bonnin, E.; Devillard, E.; Eeckhaut, V.; Rhayat, L.; Ducatelle, R.; Van Immerseel, F. Short-chain arabinoxylans prepared from enzymatically treated wheat grain exert prebiotic effects during the broiler starter period. Poult. Sci. 2018, 97, 412–424. [Google Scholar] [CrossRef]
- Kumar, S.; Chen, C.; Indugu, N.; Werlang, G.O.; Singh, M.; Kim, W.K.; Thippareddi, H. Effect of antibiotic withdrawal in feed on chicken gut microbial dynamics, immunity, growth performance and prevalence of foodborne pathogens. PLoS ONE 2018, 13, 1–23. [Google Scholar] [CrossRef]
- Neal-McKinney, J.M.; Lu, X.; Duong, T.; Larson, C.L.; Call, D.R.; Shah, D.H.; Konkel, M.E. Production of Organic Acids by Probiotic Lactobacilli Can Be Used to Reduce Pathogen Load in Poultry. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17. [CrossRef]
- Zhao, L.; Wang, G.; Siegel, P.; He, C.; Wang, H.; Zhao, W.; Zhai, Z.; Tian, F.; Zhao, J.; Zhang, H.; et al. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Torok, V.A.; Hughes, R.J.; Mikkelsen, L.L.; Perez-Maldonado, R.; Balding, K.; MacAlpine, R.; Percy, N.J.; Ophel-Keller, K. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl. Environ. Microbiol. 2011, 77, 5868–5878. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3. [Google Scholar] [CrossRef]
- Siegerstetter, S.C.; Schmitz-Esser, S.; Magowan, E.; Wetzels, S.U.; Zebeli, Q.; Lawlor, P.G.; O’Connell, N.E.; Metzler-Zebeli, B.U. Intestinal microbiota profiles associated with low and high residual feed intake in chickens across two geographical locations. PLoS ONE 2017, 12, 1–23. [Google Scholar] [CrossRef]
- Jha, R.; Singh, A.K.; Yadav, S.; Berrocoso, J.F.D.; Mishra, B. Early Nutrition Programming (in ovo and Post-hatch Feeding) as a Strategy to Modulate Gut Health of Poultry. Front. Vet. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Yadav, S.; Jha, R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J. Anim. Sci. Biotechnol. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Zhong, T.; Pandya, Y.; Joerger, R.D. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 2002, 68, 124–137. [Google Scholar] [CrossRef]
- Rodríguez, M.L.; Rebolé, A.; Velasco, S.; Ortiz, L.T.; Treviño, J.; Alzueta, C. Wheat- and barley-based diets with or without additives influence broiler chicken performance, nutrient digestibility and intestinal microflora. J. Sci. Food Agric. 2012, 92, 184–190. [Google Scholar] [CrossRef]
- Paraskeuas, V.; Mountzouris, K.C. Broiler gut microbiota and expressions of gut barrier genes affected by cereal type and phytogenic inclusion. Anim. Nutr. 2019, 5, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tellez, G.; Richards, J.D.; Escobar, J. Identification of potential biomarkers for gut barrier failure in broiler chickens. Front. Vet. Sci. 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.I.M.; Lima, F.R.; Mendonça, C.X.; Mabe, I.; Albuquerque, R.; Leal, P.M. Relative bioavailability of phosphorus in feed and agricultural phosphates for poultry. Poult. Sci. 1999, 78, 1729–1736. [Google Scholar] [CrossRef]

| Analytical Constituents (%) | Diet | ||
|---|---|---|---|
| Starter | Grower FG | Grower SG | |
| Crude fat | 3.5% | 3.1% | 3.8% |
| Crude protein | 20.5% | 19.4% | 18.0% |
| Crude fibre | 2.6% | 3.1% | 3.2% |
| Crude ash | 6.6% | 5.0% | 5.5% |
| Lysine | 1.14% | 1.13% | 0.94% |
| Methionine | 0.62% | 0.51% | 0.40% |
| Calcium | 1.00% | 0.78% | 1.00% |
| Phosphorus | 0.69% | 0.51% | 0.43% |
| Sodium | 0.15% | 0.14% | 0.17% |
| Ingredients | Corn, soy flour, wheat, soy oil, calcium carbonate, monocalcium phosphate, sodium chloride | Corn, soy flour, rice bran, calcium carbonate, sodium chloride | Wheat, soy flour, barley, soy oil, calcium carbonate, monocalcium phosphate, sodium chloride, sodium bicarbonate |
| Fast-Growing (FG) | Slow-Growing (SG) | |||
|---|---|---|---|---|
| Days of Life | Weight (g) | CR | Weight | CR |
| 0 | 47.20 ± 0.98 | 41.31 ± 1.24 | ||
| 7 | 184.80 ± 8.92 | 1.16 ± 0.10 | 146.05 ± 6.25 | 1.26 ± 0.14 |
| 14 | 492.90 ± 44.81 | 1.25 ± 0.18 | 368.23 ± 43.77 | 1.29 ± 0.35 |
| 21 | 823.32 ± 41.88 | 1.23 ± 0.16 | 547.21 ± 18.42 | 1.22 ± 0.63 |
| 28 | 1503.41 ± 77.66 | 1.30 ± 0.15 | 936.98 ± 31.20 | 1.34 ± 0.44 |
| 35 | 2043.72 ± 163.78 | 2.73 ± 0.74 | 1283.64 ± 93.16 | 2.70 ± 0.83 |
| 42 | 2605.91 ± 242.06 | 3.06 ± 1.30 | 1631.83 ± 105.98 | 3.29 ± 1.07 |
| 49 | 2049.22 ± 146.00 | 3.05 ± 1.08 | ||
| 56 | 2439.40 ± 183.25 | 3.17 ± 1.76 | ||
| 63 | 2776.33 ± 181.86 | 4.34 ± 1.99 | ||
| SAMPLING TIME | FG | SG |
|---|---|---|
| Arrival day | 88.3 a | 111.9 d |
| Mid-period | 384.4 b | 373.8 e |
| End period | 420.3 c | 447.2 f |
| Breed | Fast-growing | Slow-growing | ||||
|---|---|---|---|---|---|---|
| Sampling Time | AD | MP | E | AD | MP | E |
| Actinobacteria | 0.0% | 0.3% | 0.5% | 0.2% | 0.3% | 0.4% |
| Bacteroidetes | 5.0% a | 1.9% b | 5.7% c | 5.7% l | 1.9% m | 9.3% n |
| Cyanobacteria | 0.0% d | 0.5% d | 0.7% e | 0.0% | 0.4% | 1.1% |
| Firmicutes | 58.6% f | 95.1% g | 90.3% h | 61.1%o | 95.2% p | 85.6% q |
| Proteobacteria | 36.4% i | 1.3% j | 1.5% k | 32.8% r | 1.2% s | 1.7% s |
| Tenericutes | 0.0% | 0.3% | 0.6% | 0.2% | 0.4% | 1.1% |
| Unassigned; NA | 0.0% | 0.6% | 0.8% | 0.0% | 0.6% | 0.8% |
| Phylum | Family | Genus | AD | MP | E |
|---|---|---|---|---|---|
| Unassigned | 0.0% | 0.6% | 0.8% | ||
| Bacteroidetes | Bacteroidaceae | Bacteroides | 1.5% | 0.5% | 3.1% |
| Porphyromonadaceae | Parabacteroides | 1.2% | 0.4% | 0.7% | |
| Rikenellaceae | - | 2.0% | 1.1% | 1.2% | |
| Odoribacteraceae | Butyricimonas | 0.3% | 0.0% | 0.7% | |
| Cyanobacteria | - | - | 0.0% | 0.5% | 0.7% |
| Firmicutes | Planococcaceae | - | 0.0% | 0.5% | 0.4% |
| Enterococcaceae | - | 3.7% | 0.0% | 0.0% | |
| Enterococcus | 3.0% | 0.2% | 0.1% | ||
| Lactobacillaceae | Lactobacillus | 0.9% | 3.9% | 2.8% | |
| - | - | 0.2% | 0.5% | 0.6% | |
| - | - | 13.7% | 29.4% | 28.9% | |
| Christensenellaceae | - | 0.0% | 0.2% | 0.6% | |
| Clostridiaceae | - | 0.6% | 0.0% | 0.3% | |
| - | 5.6% | 0.2% | 0.2% | ||
| Clostridium | 4.1% | 0.5% | 0.5% | ||
| Lachnospiraceae | - | 4.9% | 10.4% | 10.2% | |
| Blauria | 0.7% | 2.0% | 2.1% | ||
| Coprococcus | 1.6% | 4.0% | 3.5% | ||
| Dorea | 0.2% | 1.4% | 1.1% | ||
| Epulopscium | 2.6% | 0.0% | 0.0% | ||
| [Ruminococcus] | 2.5% | 3.3% | 2.9% | ||
| Ruminococcaceae | - | 5.7% | 18.1% | 17.7% | |
| Anaerotruncus | 0.0% | 0.5% | 0.4% | ||
| Faecalibacterium | 0.9% | 1.5% | 2.0% | ||
| Oscillospira | 3.5% | 9.6% | 8.8% | ||
| Ruminococcus | 2.1% | 5.0% | 4.4% | ||
| Erysipelotrichaceae | - | 0.9% | 0.9% | 0.4% | |
| Coprobacillus | 0.4% | 0.9% | 0.5% | ||
| cc_115 | 0.0% | 0.9% | 0.6% | ||
| Proteobacteria | Enterobacteriaceae | - | 36.4% | 1.3% | 1.5% |
| Phylum | Family | Genus | AD | MP | E |
|---|---|---|---|---|---|
| Unassigned | 0.0% | 0.6% | 0.8% | ||
| Bacteroidetes | Bacteroidaceae | Bacteroides | 2.6% | 0.4% | 4.1% |
| Porphyromonadaceae | Parabacteroides | 1.0% | 0.5% | 1.1% | |
| Rikenellaceae | - | 2.0% | 1.1% | 3.1% | |
| Odoribacteraceae | Butyricimonas | 0.0% | 0.0% | 1.1% | |
| Cyanobacteria | 0.0% | 0.4% | 1.1% | ||
| Firmicutes | Planococcaceae | - | 0.2% | 0.5% | 0.4% |
| Enterococcaceae | - | 3.6% | 0.0% | 0.0% | |
| Enterococcus | 1.0% | 0.2% | 0.4% | ||
| Lactobacillaceae | Lactobacillus | 1.2% | 3.4% | 2.9% | |
| - | - | 0.4% | 0.6% | 0.3% | |
| - | - | 14.6% | 29.9% | 30.0% | |
| Clostridiaceae | - | 4.8% | 0.2% | 0.3% | |
| Clostridium | 2.7% | 0.4% | 0.4% | ||
| Lachnospiraceae | - | 6.5% | 10.3% | 8.6% | |
| Blauria | 0.8% | 1.8% | 1.5% | ||
| Coprococcus | 1.6% | 3.8% | 3.2% | ||
| Dorea | 0.8% | 1.3% | 0.7% | ||
| Epulopscium | 2.4% | 0.0% | 0.0% | ||
| Ruminococcus | 2.1% | 3.3% | 2.3% | ||
| Ruminococcaceae | - | 7.5% | 18.4% | 17.0% | |
| Anaerotruncus | 0.0% | 0.5% | 0.3% | ||
| Faecalibacterium | 1.5% | 1.8% | 1.5% | ||
| Oscillospira | 5.8% | 9.6% | 7.7% | ||
| Ruminococcus | 1.7% | 5.1% | 3.6% | ||
| Erysipelotrichaceae | - | 1.0% | 0.9% | 0.6% | |
| Coprobacillus | 0.4% | 0.9% | 0.5% | ||
| cc_115 | 0.0% | 0.8% | 0.6% | ||
| Proteobacteria | Enterobacteriaceae | - | 32.6% | 1.2% | 0.9% |
| Tenericutes | - | - | 0.2% | 0.4% | 0.7% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montoro-Dasi, L.; Villagra, A.; de Toro, M.; Pérez-Gracia, M.T.; Vega, S.; Marin, C. Fast and Slow-Growing Management Systems: Characterisation of Broiler Caecal Microbiota Development throughout the Growing Period. Animals 2020, 10, 1401. https://doi.org/10.3390/ani10081401
Montoro-Dasi L, Villagra A, de Toro M, Pérez-Gracia MT, Vega S, Marin C. Fast and Slow-Growing Management Systems: Characterisation of Broiler Caecal Microbiota Development throughout the Growing Period. Animals. 2020; 10(8):1401. https://doi.org/10.3390/ani10081401
Chicago/Turabian StyleMontoro-Dasi, Laura, Arantxa Villagra, María de Toro, María Teresa Pérez-Gracia, Santiago Vega, and Clara Marin. 2020. "Fast and Slow-Growing Management Systems: Characterisation of Broiler Caecal Microbiota Development throughout the Growing Period" Animals 10, no. 8: 1401. https://doi.org/10.3390/ani10081401
APA StyleMontoro-Dasi, L., Villagra, A., de Toro, M., Pérez-Gracia, M. T., Vega, S., & Marin, C. (2020). Fast and Slow-Growing Management Systems: Characterisation of Broiler Caecal Microbiota Development throughout the Growing Period. Animals, 10(8), 1401. https://doi.org/10.3390/ani10081401







