Simple Summary
Intramuscular fat (IMF), which characterizes the marbling, is a key meat quality trait in the beef industry. In this study, we validated the effect of the expression levels and single nucleotide polymorphisms (SNPs) of AKIRIN2, TTN, EDG1, and MYBPC1 genes on IMF content in Chinese Qinchuan cattle. The results showed that the expression levels of these genes could affect the IMF content, and the SNP in EGD1 could affect the IMF content in Qinchuan. The information of this study may be applied to effective marker-assisted selection, to increase the levels of marbling in Qinchuan beef production.
Abstract
Marbling is characterized by the amount and distribution of intramuscular fat (IMF). The AKIRIN2, TTN, EDG1, and MYBPC1 genes are well-known marbling-related genes, which were first identified in Japanese Black beef cattle. The objectives of this study were to analyze the correlation of the expression levels of these genes in the longissimus muscle (LM) with IMF content, and the associations between the single nucleotide polymorphisms (SNPs) in these genes and IMF content in Chinese Qinchuan cattle (n = 350). The association analyses showed that the g.42041062G>T SNP in the EDG1 gene was significantly associated with IMF content in Qinchuan (p < 0.05). Further, the expressions of the EDG1 and MYBPC1 were up-regulated (p < 0.05) in LM of Qinchuan cattle group with low IMF content. Down-regulations of the AKIRIN2 and TTN genes (p < 0.05 and p < 0.01, respectively) were observed in the Qinchuan cattle group with high IMF content. These results suggest possible effects of the expression levels of selected genes on IMF content in the LM, and the g.42041062G>T SNP in the EDG1 gene might be useful as a molecular marker for IMF content in Qinchuan.
1. Introduction
Marbling is defined by the amount and distribution of intramuscular fat (IMF) in skeletal muscles, which improves the acceptability, palatability, and tenderness of the meat [1,2,3]. With the rapid development of the economy in China, the consumers’ demands for high quality beef are also growing. Thus, it is necessary to focus on important candidate functional genes and find more effective molecular markers to increase the IMF in Chinese native cattle. Qinchuan cattle, which is one of the five most well-known native yellow cattle breeds in China, have naturally good beef quality. However, the Qinchuan breed exhibits lower marbling than Japanese Black beef (JB) cattle, which could be due to lack of selection for marbling. Therefore, better knowledge of the molecular architecture of IMF content in Chinese native cattle may lead to economic benefits to the beef industry.
The akirin 2 (AKIRIN2) gene is located within genomic regions of quantitative trait loci (QTLs) for marbling in JB, Angus and Korean native (KN) cattle [4,5,6]. The titin (TTN) gene was found in the genomic regions within QTL for marbling in JB [4]. The endothelial differentiation sphingolipid G-protein-coupled receptor 1 (EDG1) gene is located within genomic regions of QTLs for marbling in JB [4], as well as in a Belgian Blue × MARC III (1⁄₄ Angus, 1⁄₄ Hereford, 1⁄₄ Red Poll, 1⁄₄ Pinzgauer) developed half-sib family, and a Piedmontese × Angus sire developed half-sib family [7]. Moreover, the myosin binding protein C1 (MYBPC1) is included in the genomic region of QTLs for marbling in JB [8], Angus [5], and IMF in a Brangus heifer population [9]. Thus, these genes could be considered important candidate genes for IMF content in beef cattle. Furthermore, the c.*188G>A, g.231054C>T, g.1471620G>T, and g.70014208A>G single nucleotide polymorphisms (SNPs) in the AKIRIN2, TTN, EDG1, and MYBPC1 genes, respectively, showed associations of these SNPs with marbling in JB [8,10,11,12] and KN [6] cattle. However, the relationships of these candidate genes with marbling, particularly with IMF in Chinese native cattle breed, have not been investigated.
Thus, this study investigated the association of the SNPs in AKIRIN2, TTN, EDG1 and MYBPC1 genes with IMF content in Qinchuan, and evaluated the effects of expression levels of these candidate genes on IMF content in the longissimus muscle (LM) of Qinchuan.
2. Materials and Methods
2.1. DNA Samples and Phenotypes
The DNA samples and phenotypes of 350 Qinchuan adult females (aged 18 to 24 months, and unrelated for at least three generations) were provided from the National Beef Cattle Improvement Center, Northwest A & F University [13,14]. These cattle were reared on the same diets, using commercial standard procedures [14]. The measurement of IMF content was described in the previous reports [13,14]. We utilized biopsies of longissimus muscle at the 12–13th ribs from the Qinchuan adult females at 24 months. After IMF content measurement by ultrasound, the five Qinchuan cattle (low group, LG) with the lowest IMF content (average IMF content was 3.44%, ranging from 2.87 to 4.57%), and five cattle (high group, HG) with the highest IMF content (average IMF content was 8.33%, ranging from 8.26 to 8.44%) were selected for sampling. There was no genetic relationship in the ten Qinchuan cattle. Muscle biopsies were immediately dipped into liquid nitrogen and stored at −80 °C until RNA extraction. The animal care and experiments were conducted according to the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, 2004) China. The protocol was approved on 1 March 2018 by the Institutional Animal Care and Use Ethics Committee of Inner Mongolia University, with the permit number for conducting animal experiments of (IMU-2018-01).
2.2. SNP Genotyping by iPLEX MassARRAY
For the population consisting of 350 Qinchuan cattle, the four target SNPs were genotyped with the MassARRAY®® SNP genotyping system (Agena Bioscience, San Diego, CA, USA). PCR and extension primers were designed from sequences containing each target mutation and ~100 upstream and downstream bases with Assay Design Suite (http://agenabio.com/assay-design-suite-20-software), using the default settings. The genotype of each SNP was analyzed using the Sequenom MassARRAY iPLEX platform (Sequenom, San Diego, CA, USA) [15]. The resulting data were analyzed using the MassARRAY Typer 4.0 Analyzer software (Agena Bioscience, San Diego, CA, USA).
2.3. Real-Time PCR
For ten samples of the HG and LG, total RNA was isolated from 50 mg of frozen LM samples using the RNeasy Fibrous Tissue kit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer’s instructions. Total RNA was quantified by absorbance at 260 nm, and the integrity of total RNA was checked by agarose gel electrophoresis and ethidium bromide staining of the 28S and 18S bands. Total RNA (2 µg) was reverse-transcribed into cDNA, using an iScript Advanced cDNA Synthesis kit for RT-qPCR (Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer’s instructions. Real-time PCR was performed using SsoAdvanced SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). The mRNA expression levels of AKIRIN2, TTN, EDG1, and MYBPC1 genes in the LM were determined with the CFX Connect Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA), using the mRNA-specific primers, as shown in Table S1. The expression of each gene was normalized against GAPDH [16]. The conditions of the real-time PCR reactions were the same as described in our previous report [16]. The relative fold change was calculated using the 2−∆∆Ct calculation [17].
2.4. Statistical Analyses
We compared the expression levels of the four genes in the LM between HG and LG by Student’s t test. The relationship between different genotypes and IMF content of Qinchuan cattle was analyzed in SPSS 24.0 (SPSS, Inc., Chicago, IL, USA). The statistical linear model for this analysis was the same as in our previous report [14]:
where Yijk = trait value per individual, μ = overall population mean per trait, Gi = fixed effect associated with genotype, Ai = fixed effect of age (months), Ak was the fixed effect due to the age (years) of dam and eijk = standard error. When the number of cattle with a given genotype was less than ten, their associations and effects could not be reliably estimated. Therefore, animals with this genotype were excluded from the analysis. The Bonferroni correction was used to adjust p values [14]. The allelic frequency of the g.1471620G>T SNP in EDG1 was compared between Qinchuan and JB [12] breeds by a χ2 test.
Yijk = μ + Gi + Ai + Ak + eijk
3. Results and Discussion
3.1. Associations between Four SNPs in Candidate Genes and IMF Content in Qinchuan Cattle
The previous studies reported that the c.*188G>A SNP in the AKIRIN2 gene, the g.231054C>T SNP in the TTN gene, and the g.70014208A>G SNP in the MYBPC1 gene had significant effects on the marbling in the JB [8,10,11] and the KN [6] cattle. However, no significant effect of these SNPs on the IMF content was detected in Qinchuan (Table 1). The JB cattle have been subjected to strong selection for high marbling over the last 50 years [18,19]. On the contrary, there was no strong selection for high marbling trait in Qinchuan. These differences in results of association analysis also could be explained by differences in the genetic background of breed and/or methods of measurement for IMF content in these studies.

Table 1.
Association of single nucleotide polymorphisms (SNPs) in the AKIRIN2, TTN, EDG1 and MYBPC1 genes with IMF content in Qinchuan cattle.
For the g.1471620G>T SNP of the EDG1 gene, the IMF content of the TT homozygotes was significantly higher than that of the GG homozygotes (p < 0.05) (Table 1), in agreement with that reported for the same SNP by Yamada et al. [12] in JB, suggesting that the T allele of the g.1471620G>T SNP has an effect on the IMF content and could be considered as a candidate molecular marker for marbling, particularly for IMF content in Qinchuan. In addition, the frequency of IMF content-related T allele of the g.1471620G>T SNP in this experimental Qinchuan population was 0.153, which was significantly lower (p < 0.001, by a χ2 test) than that in the JB (0.585, [12]). This difference of frequency of the T allele also might be owing to the different selection intensity for high marbling trait between Qinchuan and JB.
3.2. Effects of Expression Levels of Four Genes on IMF Content in Longissimus Muscle
The expression levels of the AKIRIN2, TTN, EDG1, and MYBPC1 genes in the LM of the HG and LG are shown in Figure 1. The expression levels of the AKIRIN2 and TTN genes were significantly higher in the LG than those in the HG (p < 0.05 and p < 0.01, respectively). Meanwhile, the expression levels of the EDG1 and MYBPC1 genes in the LM were significantly higher in the HG than those in the LG (p < 0.05 and p < 0.05, respectively). Furthermore, the expression patterns of the four genes in the Qinchuan were consistent with previous ddPCR results (using two high-marbled JB and two low-marbled Holstein) [20], also consistent with a qRT-PCR result for the MYBPC1 expression level in the JB [16], and a qRT-PCR result for the TTN expression level in the KN [21]. Thus, these genes could be considered as important candidate genes for IMF content in beef cattle. The three known SNPs in the AKIRIN2, TTN, and MYBPC1 genes were not associated with IMF content in this experimental Qinchuan population. There may be breed-specific SNPs of each candidate gene that affect the IMF content in the Qinchuan.
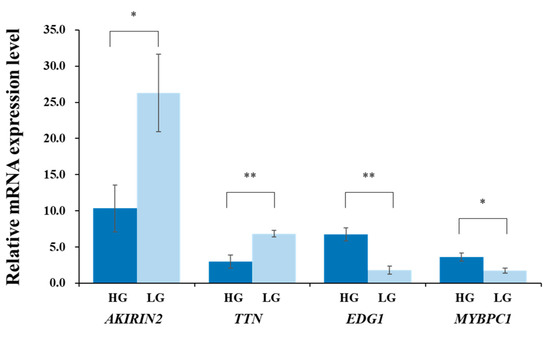
Figure 1.
Expression levels of AKIRIN2, TTN, EDG1, and MYBPC1 genes in longissimus muscle of Qinchuan cattle with high and low intramuscular fat content. High group (HG): five Qinchuan cattle with high intramuscular fat (IMF) content. Low group (LG): five Qinchuan cattle with low IMF content. Values are the means ± SE. * p < 0.05, ** p < 0.01.
On the basis of the EDG1 results from association and expression analyses, the EDG1 gene could be considered as a functional candidate gene for IMF content, and the SNP in EDG1 might be useful as a molecular marker for IMF content in Qinchuan. However, these results should be confirmed in larger cattle populations in future studies.
In addition, EDG1 is known to be involved in blood vessel formation [22]. It is likely that the high expression of EDG1 could increase the proliferation, differentiation or maturation of adipocyte-lineage cells by promoting intramuscular vascularization and then energy provision for the muscle, thereby resulting in increased IMF content. We hypothesize that the SNP has an impact on EDG1 expression and IMF content by affecting EDG1 promoter activity or mRNA stability, or both. This hypothesis requires further experimental confirmation.
4. Conclusions
In this study, we first showed that the expression levels of the AKIRIN2, TTN, EDG1, and MYBPC1 genes were associated with IMF content in Qinchuan and the T allele of the g.1471620G>T SNP in the EDG1 gene was associated with IMF content. This information may be applied to effective marker-assisted selection, to increase the marbling in Chinese Qinchuan beef cattle.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/10/8/1370/s1, Table S1: Sequences of real-time PCR primers.
Author Contributions
Conceptualization, B.T. and L.Z.; methodology, B.T., G.C., T.Y. and Y.L.; data curation, Y.L. J.L. and G.C.; writing—original draft preparation, Y.L. and G.C.; writing—review and editing, B.T. and L.Z.; project administration, B.T. and L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China (2018YFD0501700), and by the grants from the Inner Mongolia Science & Technology Plan (No. 20180260) of China, the “Grassland Meritocrat” Scholars Program (CYYC6068) of Inner Mongolia Autonomous Region, and the High-level Talents Import Program (No. 21400-5165112) of Inner Mongolia University of China, the High Level Overseas Talents Project ([2018] No.190) from Ministry of Human Resources and Social Security of China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boylston, T.D.; Morgan, S.A.; Johnson, K.A.; Busboom, J.R.; Wright, R.W.; Reeves, J.J. Lipid content and composition of wagyu and domestic breeds of beef. J. Agr. Food Chem. 1995, 43, 1202–1207. [Google Scholar] [CrossRef]
- Busboom, J.R.; Jeremiah, L.E.; Gibson, L.L.; Johnson, K.A.; Gaskins, C.T.; Reeves, J.J.; Wright, R.W. Effects of biological source on cooking and palatability attributes of beef produced for the Japanese market. Meat Sci. 1993, 35, 241–258. [Google Scholar] [CrossRef]
- Matsuishi, M.; Fujimori, M.; Okitani, K.A. Wagyu beef aroma in wagyu (Japanese black cattle) beef preferred by the Japanese over imported beef. Anim. Sci. J. 2001, 72, 498–504. [Google Scholar] [CrossRef]
- Takasuga, A.; Watanabe, T.; Mizoguchi, Y.; Hirano, T.; Ihara, N.; Takano, A.; Yokouchi, K.; Fujikawa, A.; Chiba, K.; Kobayashi, N.; et al. Identification of bovine QTL for growth and carcass traits in Japanese Black cattle by replication and identical-by-descent mapping. Mamm. Genome 2007, 18, 125–136. [Google Scholar] [CrossRef]
- McClure, M.C.; Morsci, N.S.; Schnabel, R.D.; Kim, J.W.; Yao, P.; Rolf, M.M.; McKay, S.D.; Gregg, S.J.; Chapple, R.H.; Northcutt, S.L.; et al. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim. Genet. 2010, 41, 597–607. [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.K.; Hong, M.W.; Park, S.R.; Lee, Y.S.; Kim, J.W.; Lee, H.K.; Jeong, D.K.; Song, Y.H.; Lee, S.J. Association of a single nucleotide polymorphism in the akirin 2 gene with economically important traits in Korean native cattle. Anim. Genet. 2013, 44, 750–753. [Google Scholar] [CrossRef]
- Casas, E.; Stone, R.T.; Keele, J.W.; Shackelford, S.D.; Kappes, S.M.; Koohmaraie, M. A comprehensive search for quantitative trait loci affecting growth and carcass composition of cattle segregating alternative forms of the myostatin gene. J. Anim. Sci. 2001, 79, 854–860. [Google Scholar] [CrossRef]
- Tong, B.; Sasaki, S.; Muramatsu, Y.; Ohta, T.; Kose, H.; Yamashiro, H.; Fujita, T.; Yamada, T. Association of a single-nucleotide polymorphism inmyosin-binding protein C, slow-type (MYBPC1) gene with marbling in Japanese Black beef cattle. Anim. Genet. 2014, 45, 611–612. [Google Scholar] [CrossRef]
- Peters, S.O.; Kizilkaya, K.; Garrick, D.J.; Fernando, R.L.; Reecy, J.M.; Weaber, R.L.; Silver, G.A.; Thomas, M.G. Bayesian genome-wide association analysis of growth and yearling ultrasound measures of carcass traits in Brangus heifers. J. Anim. Sci. 2012, 90, 3398–3409. [Google Scholar] [CrossRef]
- Sasaki, S.; Yamada, T.; Sukegawa, S.; Miyake, T.; Fujita, T.; Morita, M.; Ohta, T.; Takahagi, Y.; Murakami, H.; Morimatsu, F.; et al. Association of a single nucleotide polymorphism in akirin 2 gene with marbling in Japanese Black beef cattle. BMC Res. Notes 2009, 2, 131. [Google Scholar] [CrossRef]
- Yamada, T.; Sasaki, S.; Sukegawa, S.; Yoshioka, S.; Takahagi, Y.; Morita, M.; Murakami, H.; Morimatsu, F.; Fujita, T.; Miyake, T.; et al. Association of a single nucleotide polymorphism in titin gene with marbling in Japanese Black beef cattle. BMC Res. Notes 2009, 2, 78. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Sasaki, S.; Sukegawa, S.; Miyake, T.; Fujita, T.; Kose, H.; Morita, M.; Takahagi, Y.; Murakami, H.; Morimatsu, F.; et al. Novel SNP in 5′ flanking region of EDG1 associated with marbling in Japanese Black beef cattle. Anim. Sci. J. 2009, 80, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Gui, L.; Raza, S.H.A.; Zhang, S.; Khan, R.; Wang, L.; Guo, H.; Zan, L. NRF1 and ZSCAN10 bind to the promoter region of the SIX1 gene and their effects body measurements in Qinchuan cattle. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Raza, S.H.A.; Zhang, J.; Gui, L.; Rahman, S.U.; Khan, R.; Hosseini, S.M.; Kaleri, H.A.; Zan, L. Polymorphism in promoter of SIX4 gene shows association with its transcription and body measurement traits in Qinchuan cattle. Gene 2018, 656, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP genotyping using the sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2009. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Muramatsu, Y.; Ohta, T.; Kose, H.; Yamashiro, H.; Sugiyama, T.; Yamada, T. Association of the expression level of the MYBPC1 gene in skeletal muscle with marbling trait in Japanese black beef cattle. Ann. Anim. Sci. 2015, 15, 349–358. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Thomas, D.; Livak; Kenneth, J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Sasaki, Y.; Miyake, T.; Gaillard, C.; Oguni, T.; Ohtagaki, S. Comparison of genetic gains per year for carcass traits among breeding programs in the Japanese Brown and the Japanese Black cattle. J. Anim. Sci. 2006, 84, 317–323. [Google Scholar] [CrossRef]
- Tong, B.; Sasaki, S.; Muramatsu, Y.; Ohta, T.; Kose, H.; Fujita, T.; Yamada, T. The G allele at the g.70014208A > G in the MYBPC1 gene associated with high marbling in Japanese Black cattle is at a low frequency in breeds not selected for marbling. J. Genet. 2014, 93, 231–233. [Google Scholar] [CrossRef]
- Sasaki, Y.; Nagai, K.; Nagata, Y.; Doronbekov, K.; Nishimura, S.; Yoshioka, S.; Fujita, T.; Shiga, K.; Miyake, T.; Taniguchi, Y.; et al. Exploration of genes showing intramuscular fat deposition-associated expression changes in musculus longissimus muscle. Anim. Genet. 2006, 37, 40–46. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, Y.M.; Lee, S.H.; Kim, B.S.; Kim, N.K.; Choy, Y.H.; Hoon, K.; Kim, D.Y.; Im, S.K.; Oh, S.J.; et al. Identification of marbling-related candidate genes in M. Longissimus dorsi of high- and low marbled Hanwoo (Korean Native Cattle) steers. BMB Rep. 2008, 41, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wada, R.; Yamashita, T.; Mi, Y.; Deng, C.X.; Hobson, J.P.; Rosenfeldt, H.M.; Nava, V.E.; Chae, S.S.; Lee, M.J.; et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Invest. 2000, 106, 951–961. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).