Simple Summary
Dietary energy and protein play important roles in rumen fermentation. However, the impacts of dietary energy and protein on the relationships between rumen bacterial composition and ruminal metabolites were not extensively studied. In this study, we investigated the rumen fermentation status in response to different energy and protein levels, which was vital for the rumen health and resulted in the potential positive effects on the host health and production. Thereby, quantitative polymerase chain reaction analysis and 16S rRNA gene sequencing showed that the bacterial richness was significantly reduced, and the rumen bacterial composition was significantly altered with the increasing levels of dietary energy and protein. Metabolomics analysis revealed that the dominant differential metabolites were amino acids, peptides, and analogs. Moreover, high-energy and high-protein diets could enhance the ruminal antioxidative capacity by increasing the concentration of some metabolites. Correlation analysis implied that high energy and protein levels could enhance the catechol concentration by Prevotella_1 and Ruminococcus_2. This study can help to improve the dietary energy and protein use efficiency in goats.
Abstract
This study was conducted to investigate the impacts of dietary energy and protein on rumen bacterial composition and ruminal metabolites. A total of 12 ruminal samples were collected from Shaanbei white cashmere goats which were divided into two groups, including high-energy and high-protein (Group H; crude protein, CP: 9.37% in dry matter; metabolic energy, ME: 9.24 MJ/kg) and control (Group C; CP: 8.73%; ME: 8.60 MJ/kg) groups. Thereby, 16S rRNA gene sequencing and a quantitative polymerase chain reaction were performed to identify the rumen bacterial community. Metabolomics analysis was done to investigate the rumen metabolites and the related metabolic pathways in Groups C and H. The high-energy and high-protein diets increased the relative abundance of phylum Bacteroidetes and genera Prevotella_1 and Succiniclasticum, while decreasing the number of Proteobacteria (p < 0.05). The dominant differential metabolites were amino acids, peptides, and analogs. Tyrosine metabolism played an important role among the nine main metabolic pathways. Correlation analysis revealed that both Prevotella_1 (r = 0.608, p < 0.05) and Ruminococcus_2 (r = 0.613, p < 0.05) showed a positive correlation with catechol. Our findings revealed that the diets with high energy and protein levels in Group H significantly altered the composition of ruminal bacteria and metabolites, which can help to improve the dietary energy and protein use efficiency in goats.
1. Introduction
Shaanbei white cashmere goat (SWCG) is a local breed in the northern Shaanxi province of China and the total population of SWCG exceeds 10 million. SWCG is well-known for cashmere wool and meat, which are the most important economic sources of the local farmers [1,2]. Traditional grazing management is mainly dependent on natural pastures, which are limited in the extremely harsh winter. Hence, nutritional management, especially the choice of dietary nutrient density, is important to promote the growth of goats [3].
The rumen is a complex microbial ecosystem in ruminants. It can ferment feedstuffs to volatile fatty acids, microbial proteins, and vitamins, which play important roles in animal health and production [4,5]. Sometimes, it has to deal with lower quality fodder or silage [6]. Among the microbiota, bacteria are the most abundant, diverse, and metabolically active species in the rumen [7,8]. The bacterial community in the rumen are linked to various factors, such as animal diet, breed, age health and geographic region [9,10]. Diet is the major determinant of the microbial composition in the rumen [11]. For example, fibers could be degraded into glucose and xylose by Ruminococcus [12].
The functions of the rumen microbiota make ruminants highly adaptable to various diets [13]. The energy and protein in the diets are the most restrictive factors for ruminal microbial growth [14,15]. Dietary protein is utilized to synthesize microbial crude protein (MCP) for host utilization by Bacteroidetes and Prevotella [16,17]. However, protein overfeeding increases the excreted nitrogen from urine and feces, which causes environmental pollution [18] and economic losses [19]. Previous studies have reported that dietary energy can promote protein to synthesize MCP [20,21]. The effective way to improve production performance in cattle is increasing dietary energy levels under the same concentration of forage ratio [22].
Previous studies showed that the phenotypic traits of ruminants were affected by rumen microbiota, whose functions could be reflected by the ruminal metabolites [23]. However, most of the studies have only focused on the change in dietary energy [3,20] or protein [16,18], and few reports studied the effects of dietary energy and protein on the rumen bacterial composition and rumen metabolites.
In our previous study, we detected that high levels of dietary energy (metabolic energy, ME: 9.24 MJ/kg) and protein (crude protein, CP: 9.37% in dry matter (DM)) could significantly enhance the average daily weight gain (ADG), dressing percentage, and eye muscle area of SWCG [24]. The primary objective of this study was to investigate the changes in the rumen bacterial diversity by 16S rRNA gene sequencing and quantitative polymerase chain reaction (qPCR), and the metabolites and key metabolic pathways by gas chromatography tandem time-of-flight mass spectrometry (GC-TOFMS)-based metabolomics by increasing dietary energy and protein. Furthermore, we examined the relationships between ruminal bacterial communities and rumen metabolites to improve dietary energy and protein use efficiency in SWCG.
2. Materials and Methods
2.1. Ethics Statement
The use of animals and all experimental protocols (protocol number: 100403) were authorized by the Institutional Animal Care and Use Committee of Northwest A&F University (Yangling, Shaanxi, China).
2.2. Animals, Diets and Sampling
A total of 12 SWCG (age of 8 months, an average initial body weight of 24.5 ± 1.87 kg, six males and six females) were selected and fed at Diqingyuan farm (37.6° N, 108.79° E), Yulin, Shannxi Province, China. All goats were randomly allocated into two treatments based on their body weights (3 males and 3 females in each group). Each goat was housed in an individual pen, and the goats were fed individually. The diets were formulated based on the Feeding Standard of Meat-Producing Sheep and Goats (NY/T816-2004, China). Group H included high energy (metabolic energy, ME: 9.24 MJ/kg) and high protein (crude protein, CP: 9.37% in dry matter) diet. Group C included the basal diet (CP: 8.73%; ME: 8.60 MJ/kg). Rumen degradable protein (RDP) in Groups C and H was 3.62% and 4.04%, respectively. Rumen degradable starch (RDS) in Groups C and H was 11.91% and 12.54%, respectively. Also, the ratio of dietary energy to protein and the ratio of dietary forage to concentrate in the two groups were not changed (Table 1). Corn stalk and alfalfa were used to adjust the concentrate: forage ratio and dietary energy: protein ratio.

Table 1.
Composition and nutrient contents of the experimental diets (DM basis).
All animals were fed at 9:00 am and 4:00 pm daily. The experiment lasted for 65 days, including 10 days for acclimation period. Average daily feed intakes (ADFI) of per goat in Groups C and H was 1.13 kg/d and 1.14 kg/d, respectively. Hence, the ADFI of CP for each sheep in Groups C and H was 98.6 g and 106 g, respectively. After slaughtering the goats, the rumen was opened from the ventral sac. The rumen content of each goat was first homogenized by hand using sterile gloves, and then approximately 50 mL contents were collected. Rumen sample of each goat was individually filtered through four layers of sterile cheesecloth to separate rumen fluid from rumen solid phase. Then, the liquid samples were immediately frozen in liquid nitrogen and stored at −80 °C for further studies.
2.3. DNA Extraction, 16S rRNA Gene Amplicon and Sequencing
Bacterial genomic DNA was extracted from rumen fluid samples using a stool DNA kit (OMEGA Bio-Tek, Norcross, GA, USA). The extracted DNA was checked on 1% agarose gel, and DNA concentration and purity were determined with NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Total DNA from rumen content samples was used to construct 16S rRNA libraries by the NEXTflex Rapid DNA-Seq Kit (Bioo Scientific Cor. Austin, TX, USA) [25]. The universal primers [V338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and V806R (5′-GGACTACHVGGGTWTCTAAT-3′)] targeting the V3-V4 region of bacteria were used. PCR products were mixed with equimolar ratios and purified by Qiagen Gel Extraction Kit (Qiagen, Dusseldorf, Germany). Finally, Illumina MiSeq platform was used for Paired-End sequencing at Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
2.4. Sequence Processing
The raw data were merged by FLASH (version 1.2.11) [26] and filtered to obtain the clean data by the QIIME 1.9.1 [27]. Briefly, (i) 300 bp reads were truncated at any site with an average quality sore <20 over a 50 bp sliding window; (ii) both truncated reads shorter than 50 bp or containing ambiguous characters were discarded; (iii) overlapping sequences with at least a 10 bp overlap were assembled and the maximum mismatch ratio of overlap region was 0.2; (iv) samples were distinguished by the barcodes and primers. Barcode allowed mismatch was 0 and the maximum nucleotide mismatch in primer was 2. Operational taxonomic units (OTUs) were clustered with 97% similarity cut-off using UCLUST [28]. The indices of Alpha diversity were analyzed by MOTHUR (version v.1.30.2) [29]. Beta diversity was calculated by weighted UniFrac distance. Rarefaction curves, Venn diagram, bar and heatmap graphs were visualized by R software (version 3.1.2). The statistical significance of grouping was assessed by the analysis of similarity (ANOSIM) in QIIME 1.9.1 and it was determined by calculating p-value with 999 permutations [29,30]. Significant interactions between rumen bacteria were shown using Networkx. The raw reads have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (Accession number: SRP258202).
2.5. Prediction of Rumen Bacterial Function
The function of detected rumen bacteria was predicted via the phylogenetic analysis of communities by reconstruction of unobserved states (PICRUSt2) [31]. The predicted sequences were aligned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [32].
2.6. qPCR Analysis between Different Groups
Changes in phylum Bacteroidetes and genus Prevotella between different treatments were verified by absolute qPCR using iCycle thermos cycler (Bio-Read-CFX, CA, USA). The primers and annealing temperature were shown in Table S1. The external standard curves were constructed by 10-fold serial dilution of plasmid DNA containing the cloned marker loci. All standard curves met the required efficient standards (R2 > 0.99, 90% < E < 120%). The reaction mixture and conditions were according to our previous study [33].
2.7. Metabolomics Analysis by GC-TOFMS
The ruminal fluid was used for metabolomics analysis. For each sample, 200 μL rumen fluid sample, 200 μL liquid methanol and 20 μL L-2-Chlorophenylalanine (CAS#: 103616-89-3, ≥98%) (1 mg/mL in H2O) as an internal standard were sequentially added to the 1.5 mL Eppendorf (EP) tubes. The mixture was vortexed for 10 s and then centrifuged at 13,000 rpm for 15 min at 4 °C. After centrifugation, 370 μL of the supernatant was transferred to a 2 mL GC/MS glass vial and dried in a vacuum concentrator without heating. After evaporation, 80 μL of methoxy amination hydrochloride (20 mg/mL in pyridine) was added to the sample and incubated for 30 min at 80 °C. Meanwhile, 100 μL of the N,O-Bis (trimethylsilyl) trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) was added to the sample aliquots. The mixture was incubated at 70 °C for 1.5 h.
Derivatized samples were analyzed using the Agilent 7890B gas chromatograph system (Agilent, Santa Clara, CA, USA) coupled with the LECO Chroma TOF PEGASUS HT (LECO, St. Joseph County, MI, USA) [34]. Injecting 1 μL aliquot of the analyte into splitless mode, helium was used as the carrier gas. The injection, transfer line and ion source temperatures were 280, 270, and 220 °C, respectively. The mass spectrometry data were performed in full-scan mode with 50–500 m/z at 20 scans/s after 6.1 min of solvent delay.
The extraction of raw peaks, filtering and calibration of the baseline data, peak alignment, deconvolution analysis, peak identification, and peak area integration were performed by Chroma TOF 4.3X software [35]. The content of each component was calculated by the peak area normalization method. Principal component analysis (PCA) and orthogonal correction partial least squares discriminant analysis (OPLS-DA) were conducted using SIMCA software (V14.1).
2.8. Statistical Analysis
Statistical analysis was performed by Statistical Package for the Social Sciences (SPSS) version 20.0 (SPSS Inc., Chicago, IL, USA). Significant differences between Groups C and H were calculated using the Student’s t-test (for normally distributed data) and the Mann–Whitney U test (for non-normally distributed data). For the GC-TOFMS data, differential metabolites between two groups were identified combining variable importance in the projection (VIP) >1 and p < 0.05. Significant correlations between rumen bacteria and metabolite variables were assessed by Spearman correlation analysis, if the correlation coefficients (r, in absolute values) were above 0.55 [36,37]. The statistical significance was set at p < 0.05.
3. Results
3.1. Diversity, Richness and Similarity of the Ruminal Bacterial Communities
A total of 826,727 16S rRNA gene sequences were obtained from 12 different samples with 61,658 rarefied sequencing reads per sample after quality control and chimaera removal. In this study, the average sequences were 68,110 ± 4362 (minimum: 61,658; maximum: 71,473) in Group C and 69,677 ± 3532 (minimum: 67,740; maximum: 72,873) in Group H. Additionally, there was no significant difference in sequencing reads between Groups C and H. Group C exhibited the highest number of unique sequences (667 OTUs), followed by Group H (35 OTUs). Approximately 71% of the total OTUs (1704 OTUs) were shared by two groups (Figure 1A). The rarefaction curves (Figure 1B) reached the saturation plateau and the indices of Good’s coverage were above 0.99 (Supplementary Table S2), indicating that the sequencing depth was reasonable. ACE (Figure 1C) and Chao (Figure 1D) indices were significantly decreased when the goats were fed with high energy and protein diets in Group H (p < 0.05), while Shannon and Simpson indices had no significant effects (p > 0.05) (Supplementary Table S2).

Figure 1.
16S rRNA gene sequences in different dietary groups. A Venn diagram illustrating the overlap of bacterial OTUs at a 3% dissimilarity level for Groups C and H. (A). The samples of Group C included the goats fed with a typical total mixed ration (TMR), and the samples of Group H included the goats fed with the high energy and high protein diets. Rarefaction analysis of different samples (B). Differences in ACE indices (C) and Chao indices (D) between Groups C and H.
ANOSIM (p < 0.05) was performed to indicate the statistical difference between the treatments. Table 2 showed that the rumen bacterial community structures at phylum (p = 0.037), genus (p = 0.009), and OTU levels (p = 0.014) were significantly different between Groups C and H.

Table 2.
Analysis of similarities (ANOSIM) for rumen microbial composition at the phylum, genus and OTU level.
3.2. Composition and Differences of Ruminal Bacterial Communities
A total of 30 phyla were detected by taxonomic analysis. The top five prominent phyla in Groups H and C were Bacteroidetes (abundances of 63.76% and 54.46%, respectively), Firmicutes (21.20% and 19.04%), Proteobacteria (8.40% and 19.13%), Fibrobacteres (2.76% and 2.29%), and Kiritimatiellaeota (1.24% and 1.90%), which are accounted for more than 96% (Figure 2A, Supplementary Table S3). With the increase of energy and protein levels in diets, the abundance of Bacteroidetes increased significantly (p < 0.05), while the abundance of Proteobacteria significantly decreased (p < 0.05) (Figure 2B, Supplementary Table S3).
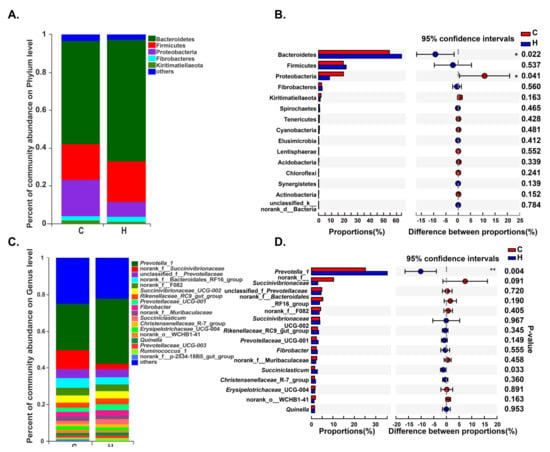
Figure 2.
Distribution of bacteria in different groups. The color-coded bar plots represent the average distribution of bacterial phyla (A) and genera (C), respectively. Only the dominant bacteria (with a relative abundance ≥1%) among rumen bacteria are shown. Extended error bar plots illustrate the mean proportions and differences in the phyla (B) or genera (D) in rumen samples. * indicates p < 0.05.
When sequences were analyzed at a lower taxonomical level, more detailed information about rumen bacteria was found. A total of 539 bacterial genera were detected. Within Group C, the most abundant sequences were those related to Prevotella_1 (abundance of 25.17%), norank_f__Succinivibrionaceae (10.35%), norank_f__Bacteroidales_RF16_group (5.33%), unclassified_f__Prevotellaceae (4.85%), norank_f__F082 (4.31%) and Succinivibrionaceae_UCG-002 (3.84%). Within Group H, the dominant taxa were associated with Prevotella_1 (35.36%), unclassified_f__Prevotellaceae (4.53%) Succinivibrionaceae_UCG-002 (3.94%), norank_f__Bacteroidales_RF16_group (3.79%), norank_f__F082 (3.64%), and Rikenellaceae_RC9_gut_group (3.36%) (Figure 2C, Supplementary Table S4). In addition, the relative abundances of genera Prevotella_1 and Succiniclasticum were significantly increased when energy and protein levels in diets were increased (p < 0.5) (Figure 2D, Supplementary Table S4).
3.3. qPCR Analysis
According to 16S rRNA gene sequencing data, the differences in the number of Bacteroidetes (phylum level) and Prevotella (genus level) between Groups C and H were further verified by absolute qPCR. As shown in Table 3, the number of Prevotella and Bacteroidetes in the rumen of Group H was significantly increased (p < 0.05) compared with Group C.

Table 3.
Influence of different nutrient density in the diets on the number of bacteria a.
3.4. Functional Predictions of Rumen Bacteria
The potential functions of the bacterial community in the rumen of SWCG were predicted by the PICRUSt2 based on 16S rRNA gene sequencing data. At KEGG level 1, metabolism-related pathways had the highest abundance (>50%). Compared with Group C, the rumen bacteria of Group H was predicted to have significantly higher capability of influencing genetic information processing and lower capability of influencing environmental information processing and human disease (p < 0.05) (Supplementary Table S5). At KEGG level 2, the highest relative abundance was carbohydrate metabolism. In addition, the abundances of genes belonged to carbohydrate metabolism, energy metabolism, nucleotide metabolism, glycan biosynthesis and metabolism, biosynthesis of other secondary metabolites, translation, and replication and repair were significantly higher in Group H than those in Group C. The abundances of genes involved in lipid metabolism, membrane transport, and signal transduction were significantly higher in Group C compared with Group H (Figure 3, Supplementary Table S6).

Figure 3.
Differences in bacterial metabolism function at KEGG level 2 between Groups C and H using PICRUSt2.
3.5. Metabolic Phenotype Profile of Rumen
A total of 607 valid peaks were integrated after GC-TOFMS analysis of rumen contents and the chromatograms were shown in Supplementary Figure S1A. Among these peaks, 264 metabolites were identified according to their major chemical classes, including amino acids, peptides, and analogues, fatty acids and conjugates; dicarboxylic acids and derivatives, and phenols and derivatives (Table 4, Supplementary Table S7). A global overview of the significant differences among the metabolites was first examined by PCA (Supplementary Figure S1B). These differences were further verified by the score plots of OPLS-DA (Figure 4A). The values of R2Y (0.94) and Q2intercept (−0.37) indicated the robustness of the OPLS-DA models (Figure 4B).

Table 4.
Significant differential metabolites between Groups C and H (VIP > 1.0; p < 0.05).
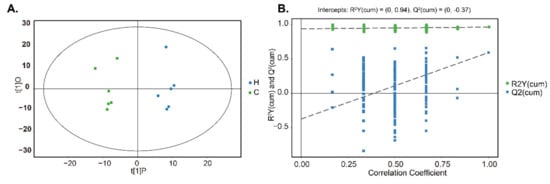
Figure 4.
Orthogonal partial least squares discriminant analysis [(O)PLS-DA] plots of rumen metabolites between Groups C and H. Score scatter plot of OPLS-DA model for Group H versus C (A). Permutation test of OPLS-DA model for Group H versus C (B).
3.6. Differences in the Ruminal Metabolites between Groups C and H
Twenty-four differential metabolites between Groups C and H were identified using VIP analysis and Student’s t-test (VIP > 1, p < 0.05) (Table 3). The main differences in ruminal metabolites between Groups C and H were the variation of amino acids, peptides, and analogs; pyridine; fatty acids and conjugates; lipids and lipid-like molecules; sugars; sugar acids and derivatives; amines. Among the metabolites, the levels of uracil, itaconic acid, 5-methoxyindole-3-acetic acid, methyl trans-cinnamate, spermidine, and catechol were higher in Group H compared with Group C. The concentrations of the remaining sixteen metabolites decreased significantly with the increase of dietary energy and protein levels.
3.7. Metabolic Pathways of Differential Metabolites
Pathway analysis is visualized in Figure 5. The varied rumen microbial metabolites between Groups C and H were identified to be mainly involved in the nine main metabolic pathways, including beta-alanine metabolism; tyrosine metabolism; pantothenate and CoA biosynthesis; sphingolipid metabolism; glutathione metabolism; glycerophospholipid metabolism; pyrimidine metabolism; tryptophan metabolism; arginine and proline metabolism. These pathways are mainly involved in amino acids metabolism, lipid metabolism, and nucleotide metabolism. Additionally, among these metabolic pathways, tyrosine metabolism had the largest impact.

Figure 5.
Metabolome view map of the deferential metabolites (VIP > 1, p < 0.05) identified in the rumen from the goats fed with the diets with different energy and protein levels. The large size indicates high pathway enrichment, and dark color indicates high pathway impact values.
3.8. Correlation Analysis between Rumen Bacteria and Rumen Metabolites
Based on Spearman correlation analysis (|r| > 0.55 and p < 0.05), we constructed the correlation networks between the bacterial genera in Groups C and H, respectively. As shown in Figure S2A and Figure S2B, 171 and 79 edges were observed in Groups C and H, respectively, which indicated that the relationships between the bacterial genera in Group C were more complex than those in Group H. The comprehensive relationships between ruminal bacterial genera were observed in this study (Table S8). Of these, Prevotella_1 was positively correlated with Succiniclasticum (r = 0.580, p < 0.05) and Ruminococcus_2 (r = 0.651, p < 0.05). Selenomonas_1 was positively correlated with Prevotellaceae_UCG-004 (r = 0.78, p < 0.01)
We determined the relationships between the differential metabolites and the top 50 bacterial communities at the genus level (Figure 6 and Supplementary Table S9). Prevotella_1 was positively correlated with 5-methoxyindole-3-acetic acid (r = 0.601, p < 0.05) and catechol (r = 0.608, p < 0.05), but negatively correlated with aconitic acid (r = −0.594, p < 0.05), 4-hydroxyphenylacetic acid (r = −0.643, p < 0.05) and phosphate (r = −0.720, p < 0.01). Succiniclasticum was negatively correlated with phosphate (r = −0.62, p < 0.05) and 2,8-dihydroxyquinoline (r = −0.65, p < 0.05). Ruminococcus_2 was positively correlated with uracil (r = 0.578, p < 0.05), catechol (r = 0.613, p < 0.05) and itaconic acid (r = 0.578, p < 0.05), while negatively correlated with 4-hydroxyphenylacetic acid (r = −0.75, p < 0.01). In addition, 5-oxoproline had high positive correlation with Lachnospiraceae_ND3007_group (r = 0.608, p < 0.05). Also, spermidine was positively correlated with Selenomonas_1 (r = 0.678, p < 0.05), Ruminococcaceae_NK4A214_group (r = 0.629, p < 0.05), Lachnospiraceae_NK3A20_group (r = 0.722, p < 0.01), Prevotellaceae_UCG−004 (r = 0.615, p < 0.05) and Prevotellaceae_NK3B31_group (r = 0.615, p < 0.05), while was negatively correlated with norank_Gastranaerophilales(r = −0.657, p < 0.05), norank_Clostridiales_vadinBB60_group (r = −0.601, p < 0.05), norank_WCHB1-41 (r = −0.706, p < 0.05) and Ruminococcaceae_UCG−002 (r = −0.650, p < 0.05). Both Butyrivibrio_2 and norank_Lachnospiraceae were negatively correlated with 5-methoxyindole-3-acetic acid (r = −0.606, p < 0.05; r = −0.685, p < 0.05), respectively.
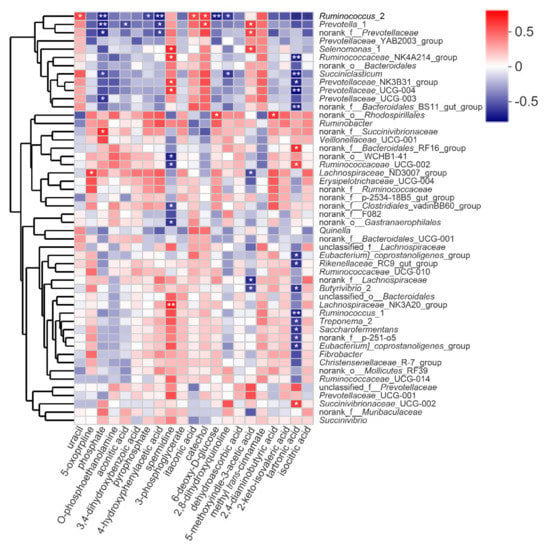
Figure 6.
Correlation between bacterial genera and deferential metabolites affected by diets with different energy and protein levels. Correlations are indicated by red and blue colors; 1 indicates a perfectly positive correlation (dark red) and close to −1 indicates a perfectly negative correlation (dark blue). Weak correlation is indicated by white colors. * and ** indicate p < 0.05 and p ≤ 0.01, respectively.
4. Discussion
4.1. Comparison of the Composition and Differences of Ruminal Bacterial Communities
Diets with high energy and protein levels in Group H effectively promoted the growth performance and carcass characteristics of goats [24]. Meanwhile, the digestion and absorption of these diets were closely related to the rumen bacteria [5]. A previous study reported that the changes in the ruminal microbiota could promote the ADG of goats [22]. Therefore, we determined the differences of rumen bacterial communities of SWCG with simultaneous changes in dietary energy and protein levels in this study.
In this study, 16S rRNA gene sequencing was used to assess the rumen bacterial community in SWCG. Liu et al. [38] and Tapio et al. [39] reported that the richness of the bacterial community was influenced by diets. Our results also revealed low bacterial richness (ACE and Chao indices) with the increasing levels of dietary energy and protein, while no significant changes were detected in the bacterial diversity (Shannon and Simpson indices). In line with the previous studies [40,41], this study revealed that Bacteroidetes, Firmicutes, and Proteobacteria were the most dominant phyla in the two groups. These bacterial phyla were the core microbiota in the rumen and their structural compositions were unchanged regardless of feeding different types of diets [42]. Among the thirty phyla, the abundance of Bacteroidetes increased significantly in Group H, which might be related to the protein degradation function of this phylum [43] and the high level of RDP in Group H.
The effects of dietary nutrient density on the bacterial population at the genus level were also detected in this study. Similar to the results of decreased bacterial richness in Group H, the high energy and protein levels of diets reduced the complexity of rumen bacterial interactions. Among the genera, Prevotella_1, belonging to the Bacteroidetes, was the most abundant bacteria in both the rumens of goats fed with different diets, which was consistent with the previous reports [3,22]. In addition, the increased population of Prevotella_1 in Group H, might be associated with the starch and protein degrading function of this genus [3,17] and the higher RDS and RDP levels in Group H. This result was also in line with the study of Wang, et.al. [3] who reported that the number of Prevotella_1 was increased when the animals fed with the high protein diet. When ruminants were fed with the high-energy diets rich in starch, the increasing number of Succiniclasticum stabilized the rumen environment by degrading succinate to propionate [44], which might be the reason for the higher number of Succiniclasticum in Group H with higher RDS levels. Bacteroidetes and Prevotella were selected in this study to verify the differences between two groups by absolute qPCR and the results agreed with those by 16S rRNA gene amplicon sequencing. Furthermore, the previous studies have reported that the level of dietary protein was positively correlated with the relative abundance of Prevotella [45,46] and the increasing levels of protein could promote the growth of cellulolytic bacteria [18]. Hence, the cellulolytic bacteria-Succiniclasticum and Ruminococcus_2 [12] were positively associated with Prevotella_1 in our study. The differences in rumen bacteria in this study suggested that high energy and protein levels in the diets might increase the number of protein-degrading bacteria.
4.2. Functional Prediction of the Ruminal Bacteria in SWCG
Whether the changes in the bacterial community structures would lead to functional differences were detected by PICRUST2. Consistent with Miao et al. [47] and He et al. [48], we found that the abundances of metabolism were the highest in the rumen at KEGG level 1. The different abundances of environmental information processing and genetic information processing between two treatments suggested that varying levels of energy and protein in the diets could influence the biochemical processes of ruminal bacteria in goats at molecular and cellular levels [49]. Liu et al. [50] also reported that the KEGG pathways involved in carbohydrate metabolism were highly enriched in the microbiota of individuals fed with high energy diets, which was consistent with our study. Furthermore, the increasing number of Prevotella_1 in Group H of this study and the involvement of the genus in energy metabolism, nucleic acid metabolism and glycan biosynthesis and metabolism [46] might indicate the increase of the above pathways in Group H. Although the PICRUSt2 approach was utilized to predict the rumen bacterial functions, this method did not accurately detect the related functions due to the limited number of sequencing studies in ruminants [51].
4.3. Comparison of the Composition and Differences of Ruminal Metabolites
Microbiota interacts with numerous physiological functions in the host through its metabolic products [52]. Thus, we used the GC-TOFMS analysis to explore the metabolic functions of ruminal microbiota. The main metabolites were amino acids, peptides, and analogues in this study, which was consistent with the previous reports [38,53]. Amino acids in the rumen are the key precursors for protein and polypeptides synthesis and are mainly obtained from the dietary proteins and microproteins [54].
According to the OPLS-DA results, a clear difference of ruminal metabolites was demonstrated between Groups C and H. These results confirmed that ruminal metabolites were closely related to the composition of diets [55].
Uracil concentration in the rumen was increased with the increase density of starch in diets [56]. Similar to our study, the higher level of RDS in Group H promoted the level of uracil. 5-oxoproline (pyroglutamic acid) could be the intermediate product in the glutathione cycle and its concertation was negatively correlated with the concentration of antioxidant-glutathione [57,58,59]. Based on the correlation analysis, Lachnospiraceae_ND3007_group might decrease the concentration of 5-oxoproline and more glutathione was produced in Group H to promote antioxidative capacity. Microbiota degrade dietary protein to tryptophan, which can be later converted into melatonin [60]. Melatonin (N-acetyl-5-methoxytryptamine) as an effective antioxidant [61] could be ultimately oxidized to 5-methoxyindole-3-acetic acid [62]. Hence, the level of 5-methoxyindole-3-acetic acid in Group H with higher RDP density was significantly higher than that in Group C. In this study, catechol as an antioxidant [63,64] was positively related to the relative abundances of Ruminococcus_2 and Prevotella_1, which might imply that high energy and protein levels in Group H could enhance the catechol concentration by these two genera. Xue et al. [65] reported that the content of spermidine increased in the group of high concentrate diets (rich in starch), which is in line with the results of our study. Furthermore, correlation analysis in this study revealed that spermidine had high positive relationships with Selenomonas_1, Ruminococcaceae_NK4A214_group, Lachnospiraceae_NK3A20_group, Prevotellaceae_UCG-004, and Prevotellaceae_NK3B31_group. Spermidine is an organic compound widely used as an antioxidant [66]. The upregulation of spermidine observed in Group H of this study might enhance the antioxidative capacity in the rumen of goats by the above genera. These data implied that diets with high energy and protein levels could improve the ruminal antioxidative capacity. However, the potential mechanisms involved in the ruminal antioxidative capacity and the interactions between ruminal bacteria and metabolites are needed to be studied in the future research.
Based on the metabolomic analysis, we found that significantly different metabolites were involved in lipid metabolism and nucleotide metabolism. This result was also identified by PICRUSt2 analysis. Tyrosine metabolism played an important role among those nine main metabolic pathways in this study, which was also detected by Ferguson et al. [67]. Furthermore, the enriched abundances of beta-alanine, arginine, and proline metabolism in this study were related to their functions. Beta-alanine could be metabolized into acetic acid and its concentration is positively associated with the amount of starch and readily available carbohydrate [38]. Arginine and proline involved in RNA synthesis and protein glycosylation are necessary for cellular function [66]. Additionally, we observed that the changes in the concentration of differential metabolites were correlated with pyrimidine metabolism, which was associated with the dietary protein. Dietary nitrogen from protein could be degraded and reused by the microbiota in order to synthesize microbial nucleic acids [38,68].
5. Conclusions
In this study, 16S rRNA gene sequencing and GC-TOFMS-based metabolomics were used to investigate the changes in rumen bacteria and metabolites in response to the diets with simultaneous changes of dietary energy and protein levels in SWCG. We observed that the bacterial richness was significantly reduced and the rumen bacterial composition was significantly altered with the increasing levels of dietary energy and protein. Metabolomics analysis revealed that the dominant differential metabolites were amino acids, peptides and analogs. Of these, the number of Prevotella_1 was significantly increased in the high energy and protein dietary and this genus could promote the catechol synthesis. Moreover, some metabolites could enhance the ruminal antioxidative capacity in Group H, which might modulate the antioxidant activity in the host. In this study, combining with the above results and the previous results of growth performance and carcass characteristics, high energy (ME, 9.24 MJ/kg) and protein (CP, 9.37%) density in diets might be the optimal dietary composition for goats. Moreover, the rumen bacterial community in the solid and epimural fractions of rumen needs to be further studied, which are necessary to gain a more complete understanding of the complexity of the rumen ecosystem [69].
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/10/7/1193/s1 Table S1. The PCR primers used in this study. Table S2. Number of diversity estimates based on the 16S rRNA gene libraries from the sequencing analysis. Table S3. The phyla of the rumen bacteria in goats. Table S4. Distribution of genera in different groups. Table S5. Predicted functions at level 1 of the rumen bacterial microbiota. Table S6. Predicted functions at level 2 of the rumen bacterial microbiota. Table S7. The mean of relative quantitative values of differential metabolites (VIP > 1.0; p < 0.05). Table S8. Sheet 1: The correlation between bacterial genera; sheet 2: The p-value between bacterial genera. Table S9. Sheet 1: The correlation between bacterial genera and differential metabolites; sheet 2: The p-value among bacterial genera and differential metabolites. Figure S1. Metabolic phenotype profile of rumen. (A) GC-TOF/MS total ion current chromatograms of rumen contents from Groups C and H; (B) PCA plot of ruminal metabolites of Groups C and H. Figure S2. Correlation analysis of the top 50 bacterial genera in Group C (A.) and H (B.), respectively. Nodes represent bacterial genera, and edges represent significant interactions among nodes (the absolute Spearman coefficients were above 0.55). The node color corresponds to the phylum taxonomic classification. The edge color represents positive (red) and negative (green) correlations.
Author Contributions
Conceptualization, Y.Y. and Y.C.; methodology, Y.W.; software, P.T. and J.L.; formal analysis, Y.W., P.T., and Y.X.; writing—original draft preparation, Y.W.; writing—review and editing, Y.Y.; supervision, Y.Y.; project administration, Y.C.; funding acquisition, Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agriculture Research System of China, grant number CARS-39-12 and National Key Research and Development Program, grant number 2018YFD0501905.
Acknowledgments
We would like to thank Wang Guojun for helping us to feed the animals and Diqingyuan farm for providing experimental site.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Han, X.; Li, B.; Wang, X.; Chen, Y.; Yang, Y. Effect of dietary concentrate to forage ratios on ruminal bacterial and anaerobic fungal populations of cashmere goats. Anaerobe 2019, 59, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ge, W.; Luo, Z.; Guo, Y.; Jiao, B.; Qu, L.; Zhang, Z.; Wang, X. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus). BMC Genom. 2017, 18, 1–13. [Google Scholar] [CrossRef]
- Wang, H.; He, Y.; Li, H.; Wu, F.; Qiu, Q.; Niu, W.; Gao, Z.; Su, H.; Cao, B. Rumen fermentation, intramuscular fat fatty acid profiles and related rumen bacterial populations of Holstein bulls fed diets with different energy levels. Appl. Microbiol. Biot. 2019, 103, 4931–4942. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, P.K.; Salem, A.Z.M.; Jena, R.; Kumar, S.; Singh, R.; Puniya, A.K. Rumen Microbiology: An Overview. In Rumen Microbiology: From Evolution to Revolution; Puniya, A.K., Singh, R., Kamra, D.N., Eds.; Springer: New Delhi, India, 2015; pp. 3–16. [Google Scholar]
- Fijałkowska, M.; Przemieniecki, S.W.; Purwin, C.; Lipiński, K.; Kurowski, T.P.; Karwowska, A. The effect of an additive containing three Lactobacillus species on the fermentation pattern and microbiological status of silage. J. Sci. Food Agric. 2019, 100, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Pitta, D.W.; Indugu, N.; Kumar, S.; Vecchiarelli, B.; Sinha, R.; Baker, L.D.; Bhukya, B.; Ferguson, J.D. Metagenomic assessment of the functional potential of the rumen microbiome in Holstein dairy cows. Anaerobe 2016, 38, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Deusch, S.; Camarinha-Silva, A.; Conrad, J.; Beifuss, U.; Rodehutscord, M.; Seifert, J. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Prajapati, V.S.; Purohit, H.J.; Raje, D.V.; Parmar, N.; Patel, A.B.; Jones, O.A.H.; Joshi, C.G. The effect of a high-roughage diet on the metabolism of aromatic compounds by rumen microbes: A metagenomic study using Mehsani buffalo (Bubalus bubalis). Appl. Microbiol. Biot. 2016, 100, 1319–1331. [Google Scholar] [CrossRef]
- Li, H.; Zhou, R.; Zhu, J.; Huang, X.; Qu, J. Environmental filtering increases with elevation for the assembly of gut microbiota in wild pikas. Microb. Biotechnol. 2019, 12, 976–992. [Google Scholar] [CrossRef]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef]
- Wang, L.; Liu, K.; Wang, Z.; Bai, X.; Peng, Q.; Jin, L. Bacterial community diversity associated with different utilization efficiencies of nitrogen in the gastrointestinal tract of goats. Front. Microbiol. 2019, 10, 239. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Kelly, W.J.; Janssen, P.H.; Attwood, G.T. Rumen microbial (Meta) genomics and its application to ruminant production. Animal 2013, 7, 184–201. [Google Scholar] [CrossRef]
- Lv, X.; Cui, K.; Qi, M.; Wang, S.; Diao, Q.; Zhang, N. Ruminal microbiota and fermentation in response to dietary protein and energy levels in weaned lambs. Animals 2020, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.H.; Davis, C.L. Some aspects of feeding high producing dairy cows. J. Dairy Sci. 1980, 63, 873–885. [Google Scholar] [CrossRef]
- He, Y.; Yu, Z.; Qiu, Q.; Shao, T.; Niu, W.; Xia, C.; Wang, H.; Su, H.; Cao, B. Effects of dietary protein levels and calcium salts of long-chain fatty acids on nitrogen mobilization, rumen microbiota and plasma fatty acid composition in Holstein bulls. Anim. Feed Sci. Technol. 2018, 246, 1–10. [Google Scholar] [CrossRef]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, C.; Li, H.Q.; Guo, G.; Huo, W.J.; Pei, C.X.; Zhang, S.L.; Wang, H. Effects of dietary protein levels and rumen-protected pantothenate on ruminal fermentation, microbial enzyme activity and bacteria population in Blonde d’Aquitaine× Simmental beef steers. Anim. Feed Sci. Technol. 2017, 232, 31–39. [Google Scholar] [CrossRef]
- Broderick, G.A. Effects of varying dietary protein and energy levels on the production of lactating dairy cows. J. Dairy Sci. 2003, 86, 1370–1381. [Google Scholar] [CrossRef]
- Lu, Z.; Xu, Z.; Shen, Z.; Tian, Y.; Shen, H. Dietary energy level promotes rumen microbial protein synthesis by improving the energy productivity of the ruminal microbiome. Front. Microbiol. 2019, 10, 847. [Google Scholar] [CrossRef]
- Cutrim, D.O.; Alves, K.S.; Dos Santos, R.D.C.; Da Mata, V.J.V.; Oliveira, L.R.S.; Gomes, D.Í.; Mezzomo, R. Body composition and energy and protein nutritional requirements for weight gain in Santa Ines crossbred sheep. Trop. Anim. Health Prod. 2016, 48, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Wang, Y.; Wang, G.; Chen, Y.; Yang, Y. Effects of dietary energy and protein levels on growth performance, serum biochemical indexes, slaughter performance and meat quality of white cashmere goats in Shaanbei white cashmere goats. Chin. J. Anim. Nutr. 2018, 30, 2194–2201. [Google Scholar] [CrossRef]
- Palomo, A.; Jane Fowler, S.; Gülay, A.; Rasmussen, S.; Sicheritz-Ponten, T.; Smets, B.F. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 2016, 10, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, W.; Bi, Y.; Tu, Y.; Beckers, Y.; Du, H.; Diao, Q. Early feeding regime of waste milk, milk, and milk replacer for calves has different effects on rumen fermentation and the bacterial community. Animals 2019, 9, 443. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019, 10, 672295. [Google Scholar]
- Fu, L.; Jiang, B.; Liu, J.; Zhao, X.; Liu, Q.; Hu, X. Genome sequence analysis of a flocculant-producing bacterium, Paenibacillus shenyangensis. Biotechnol. Lett. 2016, 38, 447–453. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, P.; Wang, L.; Zhao, Z.; Chen, Y.; Yang, Y. Bacterial community diversity associated with different levels of dietary nutrition in the rumen of sheep. Appl. Microbiol. Biotechnol. 2017, 101, 3717–3728. [Google Scholar] [CrossRef]
- Li, Q.; Kang, J.; Ma, Z.; Li, X.; Liu, L.; Hu, X. Microbial succession and metabolite changes during traditional serofluid dish fermentation. LWT-Food Sci. Technol. 2017, 84, 771–779. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Wang, Y.; Zhao, Y.; Nan, X.; Hua, D.; Sun, F.; Yang, L.; Jiang, L.; Xiong, B. Ruminal methanogenic responses to the thiamine supplementation in high-concentrate diets. Animals 2020, 10, 935. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Wang, Y.; Li, S.; Cao, Z.; Ji, S.; He, Y.; Zhang, H. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front. Microbiol. 2017, 8, 2206. [Google Scholar] [CrossRef]
- Liu, C.; Wu, H.; Liu, S.; Chai, S.; Meng, Q.; Zhou, Z. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Tapio, I.; Fischer, D.; Blasco, L.; Tapio, M.; Wallace, R.J.; Bayat, A.R.; Ventto, L.; Kahala, M.; Negussie, E.; Shingfield, K.J. Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. PLoS ONE 2017, 12, e0180260. [Google Scholar] [CrossRef]
- Zeng, H.; Guo, C.; Sun, D.; Seddik, H.; Mao, S. The ruminal microbiome and metabolome alterations associated with diet-induced milk fat depression in dairy cows. Metabolites 2019, 9, 154. [Google Scholar] [CrossRef]
- Zhang, R.; Ye, H.; Liu, J.; Mao, S. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 2017, 101, 6981–6992. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Abecia, L.; Angarita, E.; Aravena, P.; Nora Arenas, G.; Ariza, C.; et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- Solden, L.M.; Hoyt, D.W.; Collins, W.B.; Plank, J.E.; Daly, R.A.; Hildebrand, E.; Beavers, T.J.; Wolfe, R.; Nicora, C.D.; Purvine, S.O. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 2017, 11, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Xue, F.; Nan, X.; Tang, Z.; Wang, K.; Beckers, Y.; Jiang, L.; Xiong, B. Illumina Sequencing approach to characterize thiamine metabolism related bacteria and the impacts of thiamine supplementation on ruminal microbiota in dairy cows fed high-grain diets. Front. Microbiol. 2017, 8, 1818. [Google Scholar] [CrossRef]
- Du, C.; Nan, X.; Wang, K.; Zhao, Y.; Xiong, B. Evaluation of the digestibility of steam-exploded wheat straw by ruminal fermentation, sugar yield and microbial structure in vitro. RSC Adv. 2019, 9, 41775–41782. [Google Scholar] [CrossRef]
- Wang, L.; Jin, L.; Xue, B.; Wang, Z.; Peng, Q. Characterizing the bacterial community across the gastrointestinal tract of goats: Composition and potential function. MicrobiologyOpen 2019, 8, e820. [Google Scholar] [CrossRef]
- Miao, L.; Wang, P.; Hou, J.; Yao, Y.; Liu, Z.; Liu, S. Low concentrations of copper oxide nanoparticles alter microbial community structure and function of sediment biofilms. Sci. Total Environ. 2019, 653, 705–713. [Google Scholar] [CrossRef]
- He, J.; Yi, L.; Hai, L.; Ming, L.; Gao, W.; Ji, R. Characterizing the bacterial microbiota in different gastrointestinal tract segments of the Bactrian camel. Sci. Rep. 2018, 8, 654. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Kholodkevich, S.; Sharov, A.; Chen, C.; Feng, Y.; Ren, N.; Sun, K. Effects of cadmium on intestinal histology and microbiota in freshwater crayfish (Procambarus clarkii). Chemosphere 2020, 242, 125105. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X.; et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017, 23, 859–868. [Google Scholar] [CrossRef]
- Meale, S.J.; Li, S.C.; Azevedo, P.; Derakhshani, H.; DeVries, T.J.; Plaizier, J.C.; Steele, M.A.; Khafipour, E. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci. Rep. 2017, 7, 198. [Google Scholar] [CrossRef]
- Ren, W.; Wang, P.; Yan, J.; Liu, G.; Yin, Y. Melatonin alleviates weanling stress in mice: Involvement of intestinal microbiota. J. Pineal Res. 2017, 64, e12448. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, P.; Hidalgo, A.I.; Manosalva, C.; Cristi, R.; Teuber, S.; Hidalgo, M.A.; Burgos, R.A. Metabolic disturbances in synovial fluid are involved in the onset of synovitis in heifers with acute ruminal acidosis. Sci. Rep. 2019, 9, 5452. [Google Scholar] [CrossRef] [PubMed]
- Mariz, L.D.S.; Amaral, P.M.; Valadares Filho, S.C.; Santos, S.A.; Detmann, E.; Marcondes, M.I.; Pereira, J.M.V.; Silva Júnior, J.M.; Prados, L.F.; Faciola, A.P. Dietary protein reduction on microbial protein, amino acid digestibility, and body retention in beef cattle: 2. Amino acid intestinal absorption and their efficiency for whole-body deposition. J. Anim. Sci. 2018, 96, 670–683. [Google Scholar] [CrossRef]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef]
- Ametaj, B.N.; Zebeli, Q.; Saleem, F.; Psychogios, N.; Lewis, M.J.; Dunn, S.M.; Xia, J.; Wishart, D.S. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 2010, 6, 583–594. [Google Scholar] [CrossRef]
- Matsui, R.; Ferran, B.; Oh, A.; Croteau, D.; Shao, D.; Han, J.; Pimentel, D.R.; Bachschmid, M.M. Redox regulation via glutaredoxin-1 and protein S-glutathionylation. Antioxid. Redox Sign. 2020, 32, 677–700. [Google Scholar] [CrossRef]
- Zhang, R.Y.; Liu, Y.J.; Yin, Y.Y.; Jin, W.; Mao, S.Y.; Liu, J.H. Response of rumen microbiota, and metabolic profiles of rumen fluid, liver and serum of goats to high-grain diets. Animal 2019, 13, 1855–1864. [Google Scholar] [CrossRef]
- Saoi, M.; Li, A.; McGlory, C.; Stokes, T.; von Allmen, M.T.; Phillips, S.M.; Britz-McKibbin, P. Metabolic perturbations from step reduction in older persons at risk for sarcopenia: Plasma biomarkers of abrupt changes in physical activity. Metabolites 2019, 9, 134. [Google Scholar] [CrossRef]
- Tan, D.; Zheng, X.; Kong, J.; Manchester, L.C.; Hardeland, R.; Kim, S.J.; Xu, X.; Reiter, R.J. Fundamental issues related to the origin of melatonin and melatonin isomers during evolution: Relation to their biological functions. Int. J. Mol. Sci. 2014, 15, 15858–15890. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, Z.; Fang, Y.; Zheng, J.; Xu, S.; Xu, W.; Shi, L.; Mei, S.; Wu, H.; Liang, F.; et al. Melatonin suppresses microglial necroptosis by regulating deubiquitinating enzyme A20 after intracerebral hemorrhage. Front. Immunol. 2019, 10, 1360. [Google Scholar] [CrossRef]
- Lerner, A.B.; Case, J.D.; Takahashi, Y. Isolation of melatonin and 5-methoxyindole-3-acetic acid from bovine pineal glands. J. Biol. Chem. 1960, 235, 1992–1997. [Google Scholar] [PubMed]
- Morawski, F.D.M.; Deon, M.; Nicolodi, S.; de Menezes, E.W.; Costa, T.M.H.; Dias, S.L.P.; Benvenutti, E.V.; Arenas, L.T. Magnetic silica/titania xerogel applied as electrochemical biosensor for catechol and catecholamines. Electrochim. Acta 2018, 264, 319–328. [Google Scholar] [CrossRef]
- Nikolaevskaya, E.N.; Kansuzyan, A.V.; Filonova, G.E.; Zelenova, V.A.; Pechennikov, V.M.; Krylova, I.V.; Egorov, M.P.; Jouikov, V.V.; Syroeshkin, M.A. Germanium dioxide and the antioxidant properties of catechols. Eur. J. Inorg. Chem. 2019, 2019, 676–681. [Google Scholar] [CrossRef]
- Xue, F.; Pan, X.; Jiang, L.; Guo, Y.; Xiong, B. GC-MS analysis of the ruminal metabolome response to thiamine supplementation during high grain feeding in dairy cows. Metabolomics 2018, 14, 67. [Google Scholar] [CrossRef]
- Elolimy, A.; Alharthi, A.; Zeineldin, M.; Parys, C.; Loor, J.J. Residual feed intake divergence during the preweaning period is associated with unique hindgut microbiome and metabolome profiles in neonatal Holstein heifer calves. J. Anim. Sci. Biotechnol. 2020, 11, 13. [Google Scholar] [CrossRef]
- Ferguson, A.A.; Roy, S.; Kormanik, K.N.; Kim, Y.; Dumas, K.J.; Ritov, V.B.; Matern, D.; Hu, P.J.; Fisher, A.L. TATN-1 mutations reveal a novel role for tyrosine as a metabolic signal that influences developmental decisions and longevity in Caenorhabditis elegans. PLoS Genet. 2013, 9, e1004020. [Google Scholar] [CrossRef] [PubMed]
- Stentoft, C.; Rojen, B.A.; Jensen, S.K.; Kristensen, N.B.; Vestergaard, M.; Larsen, M. Absorption and intermediary metabolism of purines and pyrimidines in lactating dairy cows. Br. J. Nutr. 2015, 113, 560–573. [Google Scholar] [CrossRef]
- De Mulder, T.; Goossens, K.; Peiren, N.; Vandaele, L.; Haegeman, A.; De Tender, C.; Ruttink, T.; de Wiele, T.V.; De Campeneere, S. Exploring the methanogen and bacterial communities of rumen environments: Solid adherent, fluid and epimural. Fems Microbiol. Ecol. 2016, w251. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).