Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Procedures
2.2. Isolation and Serotyping of Salmonella
2.3. Antimicrobial Susceptibility Testing
2.4. Detection of Antimicrobial Resistance and Virulence Genes
2.5. Gel Documentation
2.6. Measurement of Biofilm Formation
2.7. Statistical Analysis
3. Results
3.1. Percentage of S. Infantis Isolated from Broiler Meat
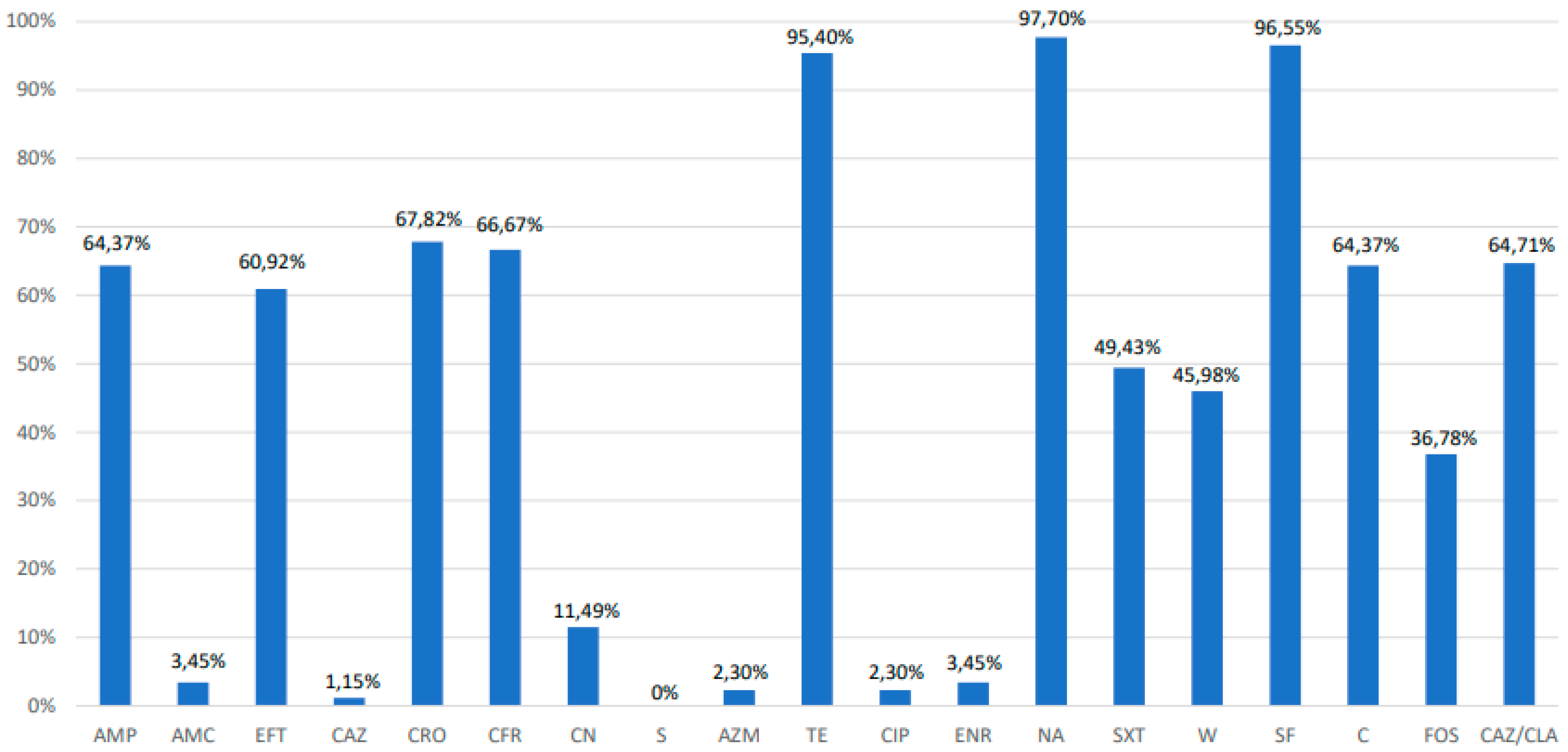
3.2. Antimicrobial Susceptibility Testing, Extended-Spectrum β-Lactamase Phenotyping and Polymerase Chain Reaction PCR Analysis of Resistance Genes
3.3. Distribution of Virulence Genes among Salmonella Infantis
3.4. Biofilm Formation
3.5. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic agents and Food-borne Outbreaks in 2014. EFSA J. 2015, 13, 4329. [Google Scholar]
- Instituto de Salud Pública de Chile. Boletín Vigilancia de Laboratorio: Salmonella spp. 2012–2016. Chile. 2017. Available online: http://www.ispch.cl/sites/default/files/BoletinSalmonella-23012017A.pdf (accessed on 15 March 2017).
- Nógrády, N.; Kardos, G.; Bistyák, A.; Turcsányi, I.; Mészáros, J.; Galántai, Z.; Juhász, A.; Samu, P.; Kaszanyitzky, J.E.; Pászti, J.; et al. Prevalence and characterization of Salmonella Infantis isolates originating from different points of the broiler chicken-human food chain in Hungary. Int. J. Food Microbiol. 2008, 127, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Aviv, G.; Tsyba, K.; Steck, N.; Salmon-Divon, M.; Cornelius, A.; Rahav, G.; Grassl, G.A.; Gal-Mor, O. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol. 2014, 16, 977–994. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Battisti, A.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef] [PubMed]
- Tate, H.; Folster, J.P.; Hsu, C.-H.; Chenb, J.; Hoffmann, M.; Li, C.; Morales, C.; Tyson, G.H.; Mukerjee, S.; Zhaoa, S. Comparative Analysis of Extended Spectrum Beta- Lactamase CTX-M-65-Producing Salmonella enterica Serovar Infantis Isolates from Humans, Food Animals, and Retail Chickens in the United States. Antimicrob. Agents Chemother. 2017, 61, e00488-17. [Google Scholar] [CrossRef]
- Nakatsuchi, A.; Inagaki, M.; Sugiyama, M.; Usui, M.; Asai, T. Association of Salmonella Serotypes with Quinolone Resistance in Broilers. Food Saf. 2018, 6, 156–159. [Google Scholar] [CrossRef]
- Rodrıíguez, I.; Barownick, W.; Helmuth, R.; Mendoza, C.; Rodicio, R.M.; Schroeter, A.; Guerra, B. Extended-spectrum b-lactamases and AmpC b-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J. Antimicrob. Chemother. 2009, 64, 301–309. [Google Scholar] [CrossRef]
- Noda, T.; Murakami, K.; Etoh, Y.; Okamoto, F.; Yatsuyanagi, J.; Sera, N.; Furuta, M.; Onozuka, D.; Oda, T.; Fujimoto, S.; et al. Increase in Resistance to Extended-Spectrum Cephalosporins in Salmonella Isolated from Retail Chicken Products in Japan. PLoS ONE 2015, 10, e0116927. [Google Scholar] [CrossRef]
- Asgharpour, F.; Rajabnia, R.; Shahandashti, E.F.; Marashi, M.A.; Khalilian, M.; Moulana, Z. Investigation of Class I Integron in Salmonella infantis and Its Association With Drug Resistance. Jundishapur J. Microbiol. 2014, 7, e10019. [Google Scholar] [CrossRef]
- Pessoa-Silva, C.L.; Toscano, C.M.; Moreira, B.M.; Santos, A.L.; Frota, A.C.; Solari, C.A.; Amorim, E.L.; Carvalho Mda, G.; Teixeira, L.M.; Jarvis, W.R. Infection due to extended-spectrum beta-lactamase-producing Salmonella enterica subsp. enterica serotype infantis in a neonatal unit. J. Pediatr. 2002, 141, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Shahada, F.; Sugiyama, H.; Chuma, T.; Sueyoshi, M.; Okamoto, K. Genetic analysis of multi-drug resistance and the clonal dissemination of beta-lactam resistance in Salmonella Infantis isolated from broilers. Vet. Microbiol. 2010, 6, 136–141. [Google Scholar]
- Velhner, M.; Kozoderović, G.; Grego, E.; Galić, N.; Stojanov, I.; Jelesić, Z.; Kehrenberg, C. Clonal spread of Salmonella enterica serovar Infantis in Serbia: Acquisition of mutations in the topoisomerase genes gyrA and parC leads to increased resistance to fluoroquinolones. Zoonoses Public Health 2014, 61, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Dohoo, I.R.; Martin, W.; Stryhn, H.E. Veterinary Epidemiologic Research; University of Prince Edward Island: Charlottetown, PE, Canada, 2003. [Google Scholar]
- ISO/TS 6579-2. Microbiology of Food and Animal Feed—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella; ISO: Genève, Switzerland, 2012. [Google Scholar]
- Anonymous. The new role of prekaging: Active, smart and intelligent. Ital. Food Bev. Technol. 2002, 27, 52–61. [Google Scholar]
- Popoff, M.Y.; Le Minor, L. Antigenic Formulas of the Salmonella Serovars; 8th Revision; WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur: Paris, France, 2001. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fourth Informational Supplement. Document M100-S23; CLSI: Wayne, PA, USA, 2013. [Google Scholar]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine. 2018. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (accessed on 2 March 2019).
- Parry, C.M.; Thieu, N.T.; Dolecek, C.; Karkey, A.; Gupta, R.; Turner, P.; Dance, D.; Maude, R.R.; Ha, V.; Tran, C.N.; et al. Clinically and microbiologically derived azithromycin susceptibility breakpoints for Salmonella enterica serovars Typhi and Paratyphi A. Antimicrob. Agents Chemother. 2015, 59, 2756–2764. [Google Scholar] [CrossRef]
- Martínez, I.; Gutierrez, L.; Tapia, G.; Ocampo, L.; Sumano, H. Serum and milk concentrations of enrofloxacin in cows intramammarily treated with a new enrofloxacin-polymorph. Med. Weter 2016, 72, 686–692. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fourth Informational Supplement. Document M100-S24; CLSI: Wayne, PA, USA, 2014. [Google Scholar]
- Eucast European Committee on Antimicrobial Susceptibility Testing. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_docume nts/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf (accessed on 2 March 2019).
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 1.0; EUCAST: Basel, Switzerland, 2016. [Google Scholar]
- Colom, K.; Perez, J.; Alonso, R.; Aranguiz, A.F.; Larino, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA_1 genes in Enterobacteriaceae. FEMS Microbial. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Wilharm, G.; Zander, E.; Wichelhaus, T.A.; Gottig, S.; Hunfeld, K.; Seifert, H.; Witte, W.; Higgins, P.G. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 2011, 66, 1998–2001. [Google Scholar] [CrossRef]
- Mulvey, M.R.; Soule, G.; Boyd, D.; Demczuk, W.; Ahmed, R. Characterization of the first extended-spectrum -lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 2003, 41, 460–462. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielay, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolonemodifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulveyet, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Torkan, S.; Bahadoranianb, M.A.; Khamesipourc, F.; Anyanwud, M.U. Detection of virulence and antibacterial resistance genes in Salmonella isolates from diarrhoeic dogs in Iran. Revue. Méd. Vét. 2015, 166, 221–228. [Google Scholar]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerr, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic resistance in the ECOR collection: Integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef]
- Malorny, B.; Hoorfar, J.; Bunge, C.; Helmuth, R. Multicenter validation of the analytical accuracy of Salmonella PCR: Towards an international standard. Appl. Environ. Microbiol. 2003, 69, 290–296. [Google Scholar] [CrossRef]
- Huehn, S.; La Ragione, R.M.; Anjum, M.; Saunders, M.; Woodward, M.J.; Bunge, C.; Helmuth, R.; Hause, E.; Malorny, B. Virulotyping and Antimicrobial Resistance Typing of Salmonella enterica Serovars Relevant to Human Health in Europe. Foodborne Pathog. Dis. 2010, 7, 523–535. [Google Scholar] [CrossRef]
- Pan, Z.; Carter, B.; Núñez-García, J.; Abuoun, M.; Fookes, M.; Ivens, A.; Woodward, M.J.; Anjum, M.F. Identification of genetic and phenotypic differences associated with prevalent and non-prevalent Salmonella Enteritidis phage types: Analysis of variation in amino acid transport. Microbiology 2009, 155, 3200–3213. [Google Scholar] [CrossRef]
- Shah, D.H.; Zhou, X.; Addwebi, T.; Davis, M.A.; Orfe, L.; Call, D.R.; Besser, T.E. Cell invasion of poultry-associated Salmonella enterica serovar Enteritidis isolates is associated with pathogenicity, motility and proteins secreted by the type III secretion system. Microbiology 2011, 157, 1428–1445. [Google Scholar] [CrossRef]
- Raffatellu, M.; Wilson, R.P.; Chessa, D.; Andrews-Polymenis, H.; Tran, Q.T.; Lawhon, S.; Khare, S.; Adams, L.G.; Baumler, A.J. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 2005, 73, 146–154. [Google Scholar] [CrossRef]
- Pascoe, B.; Méric, G.; Murray, S.; Yahara, K.; Mageiros, L.; Bowen, R.; Jones, N.H.; Jeeves, R.E.; Lappin-Scott, H.M.; Sheppard, S.K.; et al. Enhanced biofilm formation and multi-host transmission evolve from divergent genetic backgrounds in Campylobacter jejuni. Environ. Microbiol. 2015, 17, 4779–4789. [Google Scholar] [CrossRef]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.; Gonzalez, L.A.; Tablada, M.; Robledo, C.W. “InfoStat versión 2011.” Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. Available online: http://www.infostat.com.ar (accessed on 2 March 2019).
- Wajid, M.; Saleemi, M.K.; Sarwar, Y.; Ali, A. Detection and characterization of multidrug-resistant Salmonella enterica serovar Infantis as an emerging threat in poultry farms of Faisalabad, Pakistan. J. Appl. Microbiol. 2019, 147, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.K.; Murray, D.L.; Ruesch, L.A.; Thomas, M.; Lau, Z.; Scaria, J. Genotypic and Phenotypic Characterization of Salmonella Isolated from Fresh Ground Meats Obtained from Retail Grocery Stores in the Brookings, South Dakota, Area. J. Food Prot. 2018, 81, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Mancin, M.; Barco, L.; Losasso, C.; Belluco, S.; Cibin, V.; Mazzucato, M.; Bilei, S.; Carullo, M.R.; Decastelli, L.; Ricci, A.; et al. Salmonella serovar distribution from non-human sources in Italy; results from the IT-Enter-Vet network. Vet. Rec. 2018, 183, 69. [Google Scholar] [CrossRef] [PubMed]
- Hindermann, D.; Gopinath, G.; Chase, H.; Negrete, F.; Althaus, D.; Zurfluh, K.; Tall, B.D.; Stephan, R.; Nüesch-Inderbinen, M. Salmonella enterica serovar Infantis from Food and Human Infections, Switzerland, 2010–2015: Poultry-Related Multidrug Resistant Clones and an Emerging ESBL Producing Clonal Lineage. Front. Microbiol. 2017, 8, 1322. [Google Scholar] [CrossRef] [PubMed]
- Vinueza-Burgos, C.; Baquero, M.; Medina, J.; De Zutter, L. Occurrence, genotypes and antimicrobial susceptibility of Salmonella collected from the broiler production chain within an integrated poultry company. Int. J. Food Microbiol. 2019, 299, 1–7. [Google Scholar] [CrossRef] [PubMed]
- SENASA. Estudio de Prevalencia de Serotipos de Salmonella en Granjas Avícolas Tecnificadas en el Perú; SENASA: Lima, Peru, 2015.
- Medeiros, M.A.N.; de Oliveira, D.C.N.; Rodrigues, D.D.P.; de Freitas, D.R.C. Prevalence and antimicrobial resistance of Salmonella in chicken carcasses at retail in 15 Brazilian cities. Rev. Panam. Salud Publica 2011, 30, 555–560. [Google Scholar] [CrossRef] [PubMed]
- van Asten, A.J.A.M.; van Dijk, J.E. Distribution of ‘‘classic’’ virulence factors among Salmonella spp. FEMS Immunol. Med. Microbiol. 2005, 44, 251–259. [Google Scholar] [CrossRef]
- Brown, A.C.; Chen, J.C.; Francois Watkins, L.K.; Campbell, D.; Folster, J.P.; Tate, H.; Wasilenko, J.; Van Tubbergen, C.; Friedman, C.R. CTX-M-65 extended-Spectrum β-actamase–producing Salmonella enterica Serotype Infantis, United States. Emerg. Infect. Dis. 2018, 24, 2284–2291. [Google Scholar] [CrossRef]
- Karacan Sever, N.; Akan, M. Molecular analysis of virulence genes of Salmonella Infantis isolated from chickens and turkeys. Microb. Pathog. 2019, 126, 199–204. [Google Scholar] [CrossRef]
- Marin, C.; Hernandiz, A.; Lainez, M. Biofilm development capacity of Salmonella strains isolated in poultry risk factors and their resistance against disinfectants. Poult. Sci. 2009, 88, 424–431. [Google Scholar] [CrossRef]
- Díez-García, M.; Capita, R.; Alonso-Calleja, C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 2012, 31, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Schonewille, E.; Nesse, L.L.; Hauck, R.; Windhorst, D.; Hafez, H.M.; Vestby, L.K. Biofilm building capacity of Salmonella enterica strains from the poultry farm environment. FEMS Immunol. Med. Microbiol. 2012, 65, 360–365. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). 19th WHO ModelList of Essential Medicines; World Health Organization Essentialmedicines and Health Products: Geneva, Switzerland, 2015. [Google Scholar]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-type β-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report); The Centers: Atlanta, GA, USA, 2016; pp. 22–25.
- Dimitrov, T.; Udo, E.E.; Albaksami, O.; Kilani, A.A.; Shehab, E.M.R. Ciprofloxacin treatment failure in a case of typhoid fever caused by Salmonella enterica serotype Paratyphi A with reduced susceptibility to ciprofloxacin. J. Med. Microbiol. 2007, 56, 277–279. [Google Scholar] [CrossRef]
- World Organization for Animal Health (OIE). OIE List of Antimicrobial Agents of Veterinary Importance. 2019. Available online: https://www.oie.int/fileadmin/Home/esp/Our_scientific_expertise/docs/pdf/AMR/E_OIE_Lista_antimicrobianos_Julio2019.pdf (accessed on 2 March 2020).
- Tran, T.; Andres, P.; Petroni, A.; Soler-Bistué, A.; Albornoz, E.; Zorreguieta, A.; Reyes-Lamothe, R.; Sherratt, D.J.; Corso, A.; Tolmaskya, M.E.; et al. Small plasmids harboring qnrB19: A model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob. Agents Chemother. 2012, 56, 1821–1827. [Google Scholar] [CrossRef]
- Moreno-Switt, A.I.; Pezoa, D.; Sepúlveda, V.; González, I.; Rivera, D.; Retamal, P.; Navarrete, P.; Reyes-Jara, A.; Toro, M. Transduction as a potential dissemination mechanism of a clonal qnrB19-carrying plasmid isolated from Salmonella of multiple serotypes and isolation Sources. Front. Microbiol. 2019, 10, 2503. [Google Scholar] [CrossRef]
- Pallecchi, L.; Riccobono, E.; Sennati, S.; Mantella, A.; Bartalesi, F.; Trigoso, C.; Gotuzzo, F.; Bartolon, A.; Rossolini, G.M. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob. Agents Chemother. 2010, 54, 678–682. [Google Scholar] [CrossRef]
- Yue, L.; Jiang, H.X.; Liao, X.P.; Liu, J.H.; Li, S.J.; Chen, X.Y.; Chen, C.X.; Lü, D.H.; Liu, Y.H. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet. Microbiol. 2018, 132, 414–420. [Google Scholar] [CrossRef]
- Tyson, G.H.; McDermott, P.F.; Li, C.; Chen, Y.; Tadesse, D.A.; Mukherjee, S.; Bodeis-Jones, S.; Kabera, C.; Gaines, S.A.; Zhao, S.; et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 2763–2769. [Google Scholar] [CrossRef]
- SAG, Servicio Agrícola y Ganadero de Chile. Resolución Exenta Nº:4579/2018. 2018. Available online: https://www.sag.gob.cl/sites/default/files/resol_4.579-2018.pdf (accessed on 2 March 2019).
- Liu, G.; Ali, T.; Gao, J.; Ur Rahman, S.; Yu, D.; Barkema, H.W.; Huo, W.; Xu, S.; Shi, Y.; Han, B.; et al. Co-occurrence of Plasmid-Mediated Colistin Resistance (mcr-1) and Extended-Spectrum β-Lactamase Encoding Genes in Escherichia coli from Bovine Mastitic Milk in China. Microb. Drug Resist. 2019, 22. [Epub ahead of print]. [Google Scholar] [CrossRef]
- Li, Q.; Yin, J.; Li, Z.; Li, Z.; Du, Y.; Guo, W.; Bellefleur, M.; Wang, S.; Shi, H. Serotype distribution, antimicrobial susceptibility, antimicrobial resistance genes and virulence genes of Salmonella isolated from a pig slaughterhouse in Yangzhou, China. AMB Express 2019, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.B.; Butaye, P.; Cloeckaert, A.; Schwarz, S. Genes and mutations conferring antimicrobial resistance in Salmonella: An update. Microbes Infect. 2006, 8, 1898–1914. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, M.; Peighambari, S.M.; Svendsen, C.A.; Cavaco, L.M.; Agersø, Y.; Hendriksen, R.S. Molecular clonality and antimicrobial resistance in Salmonella enterica serovars Enteritidis and Infantis from broilers in three Northern regions of Iran. BMC Vet. Res. 2013, 9, 66. [Google Scholar] [CrossRef]
- Jajere, S.M. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 2019, 12, 504–521. [Google Scholar] [CrossRef]
- Mainali, C.; McFall, M.; King, R.; Irwin, R. Evaluation of antimicrobial resistance profiles of Escherichia coli isolates of broiler chickens at slaughter in Alberta, Canada. J. Food Prot. 2013, 76, 2045–2051. [Google Scholar] [CrossRef]

| Gene | Primers (5′-3′) | Product Size (pb) | Annealing T° | Reference |
|---|---|---|---|---|
| bla TEM | F: ATCAGCAATAAACCAGC R: CCCCGAAGAACGTTTTC | 516 | 54 °C | Colom et al. (2004) |
| bla CTX-M | F: ATGTGCAGYACCAGTAARGTKATGGC R: TGGGTRAARTARGTSACCAGAAYSAGCGG | 592 | 58 °C | Mulvey et al. (2003) |
| bla NDM1 | F: CTGAGCACCGCATTAGCC R: GGGCCGTATGAGTGATTGC | 621 | 52 °C | Pfeifer et al. (2011) |
| qtetA | F: GGTTCACTCGAACGACGTCA R: CTGTCCGACAAGTTGCATGA | 577 | 55 °C | Ng et al. (2001) |
| tetB | F: CCTCAGCTTCTCAACGCGTG R: GCACCTTGCTGATGACTCTT | 634 | 55 °C | Ng et al. (2001) |
| dfrA1 | F: GGAGTGCCAAAGGTGAACAGC R: GAGGCGAAGTCTTGGGTAAAAAC | 367 | 55 °C | Torkan et al. (2015) |
| qnrB | F: GATCGTGAAAGCCAGAAAGG R: ACGATGCCTGGTAGTTGTCC | 469 | 53 °C | Robicsek et al. (2006) |
| mrc1 | F: AGTCCGTTTGTTCTTGTGGC R: AGATCCTTGGTCTCGGCTTG | 320 | 58 °C | Rebelo et al. (2018) |
| mrc2 | F: CAAGTGTGTTGGTCGCAGTT R: TCTAGCCCGACAAGCATACC | 715 | 58 °C | Rebelo et al. (2018) |
| mrc3 | F: AAATAAAAATTGTTCCGCTTATG R: AATGGAGATCCCCGTTTTT | 929 | 58 °C | Rebelo et al. (2018) |
| mrc4 | F: TCACTTTCATCACTGCGTTG R: TTGGTCCATGACTACCAATG | 1116 | 58 °C | Rebelo et al. (2018) |
| mrc5 | F: ATGCGGTTGTCTGCATTTATC R: TCATTGTGGTTGTCCTTTTCTG | 1644 | 58 °C | Rebelo et al. (2018) |
| int1 | F: GGGTCAAGGATCTGGATTTCG R: ACATGGGTGTAAATCATCGTC | 483 | 62 °C | Mazel et al. (2000) |
| int2 | F: CACGGATATGCGACAAAAAGGT R: GTAGCAAACGAGTGACGAAATG | 233 | 62 °C | Mazel et al. (2000) |
| gipA | F: ACGACTGAGCAGGCTGAG R: TTGGAAATGGTGACGGTAGAC | 518 | 58 °C | Huenh et al. (2010) |
| mgtC | F: TGACTATCAATGCTCCAGTGAAT R: ATTTACTGGCCGCTATGCTGTTG | 677 | 58 °C | Huenh et al. (2010) |
| trhH | F: AACTGGTGCCGTTGTCATTG R: GATGGTCTGTGCTTGCTGAG | 418 | 53 °C | Huenh et al. (2010) |
| spvC | F: CTCCTTGCACAACCAAATGCG R: TGTCTCTGCATTTCACCACCATC | 570 | 53 °C | Huenh et al. (2010) |
| sirA | F: TGCGCCTGGTGACAAAACTG R: ACTGACTTCCCAGGCTACAGCA | 313 | 55 °C | Huenh et al. (2010) |
| pagK | F: ACCATCTTCACTATATTCTGCTC R: ACCTCTACACATTTTAAACCAATC | 151 | 60 °C | Huenh et al. (2010) |
| invA | F: GTGAAATTATCGCCACGTTCGGGCAA R: TCATCGCACCGTCAAAGGAACC | 284 | 64 °C | Malorny et al. (2003) |
| SEN1417 | F: GATCGCTGGCTGGTC R: CTGACCGTAATGGCGA | 670 | 58 °C | Pan et al. (2009) |
| sipA | F: ATGGTTACAAGTGTAAGGACTCAG R: ACGCTGCATGTGCAAGCCATC | 2055 | 53 °C | Shah et al. (2011) |
| sipD | F: ATGCTTAATATTCAAAATTATTCCG R: TCCTTGCAGGAAGCTTTTG | 1029 | 53 °C | Shah et al. (2011) |
| sopD | F: GAGCTCACGACCATTTGCGGCG R: GAGCTCCGAGACACGCTTCTTCG | 1291 | 59 | Raffatellu et al. (2005) |
| Resistance Phenotype | No. Isolates |
|---|---|
| NA | 4 |
| ENR-NA | 1 |
| TE-NA-SF | 12 |
| CRO-TE-NA-SF | 1 |
| CFR-TE-NA-SF-FOS | 1 |
| TE-NA-SXT-W-SF | 3 |
| TE-NA-SF-C-FOS | 1 |
| CFR-CIP-NA-SXT-W-SF | 1 |
| CN-TE-NA-SXT-W-SF | 4 |
| AMC-CFR-TE-NA-SXT-W-SF | 1 |
| EFT-CN-TE-NA-SXT-W-SF | 1 |
| CFR-CN-TE-NA-SXT-W-SF | 1 |
| AMP-EFT-CRO-CFR-TE-NA-SF-C | 15 |
| AMP-CN-TE-ENR-NA-SXT-W-SF | 1 |
| AMP-CRO-TE-NA-SXT-W-SF-C | 1 |
| AMP-EFT-CRO-CFR-TE-NA-SXT-SF-C | 1 |
| AMP-EFT-CRO-CFR-CN-TE-NA-SF-C | 1 |
| AMP-EFT-CRO-CFR-TE-NA-SXT-W-SF-C | 1 |
| AMP-EFT-CRO-CFR-CN-TE-ENR-NA-SF-C | 3 |
| AMP-EFT-CRO-CFR-CN-TE-NA-SF-C-FOS | 1 |
| AMP-EFT-CRO-CFR-TE-NA-SXT-W-SF-C | 1 |
| AMP-EFT-CRO-CFR-AZM-TE-NA-CIP-SXT-SF-FOS | 1 |
| AMP-EFT-CRO-CFR-CN-TE-ENR-NA-SF-C-FOS | 3 |
| AMP-EFT-CRO-CFR-CN-AZM-TE-ENR-NA-SF-C | 1 |
| AMC-EFT-CRO-CFR-TE-NA-SXT-W-SF-C-FOS | 1 |
| AMP-EFT-CRO-CFR-TE-NA-SXT-W-SF-C-FOS | 3 |
| AMP-EFT-CRO-CFR-CN-TE-NA-SXT-W-SF-FOS | 1 |
| AMP-EFT-CRO-CFR-CN-TE-NA-SXT-W-SF-C-FOS | 15 |
| AMP-AMC-EFT-CAZ-CRO-CFR-CN-TE-ENR-NA-SF-C-FOS | 1 |
| AMP-AMC-EFT-CRO-CFR-CN-TE-NA-SXT-W-SF-C-FOS | 1 |
| AMP-EFT-CRO-CFR-CN-AZM-TE-NA-SXT-W-SF-C-FOS | 1 |
| AMP-EFT-CRO-CFR-CN-TE-ENR-NA-SXT-W-SF-C-FOS | 1 |
| AMP-EFT-CRO-CFR-CN-TE-NA-ENR-SXT-W-SF-C-FOS | 1 |
| ID 1 | Virulotypes: gipA-spvC-sirA-invA-SEN1417-pagK-sipA-sipO-sopD-mgtC-trhH | No. of Strains |
|---|---|---|
| 2 | 11011111110 | 65 |
| 3 | 10011111110 | 11 |
| 6 | 11111111110 | 4 |
| 7 | 10111111110 | 4 |
| 15 | 11111111111 | 1 |
| 77 | 11011111111 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lapierre, L.; Cornejo, J.; Zavala, S.; Galarce, N.; Sánchez, F.; Benavides, M.B.; Guzmán, M.; Sáenz, L. Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile. Animals 2020, 10, 1049. https://doi.org/10.3390/ani10061049
Lapierre L, Cornejo J, Zavala S, Galarce N, Sánchez F, Benavides MB, Guzmán M, Sáenz L. Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile. Animals. 2020; 10(6):1049. https://doi.org/10.3390/ani10061049
Chicago/Turabian StyleLapierre, Lisette, Javiera Cornejo, Sebastián Zavala, Nicolás Galarce, Fernando Sánchez, María Belén Benavides, Miguel Guzmán, and Leonardo Sáenz. 2020. "Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile" Animals 10, no. 6: 1049. https://doi.org/10.3390/ani10061049
APA StyleLapierre, L., Cornejo, J., Zavala, S., Galarce, N., Sánchez, F., Benavides, M. B., Guzmán, M., & Sáenz, L. (2020). Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile. Animals, 10(6), 1049. https://doi.org/10.3390/ani10061049







