Simple Summary
Reproductive performance is an important factor, which determines productive life and drives culling decisions in dairy herds. There are strong motives for including reproductive performance in genetic selection programs of dairy cows; however, low heritability estimates reported for reproductive performance measures limit the genetic selection efficiency. More effective genetic selection could be achieved using genomic information. The aim of this study was to identify genomic region(s) associated with the length of the calving interval in Holstein cows. The accuracies of genomic estimated breeding values (GEBVs) with single-step genomic BLUP (ssGBLUP) and the pedigree-based BLUP were compared as well. The results showed that the accuracies of GEBVs using the single-step genomic BLUP were much higher than those estimated using the pedigree-based BLUP. We identified three genomic regions (BTA3, BTA6, and BTA7) associated with the length of the calving interval in Holstein dairy cows.
Abstract
The aim of the present study was to identify genomic region(s) associated with the length of the calving interval in primiparous (n = 6866) and multiparous (n = 5071) Holstein cows. The single nucleotide polymorphism (SNP) solutions were estimated using a weighted single-step genomic best linear unbiased prediction (WssGBLUP) approach and imputed high-density panel (777 k) genotypes. The effects of markers and the genomic estimated breeding values (GEBV) of the animals were obtained by five iterations of WssGBLUP. The results showed that the accuracies of GEBVs with WssGBLUP improved by +5.4 to +5.7, (primiparous cows) and +9.4 to +9.7 (multiparous cows) percent points over accuracies from the pedigree-based BLUP. The most accurate genomic evaluation was provided at the second iteration of WssGBLUP, which was used to identify associated genomic regions using a windows-based GWAS procedure. The proportion of additive genetic variance explained by windows of 50 consecutive SNPs (with an average of 165 Kb) was calculated and the region(s) that accounted for equal to or more than 0.20% of the total additive genetic variance were used to search for candidate genes. Three windows of 50 consecutive SNPs (BTA3, BTA6, and BTA7) were identified to be associated with the length of the calving interval in primi- and multiparous cows, while the window with the highest percentage of explained genetic variance was located on BTA3 position 49.42 to 49.52 Mb. There were five genes including ARHGAP29, SEC24D, METTL14, SLC36A2, and SLC36A3 inside the windows associated with the length of the calving interval. The biological process terms including alanine transport, L-alanine transport, proline transport, and glycine transport were identified as the most important terms enriched by the genes inside the identified windows.
1. Introduction
Milk yield and reproductive performance are two important factors which determine productive life and culling decisions in dairy herds [1,2]. While improvements in management and nutrition, along with intense genetic selection have increased milk yield in recent decades, reproductive efficiency is among the main causes of culling and replacement of Holstein cows worldwide [3,4,5,6,7]. Norman, et al. [8] reported that despite the continued march of increasing production per cow per year, in the US, the trend for at least some reproductive parameters dairy cows has been improved. It is well documented that declining fertility cannot be improved through improving management alone, hence genetic selection has attracted much attention [9]. Indicators including age at first calving, days from calving to first breeding, days open, the length of the calving interval, pregnancy rates, and the number of services per conception can be used to evaluate the reproductive performance in dairy cows [10,11,12]. Although studies have shown the existence of genetic variance for reproductive performance, the additive genetic variance reported accounted for only a small fraction of the total variance [13,14,15]. There are strong motives for including reproductive performance in genetic selection programs of dairy cows, but the very low heritability estimates reported for most of the considered reproductive performance measures makes selection for reproductive performance not effective enough [9,16]. However, low heritability estimates for reproductive performance does not indicate the unimportance of genetic selection. More effective genetic selection for female fertility could be achieved by using information from the whole genome and incorporating the information of quantitative trait loci (QTL) into selection decisions [9,16]. Genomic information can be used to improve the accuracy of breeding values and to perform genome-wide association studies (GWAS) with the aim of identifying genomic region(s) explaining genetic variance of traits. Although, genome-wide association studies carried out within a variety of cattle breeds identified many single nucleotide polymorphisms (SNPs) associated with the length of the calving interval (CI), as an indicator for reproductive performance, they are mainly based on the polygenic estimated breeding value (EBV), daughter yield deviation (DYD), or deregressed proof for CI [4,17,18,19]. The single-step genomic best linear unbiased prediction (ssGBLUP) approach [20,21], a quite common procedure in GWAS, has the advantage of simultaneously using the phenotypes of genotyped and nongenotyped animals, pedigrees, and genotypes; therefore, there is no need to calculate pseudo-phenotypes. It has been reported that the use of ssGBLUP procedure increased the accuracy of genetic evaluation in many contexts and species compared with pedigree-based BLUP [22,23]. However, the ssGBLUP assumes that all SNPs explain the same amount of genetic variance, which is unlikely in the case of traits whose major genes or QTL are segregating. The weighted single-step genomic BLUP (WssGBLUP) approach [24] allows the use of different weights for SNPs according to their trait-relevant importance and improves the accuracy of genetic evaluation and the precision of estimates of SNP effects .
The objective of this study was to use the WssGBLUP procedure and imputed high-density panel (777 k) genotypes to identify genomic region(s) associated with calving interval in Holstein dairy cows. The accuracies of genomic estimated breeding values (GEBVs) with the WssGBLUP procedure and the pedigree-based BLUP were compared as well.
2. Materials and Methods
2.1. Phenotypic and Genotypic Data
Data in this study were collected as part of the Genotype plus Environment (GplusE) FP7-Project (http://www.gpluse.eu). The data were records of 11,937 lactations on 6866 primiparous and 5071 multiparous Holstein cows calving between 2010 and 2018, distributed among 118 herds in four countries (Belgium, the Netherlands, Great Britain, and Denmark). The calving interval was calculated as the difference between calving dates from successive parities and was restricted to the range of 270 to 700 d. Genotyping was performed using the Illumina Bovine 10 K low-density chip (n = 20,462), Bovine SNP50K (n = 10,638) or BovineHD SNP panel (795 animals). Genotypes of animals were imputed to high density (HD) with a reference population consisting of 46 HD males and 749 HD females [25]. In total, 12,367 out of 31,895 genotyped individuals, had either phenotypic data or were in the pedigree file which was used in the association analysis. (The number of animals with records was 6866, the number of animals with records and with genotypes was 5345, the number of animals with records and no genotypes was 1521, and the number of animals with genotypes and no records was 7022). Only SNPs located on Bos taurus autosomes (BTA) were considered. SNP markers with minor allele frequency less than 5% were excluded. Finally, 566,345 out of 730,539 SNPs were available for the association analysis. Ethics approval and consent to participate were not applicable to this study.
2.2. Variance Components Estimation
The pedigree consisted of 43,181 individuals (12,367 and 6866 out of 43,181 animals had genotype and phenotype data, respectively). The genetic analyses were carried out through the average information restricted maximum likelihood (AIREML) via AIREMLf90 from the BLUPF90 software package [26]. A linear single-trait animal model was used for the length of the CI in primiparous cows. The linear model included fixed effect of herd-year-season of calving (HYS), and country, covariate effects of age at first calving in both linear and quadratic forms, as well as animal and residual random effects. The complete model for primiparous cows can be represented as follows:
where yijk represents the length of the CI for animal k, µ is the overall mean, HYSi is the fixed effect of ith herd-year-season of calving, conj is the fixed effect of jth country, b1 and b2 are the linear and quadratic regression coefficients of the length of the CI on the age at first calving, agek is the age at first calving of kth cows, ak is the additive genetic effect, and eijk is the random residual error. The additive genetic and residual variances were obtained as follows:
where a is the vector of direct additive genetic effects, e is a vector of residual effects, and are, respectively, total additive genetic and residual variances, and H is the genetic relationship matrix combining SNP information and pedigree data (A) [20]:
where is the inverse of the pedigree-based relationship matrix for all animals; is the inverse of the numerator relationship matrix for the genotyped individuals; and G is the genomic relationship matrix [27].
where Z is the matrix of gene content adjusted for allele frequencies (0, 1, or 2 for aa, Aa, and AA, respectively); D is a diagonal matrix of weights for SNP variances; M is the number of SNPs, and pi is the estimated minor allele frequency at ith locus. The genetic analyses for the length of the CI in multiparous cows were carried out using a linear single-trait repeatability animal model, which was the same as the model used for primiparous cows but here, the fixed effect of parity was included into the model. In addition, a third random effect representing the environmental permanent effect associated with animals having repeated records was included into the model. This effect, assumed to be uncorrelated with additive genetic effects, allowed for the partitioning of the environmental variance into permanent and temporary components. The complete model for multiparous cows can be represented as follows:
Most of the terms in this model were defined as for the linear single-trait animal model except for pk, the effect of lactation number, and pel, is the permanent environmental effect of cow l.
Pedigree-based (co)variance components and breeding values were estimated considering the same linear animal model used to estimate the (co)variance components mentioned before. In the pedigree-based BLUP the genomic information was excluded, and the direct additive genetic effects were estimated using the pedigree-based relationship matrix.
2.3. Weighted Single-Step Genome-Wide Association Study
The analyses were performed using the weighted single-step genome-wide association study (WssGWAS) methodology [24], considering the same linear animal model used to estimate the (co)variance components mentioned before. The animal effects were decomposed into those for genotyped (ag) and ungenotyped animals (an). The animal effects of genotyped animals are a function of the SNP effects, , where Z is a matrix relating genotypes of each locus and u is a vector of the SNP marker effect. The variance of animal effects was assumed as follows:
where D is a diagonal matrix of weights for variances of markers (at iteration 1, SNP weights in the D matrix are equal to 1) and is the genetic additive variance captured by each SNP marker when the weighted relationship matrix (G*) was built with no weight.
The SNP effects were obtained using following equation:
where λ was defined by VanRaden [27] as a normalizing constant, as described below:
The following iterative process [24] was used to estimate the SNP effects. Step 1. D = I in the first step. Step 2. Calculate the G matrix. Step 3. Calculate GEBVs for the entire data set using ssGBLUP. Step 4. Estimate SNP effects from solutions of genomic breeding values in the previous step: (û): . Step 5. Estimate of the effect of each SNP: , where i is the ith SNP. Step 6. Normalize the vector of variances of SNP effects to get the SNP weights (this normalization process ensures that the sum of the variances remain constant and equal to the number of SNP). Step 7. Use SNP weights to construct the D matrix; exit or loop to step 2. The effects of markers were obtained by five iterations from Steps 2 to 7. The accuracies of genomic estimated breeding values (GEBVs) with ssGBLUP and the pedigree-based BLUP were estimated using following formula.
where PEV is the prediction error variance, and is the additive genetic variance of the trait. The percentage of genetic variance explained by ith genomic region was estimated using the following formula.
where is the genetic value of the ith region that consists of n consecutive SNP (n = 1, 5, 10, 20, and 50), is the total genetic variance, Zj is the vector of SNP content of the jth SNP for all individuals, and is the marker effect of the jth SNP within the ith genomic region. The results were presented by the proportion of additive genetic variance explained by each genomic region of n consecutive SNP.
2.4. Gene Prospection
The chromosome segments associated with the length of the calving interval were selected to explore and determine potential quantitative trait loci (QTL). The database (version UMD3.1) including gene locations, start positions and end sites for all bovine genes (http://www.ensembl.org/index.html) was used for identification of genes. The list of genes inside the genomic region(s) associated with the length of the calving interval, considered as positional candidate genes, was uploaded to Enrichr for gene ontology (GO) enrichment analysis [28,29]. Significantly enriched biological process terms with at least two genes from the input gene list were identified based on the retrieved adjusted P value.
3. Results and Discussion
Variance Components and Accuracy of Genomic Predictions
The mean (SD) of the length of the CI in primi- and multiparous cows were 395.1 (69.1) and 396.7 (62.9) d, respectively. The median of the length of the CI in primi- and multiparous cows were 375 and 381 d, respectively. The additive and residual variances estimated using the AIREML method for the length of the CI in primiparous cows were 201.3 (SE = 57.60) and 3728.7 (SE = 85.76), respectively. The estimated additive, permanent environmental effect, and residual variances in multiparous cows were 198.3 (67.8), 71.4 (100.4), and 2953.7 (103.15), respectively. The heritability estimates for the length of the CI in primi- and multiparous cows were 0.05 (0.01) and 0.06 (0.01), respectively, which are consistent with findings from other studies on dairy cattle [17,30,31,32,33]. Generally, the large unexplained residual variation observed for the length of the CI is attributable not only to the large effect of the environmental factors on this trait but also to the low quality of the data [34].
The single-step genomic BLUP (ssGBLUP) method assumes equal variance for all single nucleotide polymorphisms, which is unlikely in the case for traits whose major genes or QTL are segregating. To overcome the limitation of ssGBLUP, unequal variance or weights for all SNP are applied in an approach called weighted single-step genomic BLUP (WssGBLUP), in which SNP effects are weighted according to their importance for the trait of interest [24]. Accuracies of GEBVs estimated for primiparous cows were 0.32, 0.33, 0.32, 0.32, and 0.32 for iterations one to five of WssGBLUP, respectively. The corresponding values for multiparous cows were 0.37, 0.38, 0.37, 0.37, and 0.37. The accuracies of genomic estimated breeding value (GEBV) provided at the second iteration of WssGBLUP improved by +5.4 to +5.7, (primiparous cows) and +9.4 to +9.7 (multiparous cows) percent points over accuracies from the pedigree-based BLUP. In this study, the influence of the number of iterations (1–5) on the accuracy of genomic predictions for the length of the calving interval was investigated. The most accurate genomic evaluation was provided at the second iteration of WssGBLUP, which was used to identify associated genomic regions using a windows-based GWAS procedure. Wang, Misztal, Aguilar, Legarra, and Muir [24] investigated the influence of the number of iterations (1–8) on the accuracy of genomic predictions and showed that the highest accuracies were obtained at the second iteration and then decreased slightly. Previous studies on dairy goats [35,36] and broiler chickens [37] have also reported that the accuracy of GEBVs estimated using WssGBLUP was maximized by the second iteration and then decreased slightly. The decline in accuracy with iteration may be a result of the continuous addition of weights to the SNP with large effects while shrinking the SNP with small influence [38]. In the window-based GWAS procedure, different window types (distinct or sliding windows) and variable window sizes (defined as the number of SNPs or the number of base pairs) can be used. The common form for declaring importance is to use a threshold on the additive genetic variance explained by individual windows [39]. However, it is unclear what window size is optimal and there is no standard to define the threshold on explained genetic variance [39]. Therefore, determining the proper window size is usually subjective and researchers often have not justified their choices or sometimes have acknowledged that their choices are arbitrary [40,41]. In the present study, sliding windows of 1, 5, 10, 20, and 50 consecutive SNPs were used to identify genomic region(s) associated with the length of the calving interval in primi- and multiparous cows and to determine if the region(s) identified may change depending on the window size. General information about the results of ssGWAS for primi- and multiparous cows are described in Tables S1–S5 and Tables S6–S10, respectively. The results of different sliding window sizes in primi- and multiparous cows showed that the identified peaks changed depending on the window size where smaller window sizes (windows of 1, 5, and 10 consecutive SNP) being accompanied with large noises (Figure 1 and Figure 2). Fragomeni, et al. [42] also reported that small window sizes are accompanied with large noises. Furthermore, it has been shown that single-SNP GWAS cannot be effective enough, because single-SNPs provide limited information about the content of flanking genomic regions [43,44,45]. In the present study, 50-adjacent SNP windows (with an average of 165 Kb widow size) that explained equal to or more than 0.20% of the genetic variance was considered as the threshold for significance. Han and Peñagaricano [46] considered 1.5 Mb SNP windows that explained more than 0.50% of the genetic variance as the threshold to declare significance. Suwannasing, et al. [47] using the Porcine SNP60k BeadChip, considered 5-adjacent SNP windows that explained more than 1% of total genetic variance as the threshold to declare significance. de Oliveira Silva, et al. [48] using the BovineHD SNP panel, considered 50-adjacent SNP windows (with average of 280 kb) that explained more than 0.50% of additive genetic variance as the threshold to declare significance. The results identified three windows (on BTA3, BTA6, and BTA7) associated with calving interval in primi- and multiparous cows (Figure 1 and Figure 2). These three regions combined explained 0.51% and 0.68 % of the total genetic variances of the length of the calving interval, respectively, in primi- and multiparous cows (Table 1). The length of the calving interval generally has a low heritability and probably that is the reason why in neither primi- nor multiparous cows windows highly associated with this trait were identified.
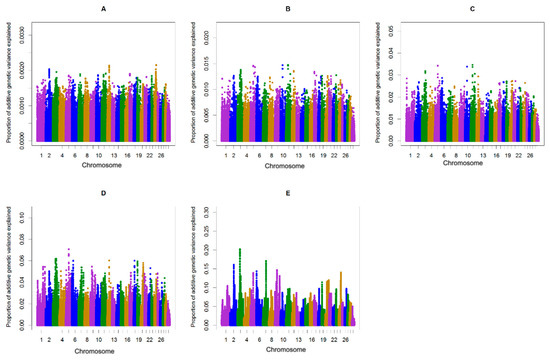
Figure 1.
Additive genetic variance explained by windows of (A) 1, (B) 5, (C) 10, (D) 20, and (E) 50 consecutive single nucleotide polymorphisms (SNPs) across chromosomes for the length of the calving interval in primiparous cows.
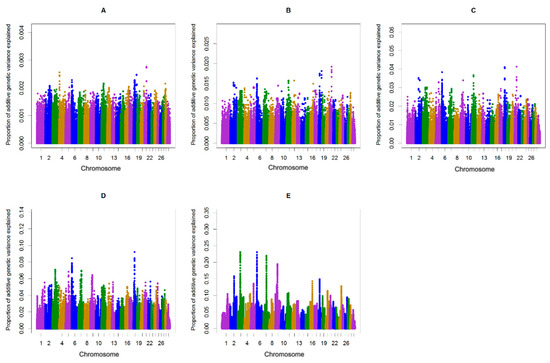
Figure 2.
Additive genetic variance explained by windows of (A) 1, (B) 5, (C) 10, (D) 20, and (E) 50 consecutive SNPs across chromosomes for the length of the calving interval in multiparous cows.

Table 1.
Identification of genes based on the additive genetic variance explained by 50-adjacent SNP windows for the length of the calving interval in primi- and multiparous Holstein cows.
Although, genome-wide association studies carried out within a variety of cattle breeds identified many single nucleotide polymorphisms (SNPs) associated with the length of the calving interval (CI), the results are inconsistent [17,18,49]. Zhou, Li, Cai, Liu, Yin, Shi, and Zhang and Zhang [49] using a single SNP regression mixed linear model, identified two SNPs (BTA19 and BAT25) associated with the length of the calving interval in Xinjiang Brown cattle. Minozzi, Nicolazzi, Stella, Biffani, Negrini, Lazzari, Ajmone-Marsan, Williams [17] using a single SNP regression mixed linear model, identified five SNPs (BTA2. BTA5, BTA8, BTA24, and BTA28) associated with the length of the calving interval in Italian Holstein Cattle. Nayeri, Sargolzaei, Abo-Ismail, May, Miller, Schenkel, Moore, and Stothard [18] using a single SNP regression mixed linear model, identified a total of eight highly significant SNPs on BTA21 associated with days open in Canadian dairy Holstein cattle.
In this study, the genomic region with the highest percentage of explained genetic variance was located on the BTA3 position 49.42 to 49.52 Mb, which overlaps among window sizes of 20 and 50 consecutive SNPs in primi- and multiparous cows. This region also overlaps with QTLs for dystocia [50,51], milk fat yield [52], and residual feed intake [53] in dairy cows. Genes including ARHGAP29, SEC24D, METTL14, SLC36A2, and SLC36A3 were identified inside the windows associated with CI (Table 1). SLC36A2 was reported to be associated with milk beta-casein percentage, milk kappa-casein percentage, and milk protein yield in Chinese Holstein cows [49].
Significantly-enriched biological processes with two genes from the input gene list are shown in Table 2. The biological process including alanine transport, L-alanine transport, and proline transport and glycine transport were identified as the most important term enriched by the identified genes inside the identified windows. Amino acid transport is defined as the directed movement of amino acids, organic acids containing one or more amino substituents, into, out of, or within a cell, or between cells, by means of some agent such as a transporter. An amino acid transporter is a membrane transport protein that transports amino acids. The amino acid transport systems in early embryos likely are regulated at the genetic level by various conditions in the female reproductive tract; however, the precise mechanisms of regulation and their physiological consequences are yet to be fully described [54]. Embryo amino acid content is determined, at least in part, by regulation of amino acid transport [55,56,57]. Lane and Gardner [58] reported that amino acids improve mouse embryo development primarily during cleavage, and support development of more viable embryos. Moore and Bondioli [59] analyzed the bovine oviductal fluid for free amino acid content and reported that glycine and alanine were the two most predominant amino acids. Elhassan, et al. [60] reported that alanine, glutamate, glycine, and taurine, are present in strikingly high concentrations in both bovine oviductal and uterine fluids, suggesting that they might play important roles in early embryo development. Moore and Bondioli [59] reported that glycine and alanine supplementation of culture medium enhances development of in vitro matured and fertilized cattle embryos and concluded that glycine and alanine have a role in early embryonic development. It has been reported that embryo growth and development rates are important indicators of embryo viability and reproductive efficiency [61].

Table 2.
Gene ontology (GO) terms enriched by the genes inside the chromosomal region of associated milk production and lactation curve parameters.
4. Conclusion
The objective of this study was to identify genomic regions associated with the length of the calving interval in Holstein cows. In this study, the length of the calving interval, a commonly used measure of reproductive performance in dairy cow breeding goals, was used as an indicator for female reproductive performance. We compared pedigree-based BLUP with the WssGBLUP for the length of the calving interval and confirmed that the WssGBLUP method improved the accuracy of GEBV particularly at the second iteration. In the present study, different sliding window sizes were evaluated to identify genomic region(s) associated with the length of the calving interval and the results showed that the identified peaks changed depending on the window size with smaller window sizes being accompanied with large noises. We identified three windows of 50 consecutive SNPs associated with the length of the calving interval. The findings of this study can be used to search for causative mutations, and for breeding through marker-assisted selection to improve the length of the calving interval in Holstein dairy cows.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/10/3/500/s1. Table S1: Results of 1-adjacent SNP windows in primiparous cows (Primiparous1.xlsx). Table S2: Results of 5-adjacent SNP windows in primiparous cows (Primiparous5.xlsx). Table S3: Results of 10-adjacent SNP windows in primiparous cows (Primiparous10.xlsx). Table S4: Results of 20-adjacent SNP windows in primiparous cows (Primiparous20.xlsx). Table S5: Results of 50-adjacent SNP windows in primiparous cows (Primiparous50.xlsx). Table S6: Results of 1-adjacent SNP windows in multiparous cows (Multiparous1.xlsx). Table S7: Results of 5-adjacent SNP windows in multiparous cows (Multiparous5.xlsx). Table S8: Results of 10-adjacent SNP windows in multiparous cows (Multiparous10.xlsx). Table S9: Results of 20-adjacent SNP windows in multiparous cows (Multiparous20.xlsx). Table S10: Results of 50-adjacent SNP windows in multiparous cows (Multiparous50.xlsx).
Author Contributions
Conceptualization, H.A., M.H., and M.S.; methodology, H.A., M.H., and M.S.; formal analysis, H.A.; data curation, H.A., J.D.K., M.S., and M.H.; writing—original draft preparation, H.A.; writing—review and editing, G.O., M.A.C., M.H., and M.S.; supervision, M.H. and G.O.; project administration, M.H. and G.O.; funding acquisition, M.A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the European Union’s Seventh Framework Program for research, technological development and demonstration under grant agreement no. 613689.
Acknowledgments
The authors would like to acknowledge the support provided by the European Commission under the Seventh Framework Program for the GplusE Projects. However, the views expressed in this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission.
List of authors within the GplusE consortium: Niamh McLoughlin, Alan Fahey, Elizabeth Matthews, Andreia Santoro, Colin Byrne, Pauline Rudd, Roisin O’Flaherty, Sinead Hallinan, Claire Wathes, Zhangrui Cheng, Ali Fouladi, Geoff Pollott, Dirk Werling, Beatriz Sanz Bernardo, Alistair Wylie, Matt Bell, Mieke Vaneetvelde, Kristof Hermans, Geert Opsomer, Sander Moerman, Jenne De Koster, Hannes Bogaert, Jan Vandepitte, Leila Vandevelde, Bonny Vanranst, Johanna Hoglund, Susanne Dahl, Soren Ostergaard, Janne Rothmann, Mogens Krogh, Else Meyer, Charlotte Gaillard, Jehan Ettema, Tine Rousing, Federica Signorelli, Francesco Napolitano, Bianca Moioli, Alessandra Crisà, Luca Buttazzoni, Jennifer McClure, Daragh Matthews, Francis Kearney, Andrew Cromie, Matt McClure, Shujun Zhang, Xing Chen, Huanchun Chen, Junlong Zhao, Liguo Yang, Guohua Hua, Chen Tan, Guiqiang Wang, Michel Bonneau, Andrea Pompozzi, Armin Pearn, Arnold Evertson, Linda Kosten, Anders Fogh, Thomas Andersen, Matthew Lucey, Chris Elsik, Gavin Conant, Jerry Taylor, Nicolas Gengler, Michel Georges, Frédéric Colinet, Marilou Ramos Pamplona, Hedi Hammami, Catherine Bastin, Haruko Takeda, Aurelie Laine, Anne-Sophie Van Laere, Martin Schulze, Sergio Palma Vera, Conrad Ferris, Cinzia Marchitelli.
Conflicts of Interest
The authors declare that they do not have any conflicts of interest.
References
- Ahlman, T.; Berglund, B.; Rydhmer, L.; Strandberg, E. Culling reasons in organic and conventional dairy herds and genotype by environment interaction for longevity. J. Dairy Sci. 2011, 94, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Monti, G.; Tenhagen, B.; Heuwieser, W. Culling Policies in Dairy Herds. A. Review. J. Vet. Med. Ser. A 1999, 46, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; Williams, E.; Evans, A. A review of the causes of poor fertility in high milk producing dairy cows. Anim. Reprod. Sci. 2011, 123, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Wang, Y.; Sahana, G.; Zhang, Q.; Liu, L.; Lund, M.S.; Su, G. Genome-wide association studies for female fertility traits in Chinese and Nordic Holsteins. Sci. Rep. 2017, 7, 8487. [Google Scholar] [CrossRef]
- Lucy, M. Reproductive loss in high-producing dairy cattle: Where will it end? J. Dairy Sci. 2001, 84, 1277–1293. [Google Scholar] [CrossRef]
- Nebel, R.; Dransfield, M.; Jobst, S.; Bame, J. Automated electronic systems for the detection of oestrus and timing of AI in cattle. Anim. Reprod. Sci. 2000, 60, 713–723. [Google Scholar] [CrossRef]
- Atashi, H.; Zamiri, M.J.; Sayyadnejad, M.B.; Akhlaghi, A. Trends in the reproductive performance of Holstein dairy cows in Iran. Trop. Anim. Health Prod. 2012, 44, 2001–2006. [Google Scholar] [CrossRef]
- Norman, H.; Wright, J.; Hubbard, S.; Miller, R.; Hutchison, J. Reproductive status of Holstein and Jersey cows in the United States. J. Dairy Sci. 2009, 92, 3517–3528. [Google Scholar] [CrossRef]
- Veerkamp, R.; Beerda, B. Genetics and genomics to improve fertility in high producing dairy cows. Theriogenology 2007, 68, S266–S273. [Google Scholar] [CrossRef]
- Pryce, J.; Coffey, M.; Brotherstone, S.; Woolliams, J. Genetic relationships between calving interval and body condition score conditional on milk yield. J. Dairy Sci. 2002, 85, 1590–1595. [Google Scholar] [CrossRef]
- Mee, J.F. Temporal trends in reproductive performance in Irish dairy herds and associated risk factors. Ir. Vet. J. 2004, 57, 158. [Google Scholar] [CrossRef] [PubMed]
- Hermans, K.; Opsomer, G.; Waegeman, W.; Moerman, S.; De Koster, J.; Van Eetvelde, M.; Van Ranst, B.; Hostens, M. Interpretation and visualisation of data from dairy herds. In Pract. 2018, 40, 195–203. [Google Scholar] [CrossRef]
- Lopez, B.I.; Son, J.-H.; Seo, K.; Lim, D. Estimation of Genetic Parameters for Reproductive Traits in Hanwoo (Korean Cattle). Animals 2019, 9, 715. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, M.; Buckley, F.; Evans, R.; Berry, D.P. Additive genetic, non-additive genetic and permanent environmental effects for female reproductive performance in seasonal calving dairy females. Ir. J. Agric. Food Res. 2016, 55, 10–23. [Google Scholar] [CrossRef][Green Version]
- Rydhmer, L.; Berglund, B. Selection for reproduction: Developments in several species. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Minas Gerais, Brazil, 13–18 August 2006; p. 11-01. [Google Scholar]
- Berglund, B. Genetic improvement of dairy cow reproductive performance. Reprod. Domest. Anim. 2008, 43, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Minozzi, G.; Nicolazzi, E.L.; Stella, A.; Biffani, S.; Negrini, R.; Lazzari, B.; Ajmone-Marsan, P.; Williams, J.L. Genome wide analysis of fertility and production traits in Italian Holstein cattle. PLoS ONE 2013, 8, e80219. [Google Scholar] [CrossRef] [PubMed]
- Nayeri, S.; Sargolzaei, M.; Abo-Ismail, M.K.; May, N.; Miller, S.P.; Schenkel, F.; Moore, S.S.; Stothard, P. Genome-wide association for milk production and female fertility traits in Canadian dairy Holstein cattle. BMC Genet. 2016, 17, 75. [Google Scholar] [CrossRef]
- Frischknecht, M.; Bapst, B.; Seefried, F.R.; Signer-Hasler, H.; Garrick, D.; Stricker, C.; Fries, R.; Russ, I.; Sölkner, J.; Bieber, A. Genome-wide association studies of fertility and calving traits in Brown Swiss cattle using imputed whole-genome sequences. BMC Genom. 2017, 18, 910. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Johnson, D.; Legarra, A.; Tsuruta, S.; Lawlor, T. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- Christensen, O.F.; Lund, M.S. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar] [CrossRef]
- Onogi, A.; Ogino, A.; Komatsu, T.; Shoji, N.; Shimizu, K.; Kurogi, K.; Yasumori, T.; Togashi, K.; Iwata, H. Whole-genome prediction of fatty acid composition in meat of Japanese Black cattle. Anim. Genet. 2015, 46, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Matilainen, K.; Koivula, M.; Strandén, I.; Aamand, G.P.; Mäntysaari, E.A. Managing genetic groups in single-step genomic evaluations applied on female fertility traits in Nordic Red Dairy cattle. Interbull Bull. 2016, 50, 71–75. [Google Scholar]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A new approach for efficient genotype imputation using information from relatives. BMC Genom. 2014, 15, 478. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D. BLUPF90 and related programs (BGF90). In Proceedings of the 7th world congress on genetics applied to livestock production, Montpellier, France, 19–23 August 2002; pp. 743–744. [Google Scholar]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Tenghe, A.; Bouwman, A.; Berglund, B.; Strandberg, E.; de Koning, D.; Veerkamp, R. Genome-wide association study for endocrine fertility traits using single nucleotide polymorphism arrays and sequence variants in dairy cattle. J. Dairy Sci. 2016, 99, 5470–5485. [Google Scholar] [CrossRef]
- Zaabza, H.B.; Gara, A.B.; Hammami, H.; Jemmali, B.; Ferchichi, M.A.; Rekik, B. Genetic parameters of reproductive traits in Tunisian Holsteins. Arch. Anim. Breed. (Arch. Tierz.) 2016, 59, 209–213. [Google Scholar] [CrossRef]
- Albarrán-Portillo, B.; Pollott, G. The relationship between fertility and lactation characteristics in Holstein cows on United Kingdom commercial dairy farms. J. Dairy Sci. 2013, 96, 635–646. [Google Scholar] [CrossRef]
- Hossein-Zadeh, N.G. Genetic parameters and trends for calving interval in the first three lactations of Iranian Holsteins. Trop. Anim. Health Prod. 2011, 43, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Miglior, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.F. A 100-Year Review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef] [PubMed]
- Teissier, M.; Larroque, H.; Robert-Granié, C. Weighted single-step genomic BLUP improves accuracy of genomic breeding values for protein content in French dairy goats: A quantitative trait influenced by a major gene. Genet. Sel. Evol. 2018, 50, 31. [Google Scholar] [CrossRef] [PubMed]
- Teissier, M.; Larroque, H.; Robert-Granie, C. Accuracy of genomic evaluation with weighted single-step genomic best linear unbiased prediction for milk production traits, udder type traits, and somatic cell scores in French dairy goats. J. Dairy Sci. 2019, 102, 3142–3154. [Google Scholar] [CrossRef]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Fernando, R.L.; Vitezica, Z.; Okimoto, R.; Wing, T.; Hawken, R.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front. Genet. 2014, 5, 134. [Google Scholar] [CrossRef]
- Zhang, X.; Lourenco, D.; Aguilar, I.; Legarra, A.; Misztal, I. Weighting strategies for single-step genomic BLUP: An iterative approach for accurate calculation of GEBV and GWAS. Front. Genet. 2016, 7, 151. [Google Scholar] [CrossRef]
- Aguilar, I.; Legarra, A.; Cardoso, F.; Masuda, Y.; Lourenco, D.; Misztal, I. Frequentist p-values for large-scale-single step genome-wide association, with an application to birth weight in American Angus cattle. Genet. Sel. Evol. 2019, 51, 28. [Google Scholar] [CrossRef]
- Myles, S.; Tang, K.; Somel, M.; Green, R.E.; Kelso, J.; Stoneking, M. Identification and analysis of genomic regions with large between-population differentiation in humans. Ann. Hum. Genet. 2008, 72, 99–110. [Google Scholar] [CrossRef]
- Beissinger, T.M.; Rosa, G.J.; Kaeppler, S.M.; Gianola, D.; De Leon, N. Defining window-boundaries for genomic analyses using smoothing spline techniques. Genet. Sel. Evol. 2015, 47, 30. [Google Scholar] [CrossRef]
- Fragomeni, B.d.O.; Misztal, I.; Lourenco, D.L.; Aguilar, I.; Okimoto, R.; Muir, W.M. Changes in variance explained by top SNP windows over generations for three traits in broiler chicken. Front. Genet. 2014, 5, 332. [Google Scholar] [CrossRef]
- Braz, C.U.; Taylor, J.F.; Bresolin, T.; Espigolan, R.; Feitosa, F.L.; Carvalheiro, R.; Baldi, F.; Lucia, G.; de Oliveira, H.N. Sliding window haplotype approaches overcome single SNP analysis limitations in identifying genes for meat tenderness in Nelore cattle. BMC Genet. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, J.; Bonham, A.J.; Wang, Y.; Deng, H. Gains in power for exhaustive analyses of haplotypes using variable-sized sliding window strategy: A comparison of association-mapping strategies. Eur. J. Hum. Genet. 2009, 17, 785. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, R.; Feng, T.; Sha, Q.; Zhang, S. A variable-sized sliding-window approach for genetic association studies via principal component analysis. Ann. Hum. Genet. 2009, 73, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Peñagaricano, F. Unravelling the genomic architecture of bull fertility in Holstein cattle. BMC Genet. 2016, 17, 143. [Google Scholar] [CrossRef]
- Suwannasing, R.; Duangjinda, M.; Boonkum, W.; Taharnklaew, R.; Tuangsithtanon, K. The identification of novel regions for reproduction trait in Landrace and Large White pigs using a single step genome-wide association study. Asian Australas. J. Anim. Sci. 2018, 31, 1852. [Google Scholar] [CrossRef]
- de Oliveira Silva, R.M.; Stafuzza, N.B.; de Oliveira Fragomeni, B.; de Camargo, G.M.F.; Ceacero, T.M.; Cyrillo, J.N.d.S.G.; Baldi, F.; Boligon, A.A.; Mercadante, M.E.Z.; Lourenco, D.L. Genome-wide association study for carcass traits in an experimental Nelore cattle population. PLoS ONE 2017, 12, e0169860. [Google Scholar] [CrossRef]
- Zhou, C.; Li, C.; Cai, W.; Liu, S.; Yin, H.W.; Shi, S.; Zhang, Q.; Zhang, S. Genome-wide association study for milk protein composition traits in a Chinese Holstein population using a single-step approach. Front. Genet. 2019, 10, 72. [Google Scholar] [CrossRef]
- Seidenspinner, T.; Bennewitz, J.; Reinhardt, F.; Thaller, G. Need for sharp phenotypes in QTL detection for calving traits in dairy cattle. J. Anim. Breed. Genet. 2009, 126, 455–462. [Google Scholar] [CrossRef]
- McClure, M.; Morsci, N.; Schnabel, R.; Kim, J.; Yao, P.; Rolf, M.; McKay, S.; Gregg, S.; Chapple, R.; Northcutt, S. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim. Genet. 2010, 41, 597–607. [Google Scholar] [CrossRef]
- Rodriguez-Zas, S.L.; Southey, B.; Heyen, D.; Lewin, H. Interval and composite interval mapping of somatic cell score, yield, and components of milk in dairy cattle. J. Dairy Sci. 2002, 85, 3081–3091. [Google Scholar] [CrossRef]
- Sherman, E.; Nkrumah, J.; Li, C.; Bartusiak, R.; Murdoch, B.; Moore, S. Fine mapping quantitative trait loci for feed intake and feed efficiency in beef cattle. J. Anim. Sci. 2009, 87, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Van Winkle, L.J. Amino acid transport regulation and early embryo development. Biol. Reprod. 2001, 64, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Van Winkle, L.J.; Campione, A.L. Amino acid transport regulation in preimplantation mouse embryos: Effects on amino acid content and pre-and peri-implantation development. Theriogenology 1996, 45, 69–80. [Google Scholar] [CrossRef]
- Van Winkle, L.J.; Dickinson, H.R. Differences in amino acid content of preimplantation mouse embryos that develop In Vitro versus In Vivo: In Vitro effects of five amino acids that are abundant in oviductal secretions. Biol. Reprod. 1995, 52, 96–104. [Google Scholar] [CrossRef]
- Van Winkle, L.J.; Christensen, H.; Campione, A. Na+-dependent transport of basic, zwitterionic, and bicyclic amino acids by a broad-scope system in mouse blastocysts. J. Biol. Chem. 1985, 260, 12118–12123. [Google Scholar]
- Lane, M.; Gardner, D. Differential regulation of mouse embryo development and viability by amino acids. Reproduction 1997, 109, 153–164. [Google Scholar] [CrossRef]
- Moore, K.; Bondioli, K.R. Glycine and alanine supplementation of culture medium enhances development of in vitro matured and fertilized cattle embryos. Biol. Reprod. 1993, 48, 833–840. [Google Scholar] [CrossRef]
- Elhassan, Y.; Wu, G.; Leanez, A.; Tasca, R.; Watson, A.; Westhusin, M. Amino acid concentrations in fluids from the bovine oviduct and uterus and in KSOM-based culture media. Theriogenology 2001, 55, 1907–1918. [Google Scholar] [CrossRef]
- Morris, D.G.; Grealy, M.; Leese, H.; Diskin, M.G.; Sreenan, J. Cattle Embryo Growth Development and Viability; Teagasc: Dublin, Ireland, 2001. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).