CRISPR/Cas9-Mediated Biallelic Knockout of IRX3 Reduces the Production and Survival of Somatic Cell-Cloned Bama Minipigs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Ethics
2.2. Reagents and Chemicals
2.3. Construction of CRISPR/Cas9 Plasmid
2.4. Cell Transfection and Selection of Gene-Edited Cell Colonies
2.5. Genotyping and Detection of Exogenous Genes Integration in Gene-Edited Cell Colonies
2.6. Off-Target Detection in Gene-Edited Cell Colonies
2.7. Production of Gene-Edited Bama Minipigs by Somatic Cell Cloning
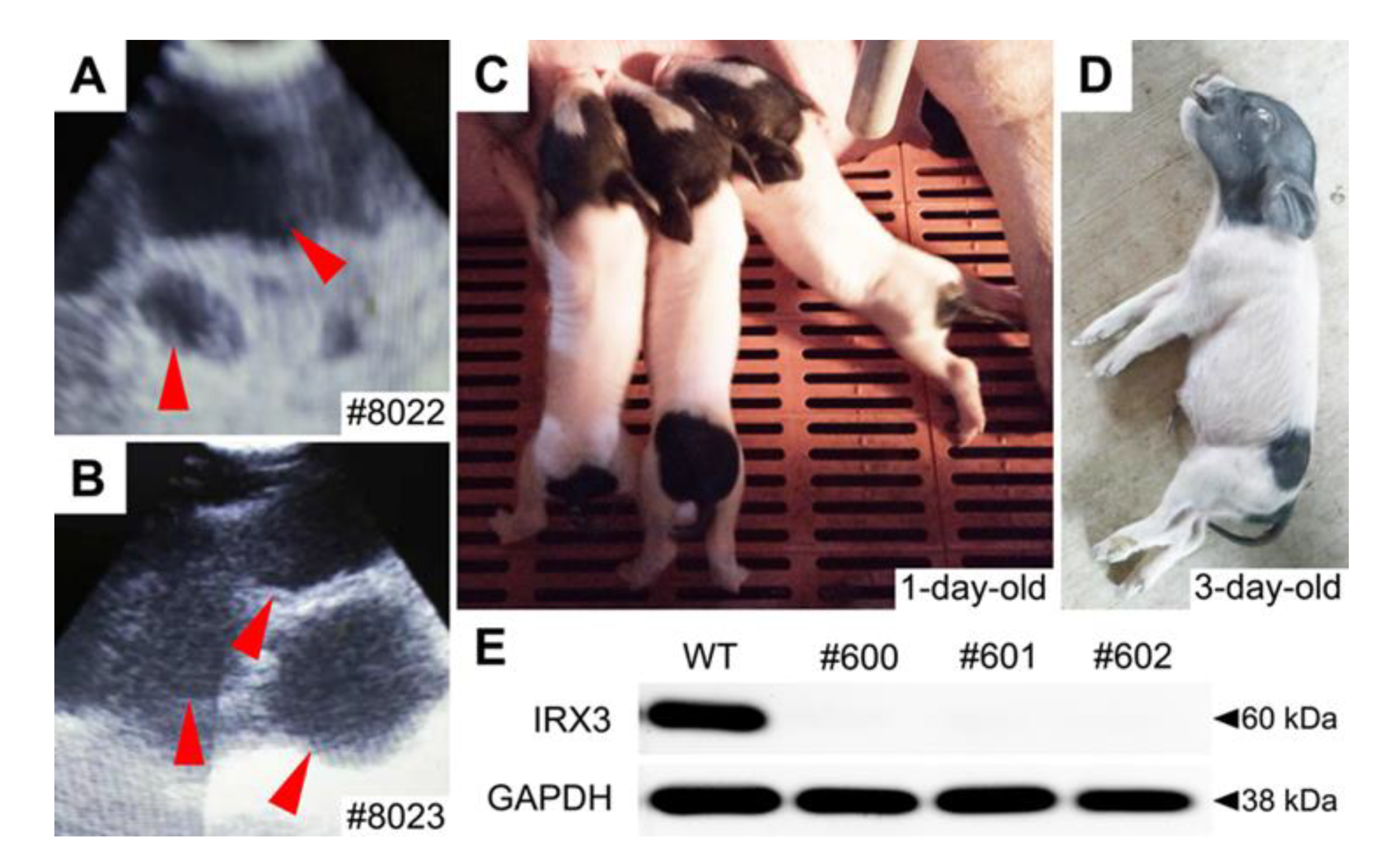
2.8. Identification of IRX3 Knockouts in Somatic Cell-Cloned Bama Minipigs
2.9. Statistical Analysis
3. Results
3.1. Design and Test of CRISPR/Cas9-Mediated IRX3 Gene Editing Target in Bama Minipig
3.2. Preparation of IRX3 Gene-Edited Bama Minipig Somatic Cells
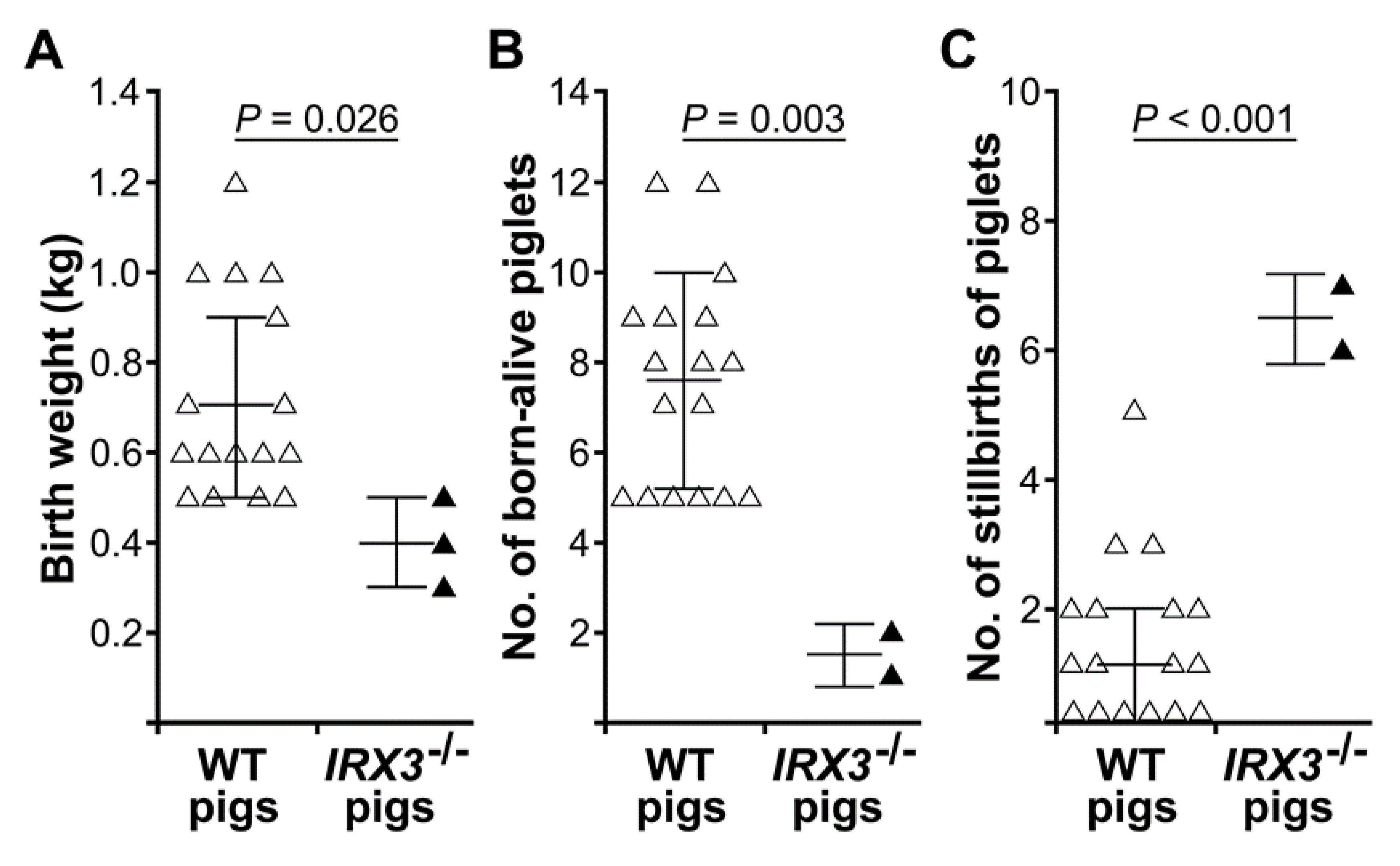
3.3. Biallelic Knockout of IRX3 Significantly Reduces the Production and Survival of Somatic Cell-Cloned Bama Minipigs
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, X.X.; Zhong, Y.Z.; Ge, Y.W.; Lu, K.H.; Lu, S.S. CRISPR/Cas9-mediated generation of Guangxi Bama minipigs harboring three mutations in α-synuclein causing Parkinson’s disease. Sci. Rep. 2018, 8, 12420. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.X.; Xu, J.; Chen-Tsai, R.Y.; Li, K. Genome editing in livestock: Are we ready for a revolution in animal breeding industry? Transgenic Res. 2017, 26, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B. Basics of genome editing technology and its application in livestock species. Reprod Domest Anim. 2017, 52 (Suppl. 3), 4–13. [Google Scholar] [CrossRef]
- Ryu, J.H.; Prather, R.S.; Lee, K. Use of gene-editing technology to introduce targeted modifications in pigs. J. Anim. Sci. Biotechno. 2018, 9, 5. [Google Scholar] [CrossRef]
- Zhao, J.G.; Lai, L.X.; Ji, W.Z.; Zhou, Q. Genome editing in large animals: Current status and future prospects. Natl. Sci. Rev. 2019, 6, 402–420. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013, 4, 220–228. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.F.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef]
- Li, W.; Teng, F.; Li, T.; Zhou, Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.Y.; Yuan, L.; Deng, J.C.; Chen, M.; Wang, Y.; Zeng, J.; Li, Z.J.; Lai, L.X. Efficient generation of myostatin gene mutated rabbit by CRISPR/Cas9. Sci. Rep. 2016, 6, 25029. [Google Scholar] [CrossRef]
- Sui, T.; Lau, Y.S.; Liu, D.; Liu, T.; Xu, L.; Gao, Y.; Lai, L.; Li, Z.; Han, R. A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis. Mod. Mech. 2018, 11, dmm032201. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.J.; Wang, X.M.; Liu, Y.Z.; Ouyang, Z.; Long, H.B.; Wei, S.; Xin, J.; Zhao, B.T.; Lai, S.; Shen, J.; et al. Generation of gene-target dogs using CRISPR/Cas9 system. Mol. Cell Biol. 2015, 7, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wang, X.M.; Shi, H.; Yan, Q.M.; Zheng, M.; Li, J.; Zhang, Q.J.; Qin, Y.M.; Zhong, Y.G.; Mi, J.; et al. Generation of ApoE deficient dogs via combination of embryo injection of CRISPR/Cas9 with somatic cell nuclear transfer. J. Genet. Genom. 2018, 45, 47–50. [Google Scholar] [CrossRef]
- Wang, K.K.; Ouyang, H.S.; Xie, Z.C.; Yao, C.G.; Guo, N.N.; Li, M.J.; Jiao, H.P.; Pang, D.X. Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 16623. [Google Scholar] [CrossRef]
- Wang, X.L.; Cao, C.W.; Huang, J.J.; Yao, J.; Hai, T.; Zheng, Q.T.; Wang, X.; Zhang, H.Y.; Qin, G.S.; Cheng, J.B.; et al. One-step generation of triple gene targeted pigs using CRISPR/Cas9 system. Sci. Rep. 2016, 5, 20620. [Google Scholar] [CrossRef]
- Bi, Y.Z.; Hua, Z.D.; Liu, X.M.; Hua, W.J.; Ren, H.Y.; Xiao, H.W.; Zhang, L.P.; Li, L.; Wang, Z.R.; Laible, G.; et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 2016, 6, 31729. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Rowland, R.R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; Mclaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef]
- Burkard, C.; Lillico, S.G.; Reid, E.; Jackson, B.; Mileham, A.J.; Ait-Ali, T.; Whitelaw, C.B.A.; Archibald, A.L. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017, 13, e1006206. [Google Scholar] [CrossRef]
- Xie, Z.C.; Pang, D.X.; Yuan, H.M.; Jiao, H.P.; Lu, C.; Wang, K.K.; Yang, Q.B.; Li, M.J.; Chen, X.; Yu, T.T.; et al. Genetically modified pigs are protected from classical swine fever virus. PLoS Pathog. 2018, 14, e1007193. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Wei, H.J.; Lin, L.; George, H.; Wang, T.; Lee, I.H.; Zhao, H.Y.; Wang, Y.; Kan, Y.N.; Shrock, E.; et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR/Cas9. Science 2017, 357, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.Y.; Shen, B.; Cui, Y.Q.; Chen, Y.C.; Wang, J.Y.; Wang, L.; Kang, Y.; Zhao, X.Y.; Si, W.; Li, W.; et al. modified cynomolgus Generation of gene- monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Liu, Z.; Wang, X.; Wang, Y.; Nie, Y.H.; Lai, L.; Sun, R.L.; Shi, L.Y.; Sun, Q.; Yang, H. Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing. Cell Res. 2018, 28, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.W.; Cai, Y.J.; Li, K.; Wei, Y.; Wang, B.A.; Sun, Y.D.; Liu, Z.; Liu, J.W.; Hu, X.D.; Wei, W.; et al. One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res. 2017, 27, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Tu, Z.C.; Liu, Z.M.; Fan, N.N.; Yang, H.M.; Yang, S.; Yang, W.L.; Zhao, Y.; Ouyang, Z.; Lai, C.D.; et al. A huntingtin knockin pig model rapitulates features of selective neurodegeneration in Huntington’s disease. Cell 2018, 173, 989–1002. [Google Scholar] [CrossRef]
- Fang, B.; Ren, X.Y.; Wang, Y.; Li, Z.; Zhao, L.H.; Zhang, M.L.; Li, C.; Zhang, Z.W.; Chen, L.; Li, X.X.; et al. Apolipoprotein E deficiency accelerates atherosclerosis development in miniature pigs. Dis. Model. Mech. 2018, 11, dmm036632. [Google Scholar] [CrossRef]
- Zheng, Q.T.; Lin, J.; Huang, J.J.; Zhang, H.Y.; Zhang, R.; Zhang, X.Y.; Cao, C.W.; Hambly, C.; Qin, G.S.; Yao, J.; et al. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proc. Natl. Acad. Sci. USA 2017, 114, 9474–9482. [Google Scholar] [CrossRef]
- Xiang, G.H.; Ren, J.L.; Hai, T.; Fu, R.; Yu, D.W.; Wang, J.; Li, W.; Wang, H.Y.; Zhou, Q. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell Mol. Life Sci. 2018, 75, 4619–4628. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, L.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO obesity variant circuitry and adipocyte browning in humans. New Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef]
- Lee, S.C.; Lee, H.; Oh, K.B.; Hwang, I.S.; Yang, H.; Park, M.R.; Ock, S.A.; Woo, J.S.; Im, G.S.; Hwang, S. Production and breeding of transgenic cloned pigs expressing human CD73. Dev. Reprod. 2017, 21, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M. Development of pig cloning studies: Past, present and future. J Anim. Feed Sci. 2004, 13, 211–238. [Google Scholar] [CrossRef]
- Callesen, M.M.; Árnadóttir, S.S.; Lyskjaer, I.; Ørntoft, M.W.; Høyer, S.; Dagnaes-Hansen, F.; Liu, Y.; Li, R.; Callesen, H.; Rasmussen, M.H.; et al. A genetically inducible porcine model of intestinal cancer. Mol Oncol. 2017, 11, 1616–1629. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. Transgenic mammalian species, generated by somatic cell cloning, in biomedicine, biopharmaceutical industry and human nutrition/dietetics–recent achievements. Pol. J. Vet. Sci. 2011, 14, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Skrzyszowska, M. The possibilities of practical application of transgenic mammalian species generated by somatic cell cloning in pharmacology, veterinary medicine and xenotransplantology. Pol. J. Vet. Sci. 2011, 14, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.A.; Lee, E.M.; Cho, B.; Alam, Z.; Kim, S.J.; Lee, S.; Oh, H.J.; Hwang, J.I.; Ahn, C.; Lee, B.C. Generation by somatic cell nuclear transfer of GGTA1 knockout pigs expressing soluble human TNFRI-Fc and human HO-1. Transgenic Res. 2019, 28, 91–102. [Google Scholar] [CrossRef]
- Lee, J.; Lee, Y.; Lee, G.S.; Lee, S.T.; Lee, E. Comparative study of the developmental competence of cloned pig embryos derived from spermatogonial stem cells and fetal fibroblasts. Reprod Domest Anim. 2019, 54, 1258–1264. [Google Scholar] [CrossRef]
- Opiela, J.; Samiec, M.; Romanek, J. In vitro development and cytological quality of inter-species (porcine bovine) cloned embryos are affected by trichostatin A-dependent epigenomic modulation of adult mesenchymal stem cells. Theriogenology 2017, 97, 27–33. [Google Scholar] [CrossRef]
- Li, Z.; He, X.; Chen, L.; Shi, J.; Zhou, R.; Xu, W.; Liu, D.; Wu, Z. Bone marrow mesenchymal stem cells are an attractive donor cell type for production of cloned pigs as well as genetically modified cloned pigs by somatic cell nuclear transfer. Cell Reprogram. 2013, 15, 459–470. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. Preimplantation developmental capability of cloned pig embryos derived from different types of nuclear donor somatic cells. Ann Anim Sci. 2010, 10, 385–398. [Google Scholar]
- Wang, X.F.; Zhu, X.X.; Liang, X.W.; Xu, H.Y.; Liao, Y.Y.; Lu, K.H.; Lu, S.S. Effects of resveratrol on in vitro maturation of porcine oocytes and subsequent early embryonic development following somatic cell nuclear transfer. Reprod Domest Anim. 2019, 54, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cui, W.; Meng, C.; Zhang, J.; Li, Y.; Qian, Y.; Xing, G.; Zhao, D.; Cao, S. MC1568 enhances histone acetylation during oocyte meiosis and improves development of somatic cell nuclear transfer embryos in pig. Cell Reprogram. 2018, 20, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Skrzyszowska, M. High developmental capability of porcine cloned embryos following trichostatin A-dependent epigenomic transformation during in vitro maturation of oocytes pre-exposed to R-roscovitine. Anim. Sci. Pap. Rep. 2012, 30, 383–393. [Google Scholar]
- Gupta, M.K.; Heo, Y.T.; Kim, D.K.; Lee, H.T.; Uhm, S.J. 5-Azacytidine improves the meiotic maturation and subsequent in vitro development of pig oocytes. Anim. Reprod Sci. 2019, 208, 106118. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. Biological transcomplementary activation as a novel and effective strategy applied to the generation of porcine somatic cell cloned embryos. Reprod Biol. 2014, 14, 128–139. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. The use of different methods of oocyte activation for generation of porcine fibroblast cell nuclear-transferred embryos. Ann. Anim. Sci. 2010, 10, 399–411. [Google Scholar]
- Bang, J.I.; Yoo, J.G.; Park, M.R.; Shin, T.S.; Cho, B.W.; Lee, H.G.; Kim, B.W.; Kang, T.Y.; Kong, I.K.; Kim, J.H.; et al. The effects of artificial activation timing on the development of SCNT-derived embryos and newborn piglets. Reprod Biol. 2013, 13, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Skrzyszowska, M. Roscovitine is a novel agent that can be used for the activation of porcine oocytes reconstructed with adult cutaneous or fetal fibroblast cell nuclei. Theriogenology 2012, 78, 1855–1867. [Google Scholar] [CrossRef]
- Song, X.; Liu, Z.; He, H.; Wang, J.; Li, H.; Li, J.; Li, F.; Jiang, Z.; Huan, Y. Dnmt1s in donor cells is a barrier to SCNT-mediated DNA methylation reprogramming in pigs. Oncotarget 2017, 8, 34980–34991. [Google Scholar] [CrossRef]
- Jin, L.; Guo, Q.; Zhang, G.L.; Xing, X.X.; Xuan, M.F.; Luo, Q.R.; Luo, Z.B.; Wang, J.X.; Yin, X.J.; Kang, J.D. The histone deacetylase inhibitor, CI994, improves nuclear reprogramming and in vitro developmental potential of cloned pig embryos. Cell. Reprogram 2018, 20, 205–213. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. Intrinsic and extrinsic molecular determinants or modulators for epigenetic remodeling and reprogramming of somatic cell-derived genome in mammalian nuclear-transferred oocytes and resultant embryos. Pol. J. Vet. Sci. 2018, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Opiela, J.; Lipiński, D.; Romanek, J. Trichostatin A-mediated epigenetic transformation of adult bone marrow-derived mesenchymal stem cells biases the in vitrodevelopmental capability, quality, and pluripotency extent of porcine cloned embryos. Biomed. Res. Int. 2015, 814686. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, H.; Yao, J.; Qin, G.; Wang, F.; Wang, X.; Luo, A.; Zheng, Q.; Cao, C.; Zhao, J. BIX-01294 increases pig cloning efficiency by improving epigenetic reprogramming of somatic cell nuclei. Reproduction 2016, 151, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Skrzyszowska, M. Can reprogramming of overall epigenetic memory and specific parental genomic imprinting memory within donor cell-inherited nuclear genome be a major hindrance for the somatic cell cloning of mammals?–a review. Ann. Anim. Sci. 2018, 18, 623–638. [Google Scholar] [CrossRef]
- Takeda, K.; Tasai, M.; Iwamoto, M.; Akita, T.; Tagami, T.; Nirasawa, K.; Hanada, H.; Onishi, A. Transmission of mitochondrial DNA in pigs and progeny derived from nuclear transfer of Meishan pig fibroblast cells. Mol. Reprod Dev. 2006, 73, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M. The role of mitochondrial genome (mtDNA) in somatic and embryo cloning of mammals. A review. J Anim Feed Sci. 2005, 14, 213–233. [Google Scholar] [CrossRef]
- St John, J.C.; Moffatt, O.; D’Souza, N. Aberrant heteroplasmic transmission of mtDNA in cloned pigs arising from double nuclear transfer. Mol. Reprod Dev. 2005, 72, 450–460. [Google Scholar] [CrossRef]
- Samiec, M. The effect of mitochondrial genome on architectural remodeling and epigenetic reprogramming of donor cell nuclei in mammalian nuclear transfer-derived embryos. J Anim Feed Sci. 2005, 14, 393–422. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.; Wu, Y.; Si, J.; Huang, Y.; Jiang, Q.; Lan, G.; Guo, Y.; Jiang, H. Scriptaid upregulates expression of development-related genes, inhibits apoptosis, and improves the development of somatic cell nuclear transfer mini-pig embryos. Cell Reprogram. 2017, 19, 19–26. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. Assessment of in vitro developmental capacity of porcine nuclear-transferred embryos reconstituted with cumulus oophorus cells undergoing vital diagnostics for apoptosis detection. Ann. Anim. Sci. 2013, 13, 513–529. [Google Scholar] [CrossRef]
- Jin, L.; Guo, Q.; Zhu, H.Y.; Xing, X.X.; Zhang, G.L.; Xuan, M.F.; Luo, Q.R.; Luo, Z.B.; Wang, J.X.; Yin, X.J.; et al. Quisinostat treatment improves histone acetylation and developmental competence of porcine somatic cell nuclear transfer embryos. Mol. Reprod Dev. 2017, 84, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Skrzyszowska, M.; Opiela, J. Creation of cloned pig embryos using contact-inhibited or serum-starved fibroblast cells analysed intra vitam for apoptosis occurrence. Ann. Anim. Sci. 2013, 13, 275–293. [Google Scholar] [CrossRef]
- Lin, T.; Lee, J.E.; Oqani, R.K.; Kim, S.Y.; Cho, E.S.; Jeong, Y.D.; Baek, J.J.; Jin, D.I. Tauroursodeoxycholic acid improves pre-implantation development of porcine SCNT embryo by endoplasmic reticulum stress inhibition. Reprod. Biol. 2016, 16, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Romanek, J.; Lipiński, D.; Opiela, J. Expression of pluripotency-related genes is highly dependent on trichostatin A-assisted epigenomic modulation of porcine mesenchymal stem cells analysed for apoptosis and subsequently used for generating cloned embryos. Anim. Sci. J. 2019, 90, 1127–1141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qu, P.; Ma, X.; Qiao, F.; Ma, Y.; Qing, S.; Zhang, Y.; Wang, Y.; Cui, W. Tauroursodeoxycholic acid (TUDCA) alleviates endoplasmic reticulum stress of nuclear donor cells under serum starvation. PLoS ONE 2018, 13, e0196785. [Google Scholar] [CrossRef] [PubMed]
- Smemo, S.; Tena, J.J.; Kim, K.H.; Gamazon, E.R.; Sakabe, N.J.; Gómez-Marín, C.; Anease, I.; Credidio, F.L.; Sobreira, D.R.; Wasserman, N.F.; et al. Obesity-associated variants within FTO form long range functional connections with IRX3. Nature 2014, 507, 371–375. [Google Scholar] [CrossRef]
- Zhu, X.X.; Nie, J.Y.; Quan, S.N.; Xu, H.Y.; Yang, X.G.; Lu, Y.Q.; Lu, K.H.; Lu, S.S. In vitro production of cloned and transgenic cloned embryos from Guangxi Huangjiang Xiang pig. Vitro Cell Dev. Biol. Ann. 2016, 52, 137–143. [Google Scholar] [CrossRef]
- Nie, J.Y.; Zhu, X.X.; Xie, B.K.; Nong, S.Q.; Ma, Q.Y.; Xu, H.Y.; Yang, X.G.; Lu, Y.Q.; Lu, K.H.; Liao, Y.Y.; et al. Successful cloning of an adult breeding boar from the novel Chinese Guike No. 1 swine specialized strain. 3 Biotech. 2016, 6, 218. [Google Scholar] [CrossRef]
- Zhu, X.X.; Zhong, Y.Z.; Ge, Y.W.; Lu, K.H.; Lu, S.S. Generation of transgenic-cloned Huanjiang Xiang pigs systemically expressing enhanced green fluorescent protein. Reprod. Domest. Anim. 2018, 53, 1546–1554. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, J.Y.; Lu, K.H.; Lu, S.S.; Zhu, X.X. Efficient generation of GHR knockout Bama minipig fibroblast cells using CRISPR/Cas9-mediated gene editing. Vitro Cell Dev Biol Ann. 2019, 55, 784–792. [Google Scholar] [CrossRef]
- Younis, S.; Schonke, M.; Massart, J.; Hjortebjerg, R.; Sundstrom, E.; Gustafson, U.; Bjornholm, M.; Krook, A.; Frystyk, J.; Zierath, J.R.; et al. The ZBED6-IGF2 axis has a major effect on growth of skeletal muscle and internal organs in placental mammals. Proc. Natl. Acad. Sci. USA 2017, 115, 2048–2057. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Liu, H.B.; Wang, M.; Li, R.Q.; Zeng, J.H.; Mo, D.; Cong, P.Q.; Liu, X.H.; Chen, Y.S.; He, Z.Y. Disruption of the ZBED6 binding site in intron 3 of IGF2 by CRISPR/Cas9 leads to enhanced muscle development in liang guang small spotted pigs. Transgenic Res. 2018, 28, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.S.; Kim, K.H.; Rosen, A.; Smyth, J.W.; Sakuma, R.; Delgado-Olguin, P.; Davis, M.; Chi, N.C.; Puviindran, V.; Gaborit, N.; et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac his-purkinje network. Proc. Natl. Acad. Sci. USA. 2011, 108, 13576–13581. [Google Scholar] [CrossRef]
- Kim, K.H.; Rosen, A.; Bruneau, B.G.; Hui, C.C.; Backx, P.H. Iroquois homeodomain transcription factors in heart development and function. Circ. Res. 2012, 110, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
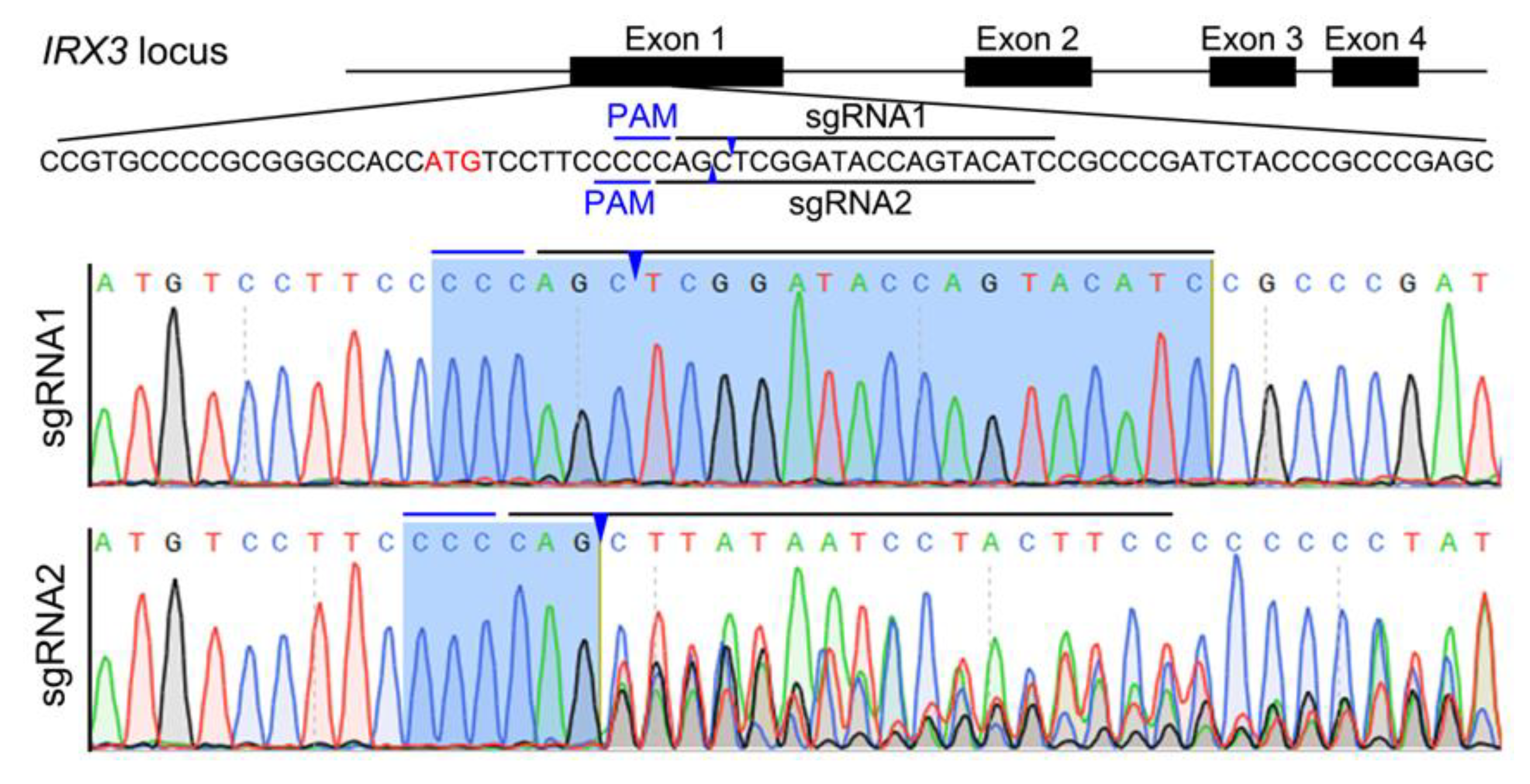
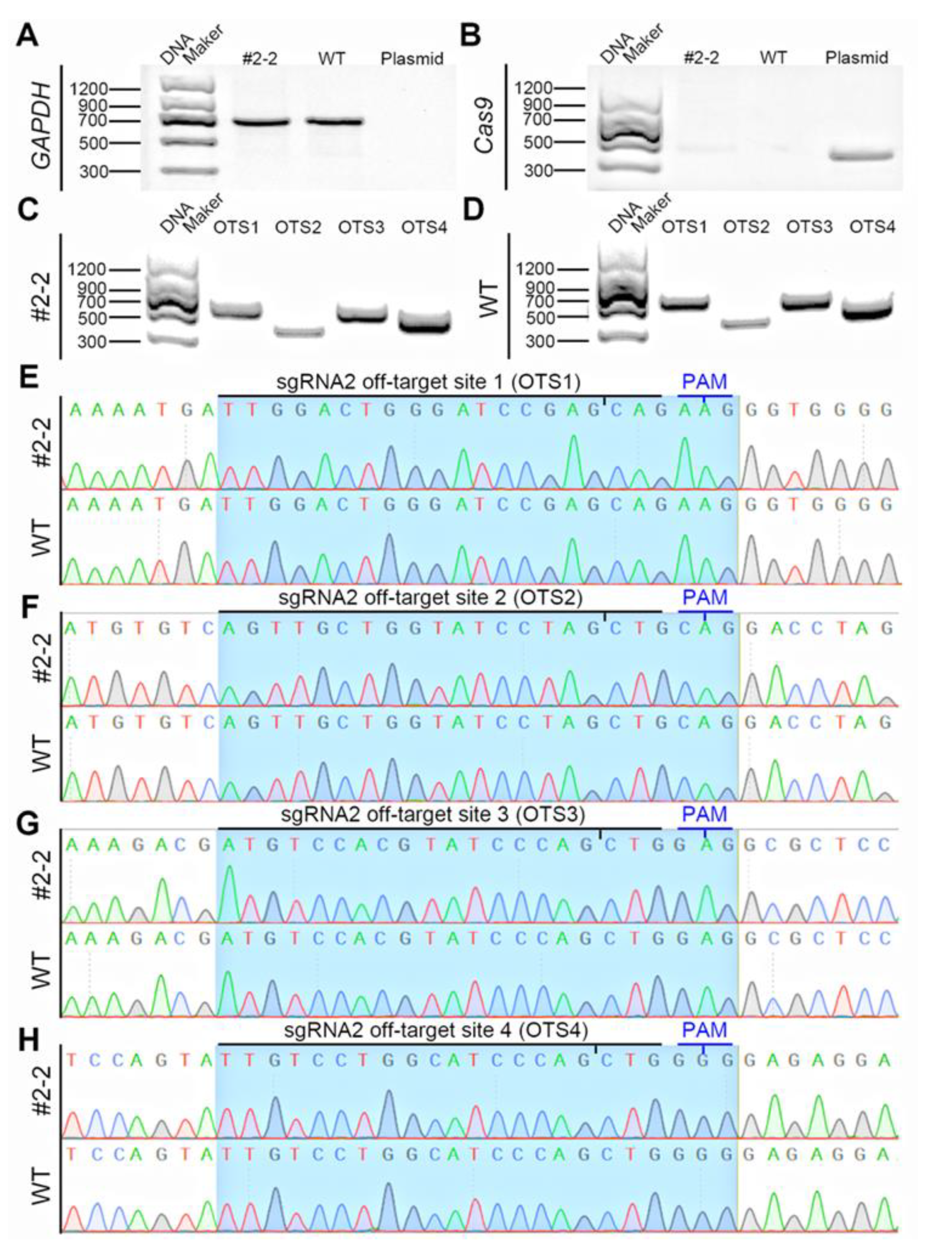

| OTS No. | Chr. | Strand | Position | Sequence * | Score | Gene | Primers for PCR and Sequencing | Amplicon (bp) |
|---|---|---|---|---|---|---|---|---|
| sgRNA2 | Chr6 | −1 | 31046354 | ATGTACTGGTATCCGAGCTGGGG | 100 | NCBI Gene ID: 100518611 | PF: AGCAGATCAATAGGCGAACG PR: CTGTCCTTCAGCTCATACTG | 617 |
| OTS1 | Chr13 | 1 | 116315529 | TTGGACTGGGATCCGAGCAGAAG | 0.49 | NCBI Gene ID: 106505748 | PF: TCAACTTCTCGCATGGTGTG PR: TGACTAGCAACTTCAGAGGC | 505 |
| OTS2 | Chr14 | −1 | 34640565 | AGTTGCTGGTATCCTAGCTGCAG | 0.40 | NCBI Gene ID: 100153965 | PF: TCTCCAGGTATTATCAGGAGT PR: CACATCCTATAAAGCTCAGTC | 347 |
| OTS3 | Chr12 | 1 | 53174151 | ATGTCCACGTATCCCAGCTGGAG | 0.27 | NCBI Gene ID: 100523637 | PF: AAGTCGGAGAGGTTGGTATC PR: TTCCTACAGAGCAGAAACCG | 500 |
| OTS4 | Chr17 | 1 | 56522870 | TTGTCCTGGCATCCCAGCTGGGG | 0.38 | None | PF: AACCTAGAGCTGTGGACAAC PR: CTCCAACACTTGTAGCCTTG | 451 |
| Colony No. | Allele | IRX3 Genotypes * | Indels |
|---|---|---|---|
| Wild type | 1/2 | ATGTCCTTCCCCCAGCTCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | WT/WT |
| #2-1 | 1 | ATGTCCTTCCCCCAG------------TACATCCGCCCGATCTACCCGCCCGAGC | -12 |
| 2 | ATGTCCTTCCCCCAGCCTCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAG | +1 | |
| #2-2 | 1 | ATGTCCTTCCCCCAG-TCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | -1 |
| 2 | ATGTCCTTCCCCCAGTCTCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAG | +1 | |
| #2-3 | 1/2 | ATGTCCTTCCCCCAG------------TACATCCGCCCGATCTACCCGCCCGAGC | -12/-12 |
| #2-4 | 1/2 | ATGTCCTTCCCCCAG------------TACATCCGCCCGATCTACCCGCCCGAGC | -12/-12 |
| #2-5 | 1/2 | ATGTCCTTCCCCCAGCTCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | WT/WT |
| #2-6 | 1 | ATGTCCTTCC------TCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | -6 |
| 2 | ATGTCCTTCC---------GATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | -9 | |
| #2-7 | 1/2 | ATG-------------TCGGATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | -13/-13 |
| #2-8 | 1 | ATGTCCTTCCCCCAG--CGGATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | -2 |
| 2 | ATGTCCTTCCCCCAG-----ATACCAGTACATCCGCCCGATCTACCCGCCCGAGC | -5 |
| Recipient Sow | Donor Cells | Gender | No. of Embryos Transferred | Day 40 Pregnancy Status * | Gestational Period(day) | No. of Piglets Delivered(Alive/Stillborn) | Cloning Efficiency(%) ** | Birth Weight(Kg) |
|---|---|---|---|---|---|---|---|---|
| #8024 | IRX-/- | ♂ | 250 | - | ||||
| #8025 | IRX-/- | ♂ | 250 | - | ||||
| #8022 | IRX-/- | ♂ | 250 | + | 117 | 1/6 | ||
| #8023 | IRX-/- | ♂ | 250 | + | 116 | 2/7 | ||
| Sum | ♂ | 1000 | 3/13 | 0.3% | 0.3; 0.4; 0.5 | |||
| #1757 | WT | ♂ | 280 | + | N/A | 9/0 | 0.6; 1.0; 0.6; 0.6; 0.7; 1.0; 0.5; 0.6; 1.2 | |
| #0205 | WT | ♂ | 230 | + | N/A | 7/1 | 1.0; 0.5; 0.6; 0.9; 0.7; 0.5; 0.5 | |
| #4993 | WT | ♂ | 230 | + | N/A | 5/2 | N/A | |
| #0207 | WT | ♂ | 212 | + | N/A | 8/0 | N/A | |
| #2020 | WT | ♂ | 246 | + | N/A | 10/2 | N/A | |
| #4998 | WT | ♂ | 243 | + | N/A | 12/1 | N/A | |
| #4990 | WT | ♂ | 240 | + | N/A | 12/0 | N/A | |
| #2021 | WT | ♂ | 230 | + | N/A | 9/0 | N/A | |
| #0706 | WT | ♂ | 230 | + | N/A | 8/2 | N/A | |
| #7388 | WT | ♂ | 235 | + | N/A | 7/1 | N/A | |
| #1801 | WT | ♂ | 220 | + | N/A | 5/0 | N/A | |
| #1123 | WT | ♂ | 220 | + | N/A | 9/0 | N/A | |
| #1215 | WT | ♂ | 220 | + | N/A | 5/5 | N/A | |
| #1802 | WT | ♂ | 220 | + | N/A | 5/3 | N/A | |
| #0207 | WT | ♂ | 280 | + | N/A | 8/0 | N/A | |
| #0208 | WT | ♂ | 280 | + | N/A | 5/3 | N/A | |
| #0307 | WT | ♂ | 280 | + | N/A | 5/2 | N/A | |
| Sum | 4096 | 129/23 | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Wei, Y.; Zhan, Q.; Yan, A.; Feng, J.; Liu, L.; Tang, D. CRISPR/Cas9-Mediated Biallelic Knockout of IRX3 Reduces the Production and Survival of Somatic Cell-Cloned Bama Minipigs. Animals 2020, 10, 501. https://doi.org/10.3390/ani10030501
Zhu X, Wei Y, Zhan Q, Yan A, Feng J, Liu L, Tang D. CRISPR/Cas9-Mediated Biallelic Knockout of IRX3 Reduces the Production and Survival of Somatic Cell-Cloned Bama Minipigs. Animals. 2020; 10(3):501. https://doi.org/10.3390/ani10030501
Chicago/Turabian StyleZhu, Xiangxing, Yanyan Wei, Qunmei Zhan, Aifen Yan, Juan Feng, Lian Liu, and Dongsheng Tang. 2020. "CRISPR/Cas9-Mediated Biallelic Knockout of IRX3 Reduces the Production and Survival of Somatic Cell-Cloned Bama Minipigs" Animals 10, no. 3: 501. https://doi.org/10.3390/ani10030501
APA StyleZhu, X., Wei, Y., Zhan, Q., Yan, A., Feng, J., Liu, L., & Tang, D. (2020). CRISPR/Cas9-Mediated Biallelic Knockout of IRX3 Reduces the Production and Survival of Somatic Cell-Cloned Bama Minipigs. Animals, 10(3), 501. https://doi.org/10.3390/ani10030501




