Rumen Methanogenesis, Rumen Fermentation, and Microbial Community Response to Nitroethane, 2-Nitroethanol, and 2-Nitro-1-Propanol: An In Vitro Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Nitrocompounds
2.2. Animals
2.3. In Vitro Experiment
2.4. Sampling
2.5. Measurement of VFA, Coenzyme, and Nitrocompound Contents
2.6. Analysis of mcrA Gene Expression
2.7. Microbial Population Analysis with Real-Time PCR
2.8. Data Calculation and Statistical Analysis
3. Results
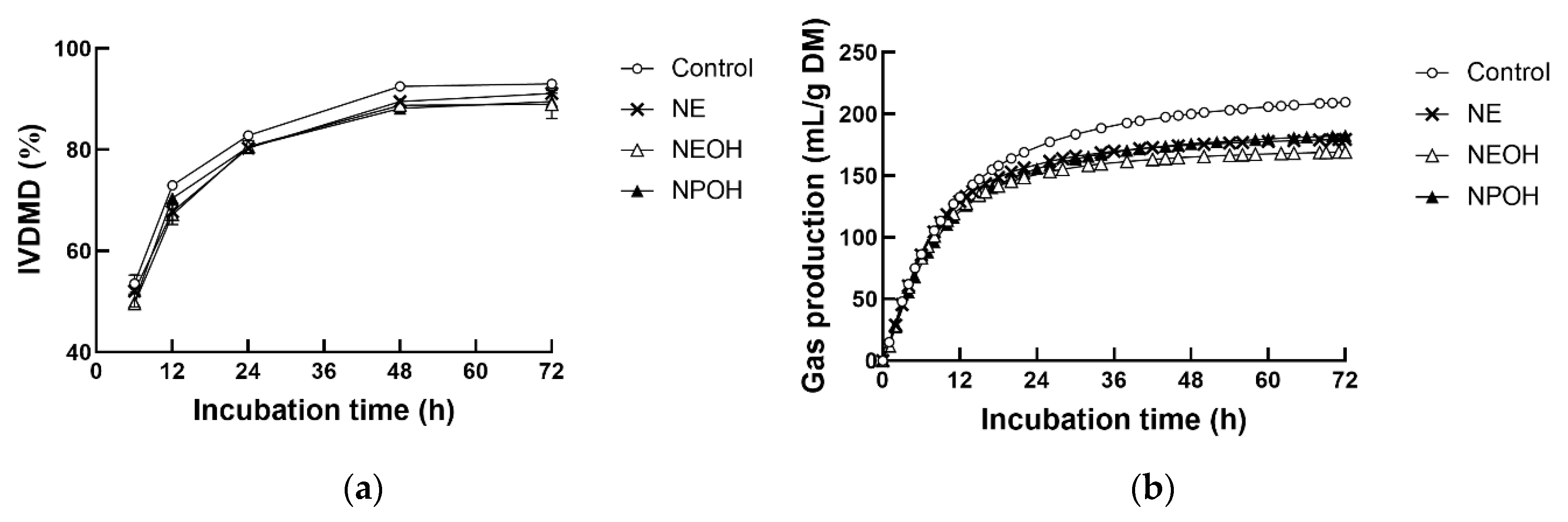
3.1. IVDMD and Gas Production Kinetics
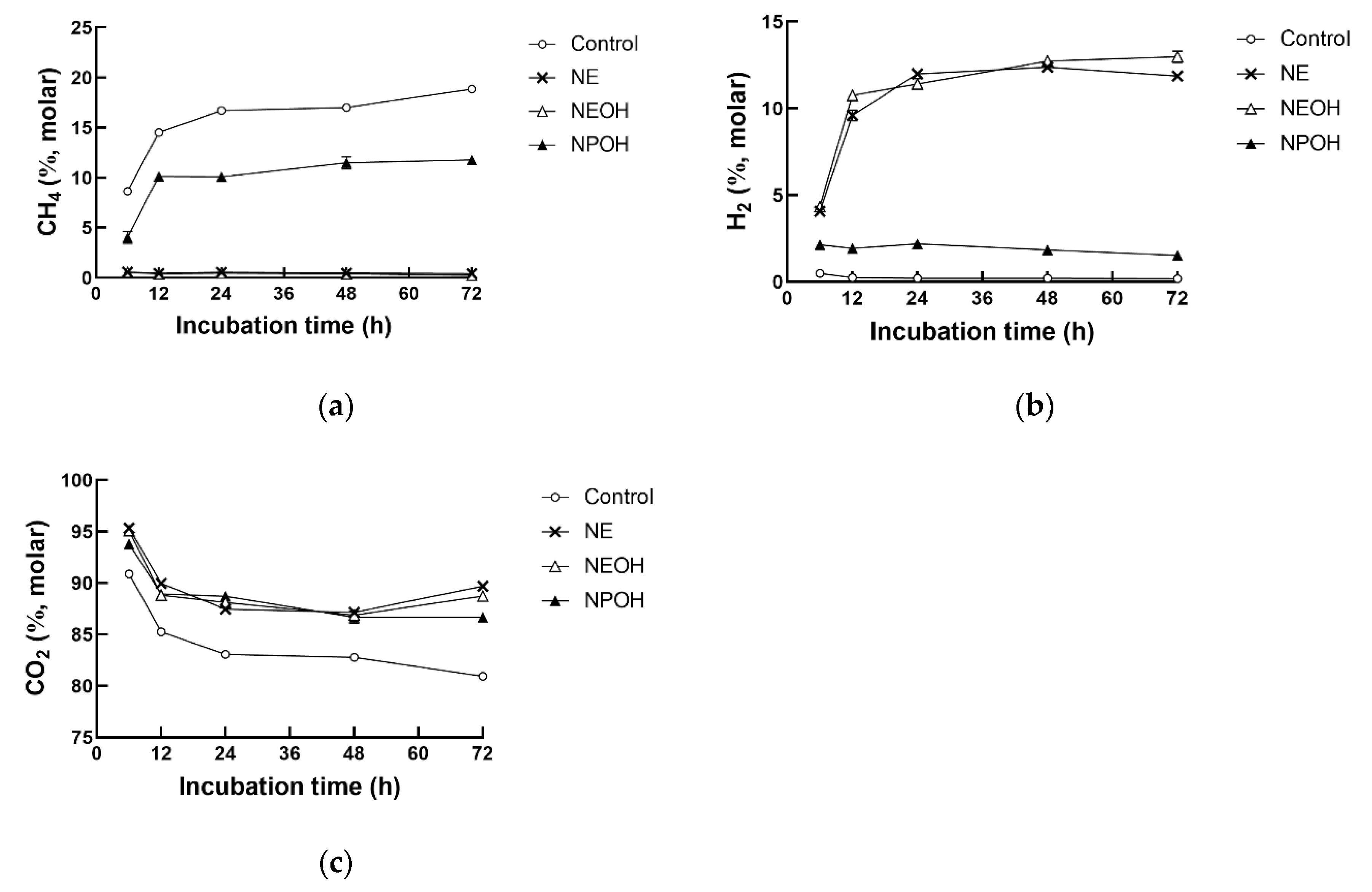
3.2. Fermentation Gas Composition
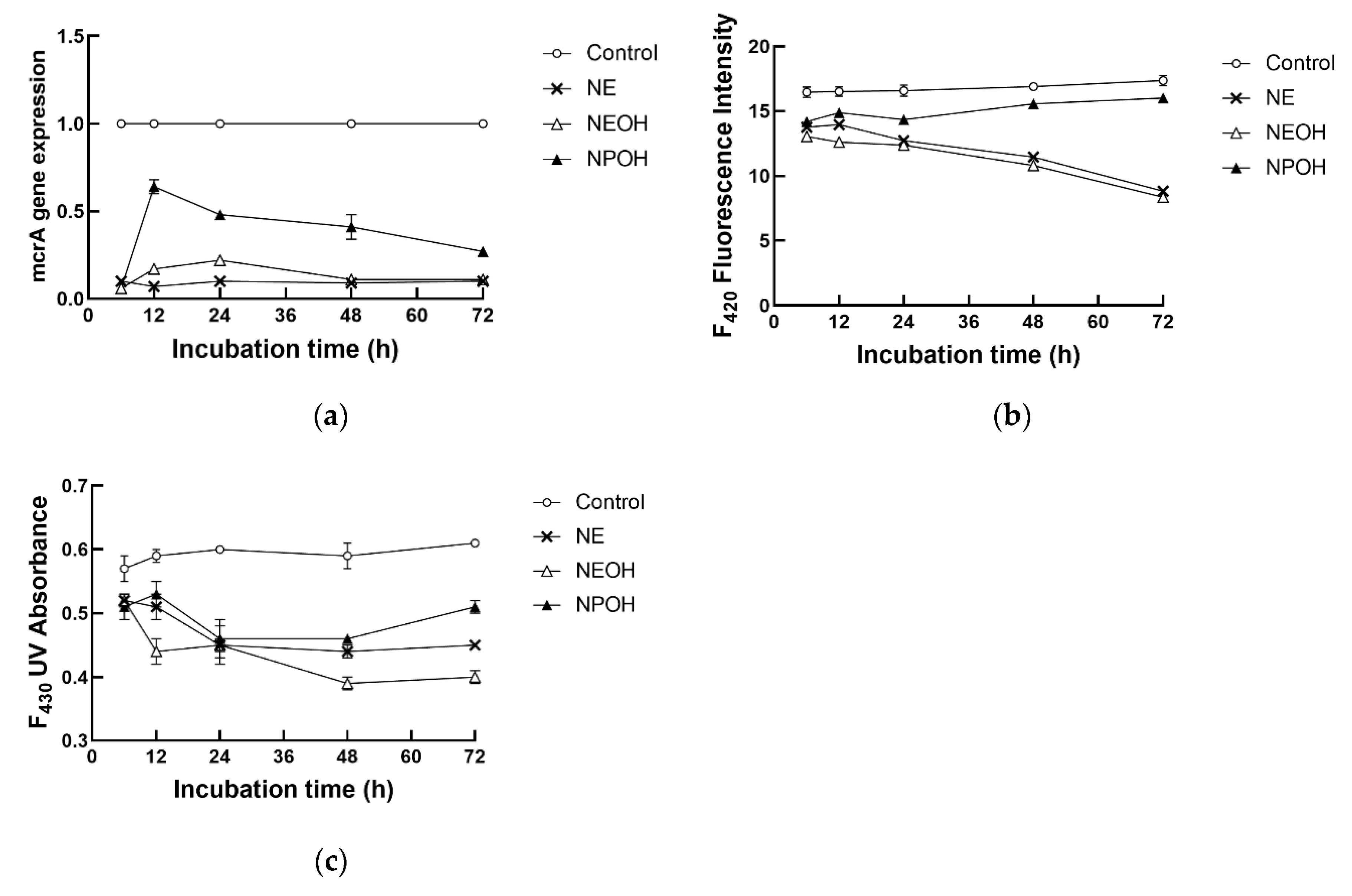
3.3. Coenzymes Related to CH4 Production
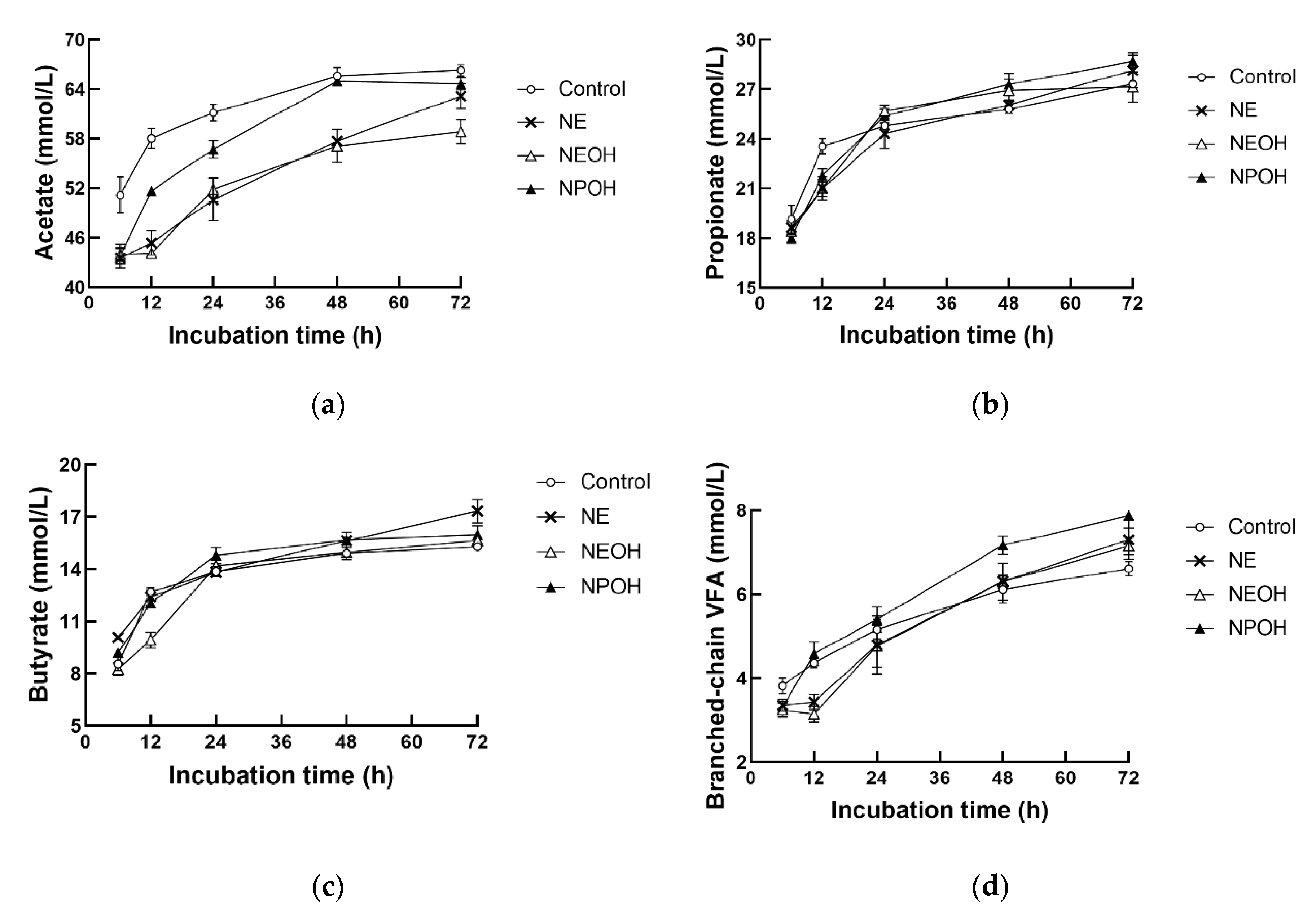
3.4. Fermentation Characteristics
3.5. Microbial Populations
3.6. Correlation between CH4 Production and mcrA Gene Expression/Coenzyme Contents
3.7. Disappearance of Nitrocompounds
4. Discussion
4.1. Effects of Nitrocompounds on IVDMD and Gas Production
4.2. Effects of Nitrocompounds on CH4 Production
4.3. Effects of Nitrocompounds on VFA Production
4.4. Effects of Nitrocompounds on Methane Production and Related Changes in Microbial Populations and Coenzyme Contents
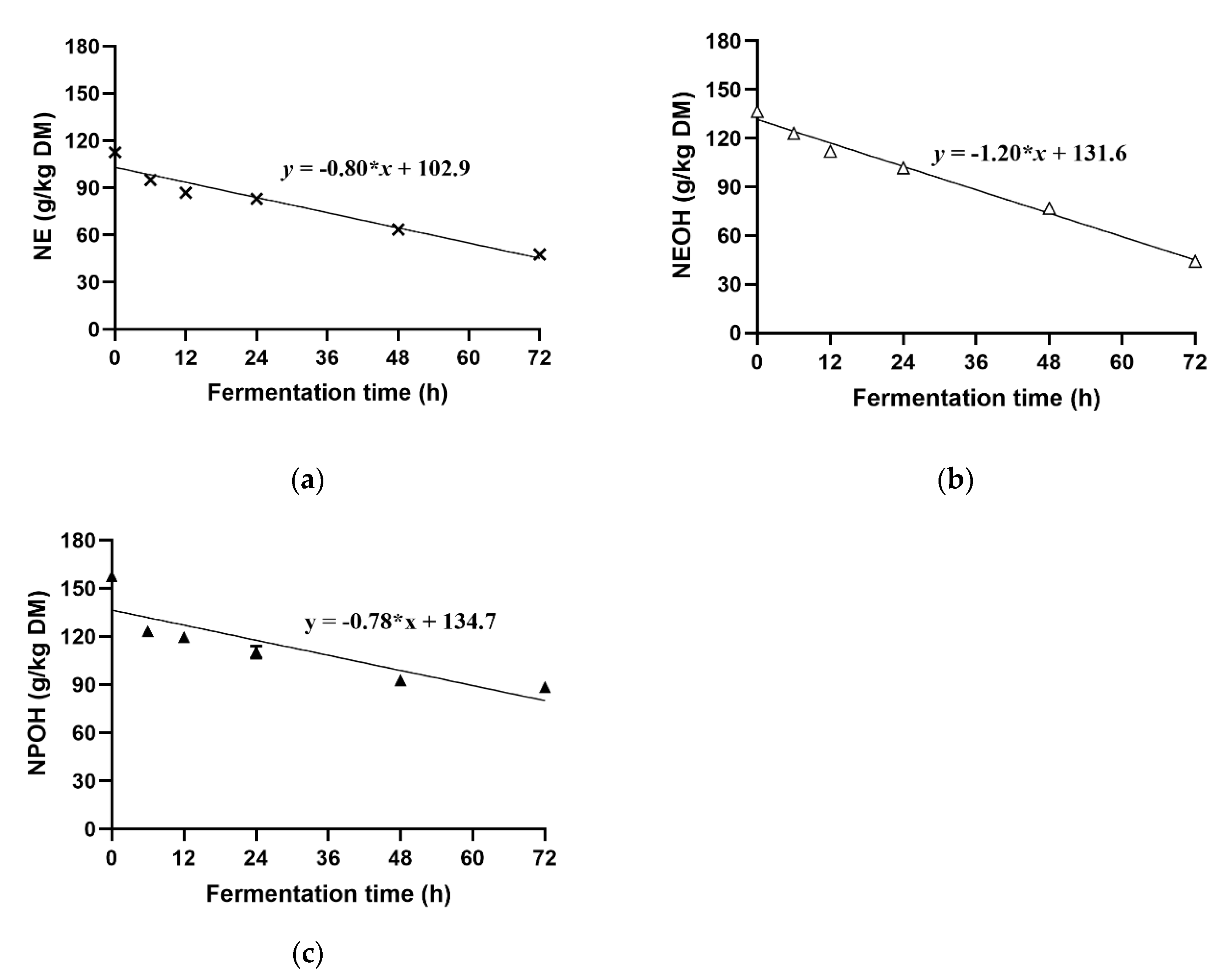
4.5. Degradation of NE, NEOH, and NPOH
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Anderson, R.C.; Majak, W.; Rassmussen, M.A.; Callaway, T.R.; Beier, R.C.; Nisbet, D.J.; Allison, M.J. Toxicity and metabolism of the conjugates of 3-nitropropanol and 3-nitropropionic acid in forages poisonous to livestock. J. Agric. Food Chem. 2005, 53, 2344–2350. [Google Scholar] [CrossRef] [PubMed]
- Latham, E.A.; Anderson, R.C.; Pinchak, W.E.; Nisbet, D.J. Insights on alterations to the rumen ecosystem by nitrate and nitrocompounds. Front. Microbiol. 2016, 7, 228. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.C.; Carstens, G.E.; Miller, R.K.; Callaway, T.R.; Schultz, C.L.; Edrington, T.S.; Harvey, R.B.; Nisbet, D.J. Effect of oral nitroethane and 2-nitropropanol administration on methane-producing activity and volatile fatty acid production in the ovine rumen. Bioresour. Technol. 2006, 97, 2421–2426. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Bañuelos, H.; Anderson, R.C.; Carstens, G.E.; Slay, L.J.; Ramlachan, N.; Horrocks, S.M.; Callaway, T.R.; Edrington, T.S.; Nisbet, D.J. Zoonotic bacterial populations, gut fermentation characteristics and methane production in feedlot steers during oral nitroethane treatment and after the feeding of an experimental chlorate product. Anaerobe. 2007, 13, 21–31. [Google Scholar] [CrossRef]
- Gutierrez-Banuelos, H.; Anderson, R.C.; Carstens, G.E.; Tedeschi, L.O.; Pinchak, W.E.; Cabrera-Diaz, E.; Krueger, N.A.; Callaway, T.R.; Nisbet, D.J. Effects of nitroethane and monensin on ruminal fluid fermentation characteristics and nitrocompound-metabolizing bacterial populations. J. Agric. Food Chem. 2008, 56, 4650–4658. [Google Scholar] [CrossRef]
- Brown, E.G.; Anderson, R.C.; Carstens, G.E.; Gutierrez-Bañuelos, H.; McReynolds, J.L.; Slay, L.J.; Callaway, T.R.; Nisbet, D.J. Effects of oral nitroethane administration on enteric methane emissions and ruminal fermentation in cattle. Anim. Feed Sci. Technol. 2011, 166–167, 275–281. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Giallongo, F.; Frederick, T.W.; Harper, M.T.; Weeks, H.L.; Brancob, A.F.; Moatec, P.J.; Deightonc, M.H.; Williamsc, R.O.S.; et al. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc. Natl. Acad. Sci. USA 2015, 112, 10663–10668. [Google Scholar] [CrossRef]
- Hendrickson, E.L.; Leigh, J.A. Roles of coenzyme F420-reducing hydrogenases and hydrogen- and F420-dependent methylenetetrahydromethanopterin dehydrogenases in reduction of F420 and production of hydrogen during methanogenesis. J. Bacteriol. 2008, 190, 4818. [Google Scholar] [CrossRef]
- Wongnate, T.; Sliwa, D.; Ginovska, B.; Smith, D.; Wolf, M.W.; Lehnert, N.; Raugei, S.; Ragsdale, S.W. The radical mechanism of biological methane synthesis by methyl-coenzyme M reductase. Science 2016, 352, 953–958. [Google Scholar] [CrossRef]
- Zheng, K.; Ngo, P.D.; Owens, V.L.; Yang, X.P.; Mansoorabadi, S.O. The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science 2016, 354, 339–342. [Google Scholar] [CrossRef]
- Zhang, D.F.; Yang, H.J. Combination effects of nitrocompounds, pyromellitic diimide, and 2-bromoethanesulfonate on in vitro ruminal methane production and fermentation of a grain-rich feed. J. Agric. Food Chem. 2012, 60, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Menke, K.H.; Steingass, H. Estimation of the energetic feed value obtained by chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Dev. 1988, 28, 7–55. [Google Scholar]
- Reuter, B.W.; Egeler, T.; Schneckenburger, H.; Schoberth, S.M. In vivo measurement of F420 fluorescence in cultures of Methanobacterium thermoautotrophicum. J. Bacteriol. 1986, 4, 325–332. [Google Scholar] [CrossRef]
- Ellefson, W.L.; Whitman, W.B.; Wolfe, R.S. Nickel-containing factor F430: Chromophore of the methylreductase of Methanobacterium. Proc. Natl. Acad. Sci. USA 1982, 79, 3707–3710. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Garcia, P.A.; Arevalos-Sanchez, M.M.; Ruiz-Barrera, O.; Anderson, R.C.; Maynez-Perez, A.O.; Rodriguez-Almeida, F.A.; Chavez-Martinez, A.; Gutierrez-Banuelos, H.; Corral-Luna, A. In vitro reduction of methane production by 3-nitro-1-propionic acid is dose-dependent. J. Anim. Sci. 2019, 97, 1317–1324. [Google Scholar] [CrossRef]
- Guo, Y.Q.; Liu, J.X.; Lu, Y.; Zhu, W.Y.; Denman, S.E.; McSweeney, C.S. Effect of tea saponin on methanogenesis, microbial community structure and expression of mcrA gene, in cultures of rumen micro-organisms. Lett. Appl. Microbiol. 2008, 47, 421–426. [Google Scholar] [CrossRef]
- Denman, S.E.; Tomkins, N.W.; McSweeney, C.S. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 2007, 62, 313–322. [Google Scholar] [CrossRef]
- Denman, S.E.; McSweeney, C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 2006, 58, 572–582. [Google Scholar] [CrossRef]
- Zhou, M.; Hernandez-Sanabria, E.; Guan, L.L. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 2009, 75, 6524–6533. [Google Scholar] [CrossRef]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef]
- France, J.; Dijkstra, J.; Dhanoa, M.S.; Lopez, S.; Bannink, A. Estimating the extent of degradation of ruminant feeds from a description of their gas production profiles observed in vitro: Derivation of models and other mathematical considerations. Br. J. Nutr. 2000, 83, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Wang, Y.L.; Wang, W.K.; Cao, Z.J.; Li, S.L.; Yang, H.J. The inhibitory action mode of nitrocompounds on in vitro rumen methanogenesis: A comparison of nitroethane, 2-nitroethanl and 2-nitro-1-propanol. J. Agric. Sci. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Van Nevel, C.J.; Demeyer, D.I. Control of rumen methanogenesis. Environ. Monit. Assess. 1996, 42, 73–97. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.C.; Callaway, T.R.; Kessel, J.A.S.V.; Soo, J.Y.; Edrington, T.S.; Nisbet, D.J. Effect of select nitrocompounds on ruminal fermentation; an initial look at their potential to reduce economic and environmental costs associated with ruminal methanogenesis. Bioresour. Technol. 2003, 90, 59–63. [Google Scholar] [CrossRef]
- Anderson, R.C.; Huwe, J.K.; Smith, D.J.; Stanton, T.B.; Krueger, N.A.; Callaway, T.R.; Edrington, T.S.; Harvey, R.B.; Nisbet, D.J. Effect of nitroethane, dimethyl-2-nitroglutarate and 2-nitro-methyl-propionate on ruminal methane production and hydrogen balance in vitro. Bioresour. Technol. 2010, 101, 5345–5349. [Google Scholar] [CrossRef]
- Anderson, R.C.; Rasmussen, M.A.; Allison, M.J. Metabolism of the plant toxins nitropropionic acid and nitropropanol by ruminal microorganisms. Appl. Environ. Microbiol. 1993, 59, 3056–3061. [Google Scholar] [CrossRef]
- Anderson, R.C.; Rasmussen, M.A. Use of a novel nitrotoxin-metabolizing bacterium to reduce ruminal methane production. Bioresour. Technol. 1998, 64, 89–95. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Cao, Z.J.; Wang, Y.L.; Wang, Y.J.; Yang, H.J.; Li, S.L. Nitrocompounds as potential methanogenic inhibitors in ruminant animals: A review. Anim. Feed Sci. Technol. 2018, 236, 107–114. [Google Scholar] [CrossRef]
- Angermaier, L.; Simon, H. On the reduction of aliphatic and aromatic nitro compounds by Clostridia, the role of ferredoxin and its stabilization. Hoppe-Seyler’s Z. Physiol. Chem. 1983, 364, 961–975. [Google Scholar] [CrossRef]
- Anderson, R.C.; Krueger, N.A.; Stanton, T.B.; Callaway, T.R.; Edrington, T.S.; Harvey, R.B.; Jung, Y.S.; Nisbet, D.J. Effects of select nitrocompounds on in vitro ruminal fermentation during conditions of limiting or excess added reductant. Bioresour. Technol. 2008, 99, 8655–8661. [Google Scholar] [CrossRef]
- Soltan, Y.A.; Hashem, N.M.; Morsy, A.S.; El-Azrak, K.M.; Sallam, S.M. Comparative effects of Moringa oleifera root bark and monensin supplementations on ruminal fermentation, nutrient digestibility and growth performance of growing lambs. Anim. Feed Sci. Technol. 2018, 235, 189–201. [Google Scholar] [CrossRef]
- Machmüller, A. Medium-chain fatty acids and their potential to reduce methanogenesis in domestic ruminants. Agric. Ecosyst. Environ. 2006, 112, 107–114. [Google Scholar] [CrossRef]
- Janssen, P.H.; Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625. [Google Scholar] [CrossRef] [PubMed]
- Wina, E.; Muetzel, S.; Hoffmann, E.; Makkar, H.P.S.; Becker, K. Saponins containing methanol extract of sapindus rarak affect microbial fermentation, microbial activity and microbial community structure in vitro. Anim. Feed Sci. Technol. 2005, 121, 159–174. [Google Scholar] [CrossRef]
- Vogels, G.D.; Hoppe, W.F.; Stumm, C.K. Association of methanogenic bacteria with rumen ciliates. Appl. Environ. Microbiol. 1980, 40, 608–612. [Google Scholar] [CrossRef]
- Dehority, B.A.; Tirabasso, P.A. Antibiosis between ruminal bacteria and ruminal fungi. Appl. Environ. Microbiol. 2000, 66, 2921–2927. [Google Scholar] [CrossRef]
- Lessner, D.J. Methanogenesis Biochemistry. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2009. [Google Scholar]
- Hallam, S.J.; Peter, R.G.; Christina, M.P.; Paul, M.R.; Edward, F.D. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 2003, 69, 5483–5491. [Google Scholar] [CrossRef]
- Gunsalus, R.P.; Wolfe, R.S. Chromophoric factors F342 and F430 of Methanobacterium thermoautotrophicum. FEMS Microbiol. Ecol. 1978, 3, 191–193. [Google Scholar] [CrossRef]
- Schönheit, P.; Keweloh, H.; Thauer, R.K. Factor F420 degradation in Methanobacterium thermoautotrophicum during exposure to oxygen. FEMS Microbiol. Ecol. 2010, 12, 347–349. [Google Scholar] [CrossRef][Green Version]
- Anderson, R.C.; Rasmussen, M.A.; Jensen, N.S.; Allison, M.J. Denitrobacterium detoxificans gen. nov.; sp. nov.; a ruminal bacterium that respires on nitrocompounds. Int. J. Syst. Evol. Microbiol. 2000, 50, 633–638. [Google Scholar] [CrossRef]
| Items 1 | Treatment 2 | SEM | p3 | |||||
|---|---|---|---|---|---|---|---|---|
| Control | NE | NEOH | NPOH | Treatment | Time | Trt × Time | ||
| Gas production kinetics | ||||||||
| A, mL/g DM | 199 a | 178 b | 170 c | 173 bc | 1.96 | <0.01 | - | - |
| c, h−1 | 0.09 b | 0.11a | 0.12 a | 0.10 b | 0.02 | <0.01 | - | - |
| T1/2, h | 3.1 | 3.1 | 3.0 | 3.1 | 0.02 | 0.07 | - | - |
| AGPR, mL/h | 13.3 a | 13.9 a | 13.9 a | 11.9 b | 0.29 | <0.01 | - | - |
| Fermentation gas composition, % | ||||||||
| CH4 | 15.1 a | 0.5 c | 0.5 c | 8.8 b | 0.39 | <0.01 | <0.01 | <0.01 |
| H2 | 0.3 d | 9.8 b | 10.5 a | 2.0 c | 0.23 | <0.01 | <0.01 | <0.01 |
| CO2 | 84.6 b | 89.4 a | 89.0 a | 89.2 a | 0.36 | <0.01 | <0.01 | 0.01 |
| Items 1 | Treatment 2 | SEM | p3 | |||||
|---|---|---|---|---|---|---|---|---|
| Control | NE | NEOH | NPOH | Treatment | Time | Trt × Time | ||
| Coenzyme content | ||||||||
| F420 | 16.7 a | 11.9 c | 11.4 c | 15.2 b | 0.23 | <0.01 | <0.01 | <0.01 |
| F430 | 0.59 a | 0.48 b | 0.44 c | 0.50 b | 0.02 | <0.01 | <0.01 | 0.01 |
| mcrA expression | 1 a | 0.11 c | 0.12 c | 0.42 b | 0.02 | <0.01 | <0.01 | <0.01 |
| Total VFA, mmol/L | 102.8 a | 93.7 c | 94.7 bc | 97.4 b | 1.64 | <0.01 | <0.01 | <0.01 |
| Acetate, mmol/L | 60.9 a | 52.1 c | 53.1 c | 54.7 b | 1.31 | <0.01 | <0.01 | 0.02 |
| Propionate, mmol/L | 24.0 | 23.1 | 24.2 | 23.9 | 0.53 | 0.33 | <0.01 | 0.02 |
| Butyrate, mmol/L | 12.8 bc | 13.5 a | 12.5 c | 13.2 ab | 0.30 | <0.01 | <0.01 | <0.01 |
| BCVFA, mmol/L | 5.1 | 5.0 | 4.9 | 5.6 | 0.31 | 0.14 | <0.01 | 0.14 |
| 2Hrec, % | 67.6 a | 47.2 c | 48.5 c | 59.3 b | 0.87 | <0.01 | <0.01 | <0.01 |
| Items | Treatment | |||||
|---|---|---|---|---|---|---|
| Control | NE | NEOH | NPOH | SEM | p | |
| Total methanogen | 0.65 a | 0.33 b | 0.41 b | 0.38 b | 0.041 | 0.04 |
| Methanobacteriales | 0.39 a | 0.21 c | 0.25 bc | 0.32 b | 0.013 | 0.02 |
| Methanomicrobiales, ×10−3 | 14.2 a | 4.6 c | 9.5 b | 5.1 c | 0.985 | 0.01 |
| Methanococcales, ×10−3 | 0.44 bc | 0.56 ab | 0.65 a | 0.38 c | 0.041 | 0.03 |
| Fungus, ×10−3 | 1.82 a | 0.37 b | 1.25 ab | 1.14 ab | 0.240 | 0.09 |
| Protozoa, ×10−3 | 0.28 | 0.29 | 0.43 | 0.22 | 0.068 | 0.29 |
| Ruminococcus albus | 0.12 b | 0.10 b | 0.18 a | 0.18 a | 0.012 | <0.01 |
| Ruminococcus flavefaciens | 0.58 a | 0.09 b | 0.17 b | 0.43 a | 0.056 | 0.01 |
| Fibrobacter succinogenes, ×10−3 | 3.33 a | 1.39 b | 0.97 b | 0.56 b | 0.232 | <0.01 |
| Items | mcrA | F420 | F430 |
|---|---|---|---|
| methane production | 0.88 * | 0.74 ** | 0.24 * |
| mcrA | 0.78 ** | 0.22 * | |
| F420 | 0.26 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Wang, Y.; Si, X.; Cao, Z.; Li, S.; Yang, H. Rumen Methanogenesis, Rumen Fermentation, and Microbial Community Response to Nitroethane, 2-Nitroethanol, and 2-Nitro-1-Propanol: An In Vitro Study. Animals 2020, 10, 479. https://doi.org/10.3390/ani10030479
Zhang Z, Wang Y, Si X, Cao Z, Li S, Yang H. Rumen Methanogenesis, Rumen Fermentation, and Microbial Community Response to Nitroethane, 2-Nitroethanol, and 2-Nitro-1-Propanol: An In Vitro Study. Animals. 2020; 10(3):479. https://doi.org/10.3390/ani10030479
Chicago/Turabian StyleZhang, Zhenwei, Yanlu Wang, Xuemeng Si, Zhijun Cao, Shengli Li, and Hongjian Yang. 2020. "Rumen Methanogenesis, Rumen Fermentation, and Microbial Community Response to Nitroethane, 2-Nitroethanol, and 2-Nitro-1-Propanol: An In Vitro Study" Animals 10, no. 3: 479. https://doi.org/10.3390/ani10030479
APA StyleZhang, Z., Wang, Y., Si, X., Cao, Z., Li, S., & Yang, H. (2020). Rumen Methanogenesis, Rumen Fermentation, and Microbial Community Response to Nitroethane, 2-Nitroethanol, and 2-Nitro-1-Propanol: An In Vitro Study. Animals, 10(3), 479. https://doi.org/10.3390/ani10030479






