Population Genomic Analysis of Two Endemic Schizothoracins Reveals Their Genetic Differences and Underlying Selection Associated with Altitude and Temperature
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Materials
2.2. DNA Extraction, SLAF Library Construction and High-Throughput Sequencing
2.3. SLAF Markers and SNP Calling
2.4. Data Processing
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Robertson, C. Ecological adaptation and ecological selection. Science 1906, 23, 307–310. [Google Scholar] [CrossRef][Green Version]
- Savolainen, O.; Pyhäjärvi, T.; Knürr, T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 595–619. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2010, 7, 1225–1241. [Google Scholar] [CrossRef]
- Han, D.L. Xinjiang Geographic Manual; Xinjiang People’s Publishing House: Urumqi, China, 1993; pp. 12–14. [Google Scholar]
- Li, J.F. Climate in Xinjiang; China Meteorological Press: Beijing, China, 1991; pp. 97–108. [Google Scholar]
- Chen, X.; Luo, G.P.; Xia, J.; Zhou, K.F.; Lou, S.P.; Ye, M.Q. Ecological response to the climate change on the northern slope of the Tianshan Mountains in Xinjiang. Sci. China. Ser. D 2005, 48, 765–777. [Google Scholar] [CrossRef]
- Cao, W.X.; Chen, Y.Y.; Wu, Y.F.; Zhu, S. Origin and evolution of Schizothoracine fishes in relation to the upheaval of the Xizang Plateau. In The Comprehensive Scientific Expedition to the Qinghai-Xizang Plateau, Collection in Studies on the Period, Amplitude and Type of the Uplift of the Qinghai-Xizang Plateau; Chinese Academy of Sciences, Ed.; Science Press: Beijing, China, 1981; pp. 118–130. (In Chinese) [Google Scholar]
- He, D.K.; Chen, Y.F.; Chen, Y.Y.; Chen, Z.M. Molecular phylogeny of the specialized schizothoracine fishes (Teleostei: Cyprinidae), with their implications for the uplift of the Qinghai-Tibetan Plateau. Chin. Sci. Bull. 2004, 49, 39–48. (In Chinese) [Google Scholar] [CrossRef]
- He, D.K.; Chen, Y.F. Molecular phylogeny and biogeography of the highly specialized grade schizothoracine fishes (Teleostei: Cyprinidae) inferred from cytochrome b sequences. Chin. Sci. Bull. 2007, 52, 777–788. (In Chinese) [Google Scholar] [CrossRef]
- Ning, W.; Chang, M.M. Pliocene cyprinids (Cypriniformes, Teleostei) from Kunlun Pass Basin, northeastern Tibetan Plateau and their bearings on development of water system and uplift of the area. Sci. China Earth. Sci. 2010, 53, 485–500. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, R.M.; Cai, L.G. Fish fauna of Xinjiang; Xinjiang Science and Technology Press: Urumqi, China, 2012; pp. 125–128. (In Chinese) [Google Scholar]
- Li, G.G.; Tang, Y.T.; Zhang, R.Y.; Zhao, K. Phylogeography of Diptychus maculatus (Cyprinidae) endemic to the northern margin of the QTP and Tien Shan region. BMC Evol. Biol. 2016, 16, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Li, G.G.; Peng, Z.G.; Zhang, R.Y.; Tang, Y.T.; Tong, C.; Feng, C.G.; Zhang, C.F.; Zhao, K. Mito-nuclear phylogeography of the cyprinid fish Gymnodiptychus dybowskii in the arid Tien Shan region of Central Asia. Biol. J. Linn. Soc. 2016, 118, 304–314. [Google Scholar] [CrossRef]
- Guo, Y.; Meng, W.; Liu, J.; Zhang, R.M.; Cai, L.L. Comparison of morphological characters of Diptychus maculates in different rivers of Xinjiang. Chin. J. Fish. 2009, 22, 1–5. (In Chinese) [Google Scholar]
- Meng, W.; Yang, T.Y.; Guo, Y.; Hai, S.; Ma, Y.W.; Ma, X.F.; Cai, L.G. Remarkable genetic divergence of Gymnodiptychus dybowskii between south and north of Tianshan Mountain in northwest China. Biochem. Syst. Ecol. 2015, 58, 48–50. [Google Scholar] [CrossRef]
- Meng, W.; Yang, T.Y.; Hai, S.; Ma, Y.W.; Cai, L.G.; Ma, X.F.; Gao, T.X.; Guo, Y. Extensive genetic divergence among Diptychus maculates populations in northwest China. Chin. J. Oceanol. Limnol. 2015, 33, 577–584. [Google Scholar] [CrossRef]
- Meng, W.; Yang, T.Y.; Liu, Y.G.; Mahmut, H.; Gao, T.X. Comparative mitogenomic and phylogentic analyses of a Schizothoracine fish, Gymnodiptychus dybowskii from two water systems in Xinjiang. Pakistan J. Zool. 2018, 50, 2119–2127. [Google Scholar] [CrossRef]
- Sun, X.W.; Liu, D.Y.; Zhang, X.F.; Li, W.B.; Liu, H.; Hong, W.G.; Jiang, C.B.; Guan, N.; Ma, C.X.; Zeng, H.P.; et al. SLAF-seq: An efficient method of large–scale De Novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Qiu, G.F.; Xiong, L.W.; Han, Z.K.; Liu, Z.Q.; Feng, J.B.; Wu, X.G.; Yan, Y.L.; Shen, H.; Huang, L.; Chen, L. A second generation SNP and SSR integrated linkage map and QTL mapping for the Chinese mitten crab Eriocheir sinensis. Sci. Rep. 2017, 7, 39826. [Google Scholar] [CrossRef]
- Ma, Y.W.; Guo, Y.; Zhang, R.M.; Tuersun, T.; Xie, C.G.; Liu, J.; Li, L. Fauna composition and distribution of aboriginal fish in the Tarim River of Xinjiang Uygur Autonomous Region. J. Fish. China 2009, 33, 949–956. (In Chinese) [Google Scholar]
- Guo, Y.; Cai, L.G.; Zhang, R.M.; Adakbek, K. The native fisher distributed and made of evolution in the Yili River (China’s Section). Arid Zone Res. 1999, 16, 31–35. (In Chinese) [Google Scholar]
- Cai, L.G.; Niu, J.G.; Liu, C.C.; Zou, M.; Xie, P.; Adakbek, K.; Liu, J.; Li, H. Species diversity and dominant fish species in different reaches of the Ili River, Xinjiang. Acta Hydrobiol. Sin. 2017, 41, 819–826. (In Chinese) [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989. [Google Scholar]
- Xu, P.; Zhang, X.F.; Wang, X.M.; Li, J.T.; Liu, G.M.; Kuang, Y.Y.; Xu, J.; Zheng, X.H.; Ren, L.F.; Wang, G.L.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 11, 1212–1219. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.X.; Cheng, T.R.; Yang, W.R.; Pan, H.T.; Zhong, J.J.; Huang, L.; Liu, E.Z. High–density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant (Prunus mume Sieb. et Zucc). DNA Res. 2015, 22, 183–191. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA 4, a molecular evolutionary genetic analysis MEGA software version 4.0. Mol. Biol. Evol. 2007, 24, 596–1599. [Google Scholar] [CrossRef]
- Foll, M.; Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef]
- Coop, G.; Witonsky, D.; Di Rienzo, A.; Pritchard, J.K. Using environmental correlations to identify loci underlying local adaptation. Genetics 2010, 185, 1411–1423. [Google Scholar] [CrossRef]
- Kevin, C.; Basile, J.; Johanna, L.; Olivier, F. LFMM 2: Fast and accurate inference of gene-environment associations in genome-wide studies. Mol. Biol. Evol. 2019, 4, 852–860. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114–138. [Google Scholar] [CrossRef] [PubMed]
- Dray, S.; Dufour, A. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Wang, I.J.; Bradburd, G.S. Isolation by environment. Mol. Ecol. 2014, 23, 5649–5662. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.H. Vegan: Community ecology package. R package version 2.2-1. J. Stat. Softw. 2015, 48, 103–132. [Google Scholar]
- Frichot, E.; Schoville, S.D.; Bouchard, G.; François, O. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 2013, 30, 1687–1699. [Google Scholar] [CrossRef]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Avise, J.C.; Arnold, J., Jr.; Ball, R.M.; Bermingham, E.; Lab, T.; Neigel, J.; Reeb, C.A.; Saunders, N.C. Intraspecific Phylogeography: The Mitochondrial DNA Bridge Between Population Genetics and Systematics. Annu. Rev. Ecol. Syst. 1987, 1, 489–522. [Google Scholar] [CrossRef]
- Yang, T.Y.; Meng, W.; Ma, Y.W.; Cai, L.G.; Guo, Y. Genetic structure analysis of Diptychus maculatus between two water systems in Xinjiang based on mitochondrial COI and Cyt b gene sequence. Freshw. Fish 2014, 44, 41–47. (In Chinese) [Google Scholar]
- Stahl, F.W. Genetic recombination. Sci. Am. 1987, 256, 90–101. [Google Scholar] [CrossRef]
- Kodric-Brown, A. Genetic introgression after secondary contact. Trends Ecol Evol. 1989, 4, 329–330. [Google Scholar] [CrossRef]
- Xie, C.G.; Ma, Y.W.; Guo, Y. Analysis of biogeography of fishes in Tarim Basin. Chin. J. Fish. 2015, 28, 40–46. (In Chinese) [Google Scholar]
- Masel, J. Genetic drift. Curr. Biol. 2011, 21, 837–838. [Google Scholar] [CrossRef] [PubMed]
- Gavrilets, S.; Hastings, A. Founder effect speciation: A theoretical reassessment. Am. Nat. 1996, 147, 466–491. [Google Scholar] [CrossRef]
- Jha, A.R.; Zhou, D.; Brown, C.D.; Kreitman, M.; Haddad, G.G.; White, K.P. Shared genetic signals of hypoxia adaptation in drosophila and in high-altitude human populations. Mol. Biol. Evol. 2016, 33, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The Genetical Theory of Natural Selection: A Complete Variorum Edition; Oxford University Press: Oxford, UK, 1999; pp. 59–71. [Google Scholar]
- Simonson, T.S.; Yang, Y.Z.; Huff, C.D.; Yun, H.X.; Qin, G.; Witherspoon, D.J.; Bai, Z.Z.; Lorenzo, F.R.; Xing, J.C.; Jorde, L.B.; et al. Genetic Evidence for High-Altitude Adaptation in Tibet. Science 2010, 329, 72–75. [Google Scholar] [CrossRef]
- Lorenzo, F.R.; Huff, C.; Myllymäki, M.; Olenchock, B.; Swierczek, S.; Tashi, T.; Gordeuk, V.; Wuren, T.; Ri-Li, G.; McClain, D.A.; et al. A genetic mechanism for Tibetan high-altitude adaptation. Nat. Genet. 2014, 46, 951–956. [Google Scholar]
- Narum, S.R.; Hess, J.E. Comparison of F (ST) outlier tests for SNP loci under selection. Mol. Ecol. Resour. 2011, 11, 184–194. [Google Scholar] [CrossRef]
- Blair, L.M.; Granka, J.M.; Feldmanm, M.W. On the stability of the Bayenv method in assessing human SNP-environment associations. Hum. Genom. 2014, 8, 1–13. [Google Scholar] [CrossRef]
- Bohonak, A.J. IBD (Isolation by Distance): A program for analyses of isolation by distance. J. Hered. 2002, 93, 153–154. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef] [PubMed]
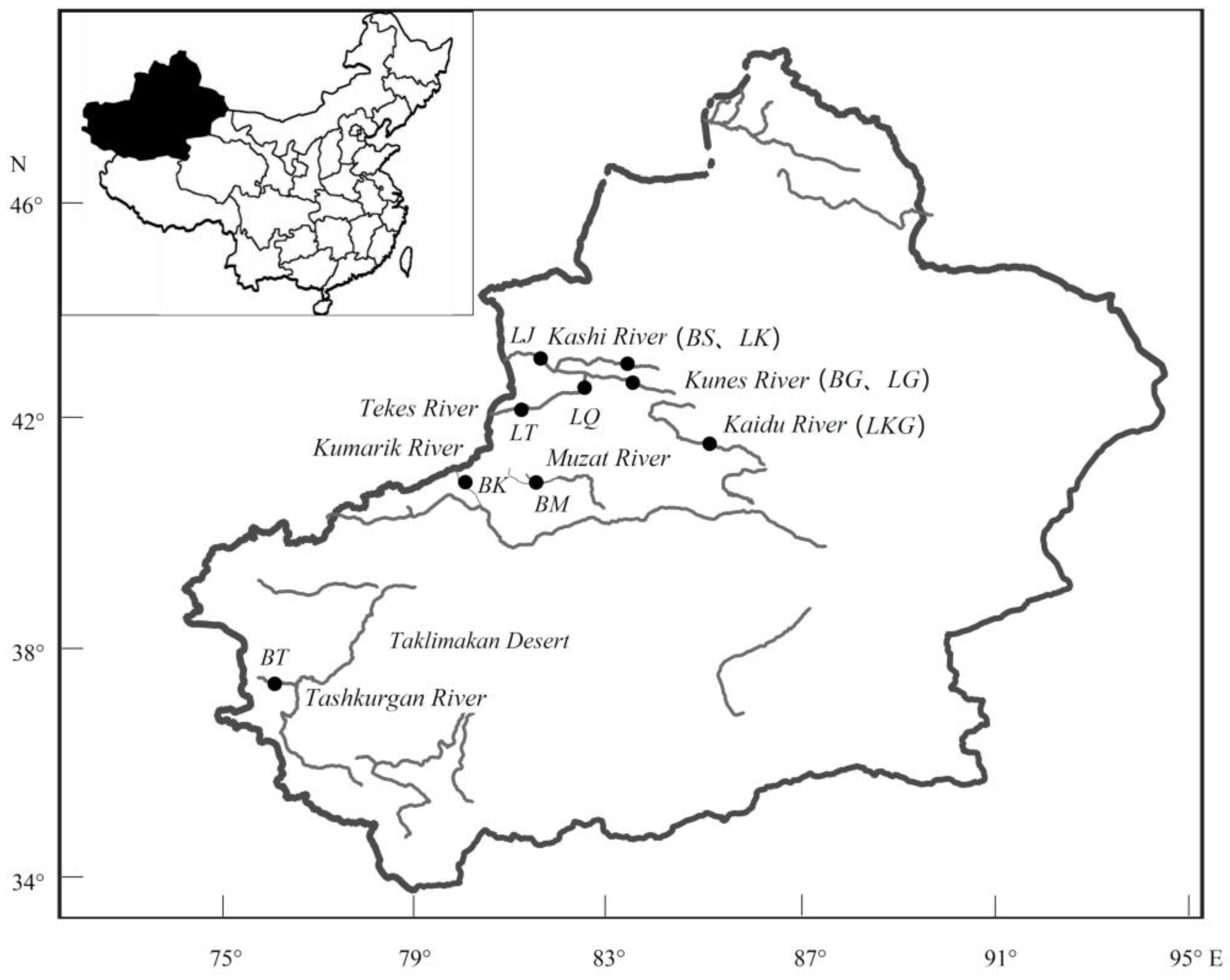
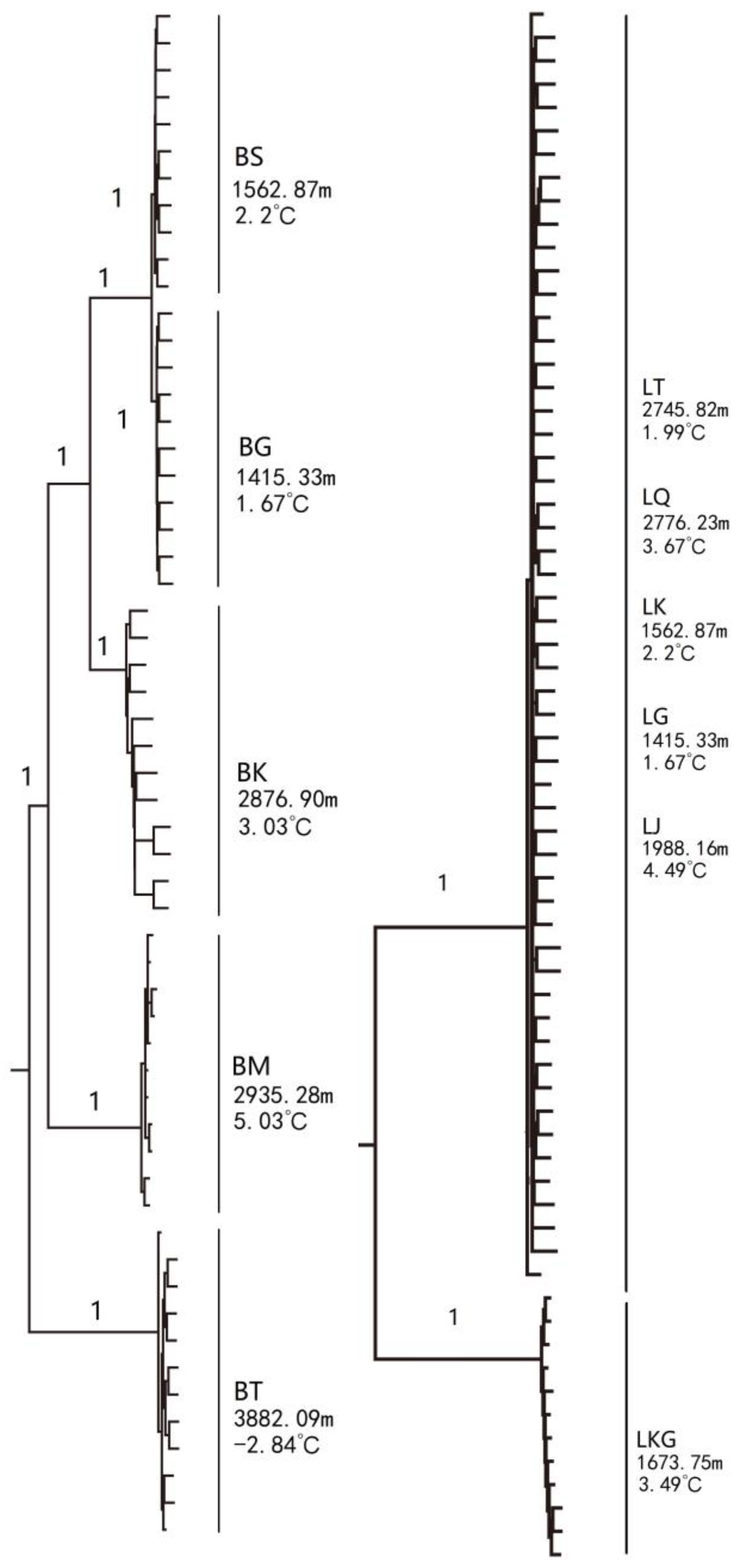
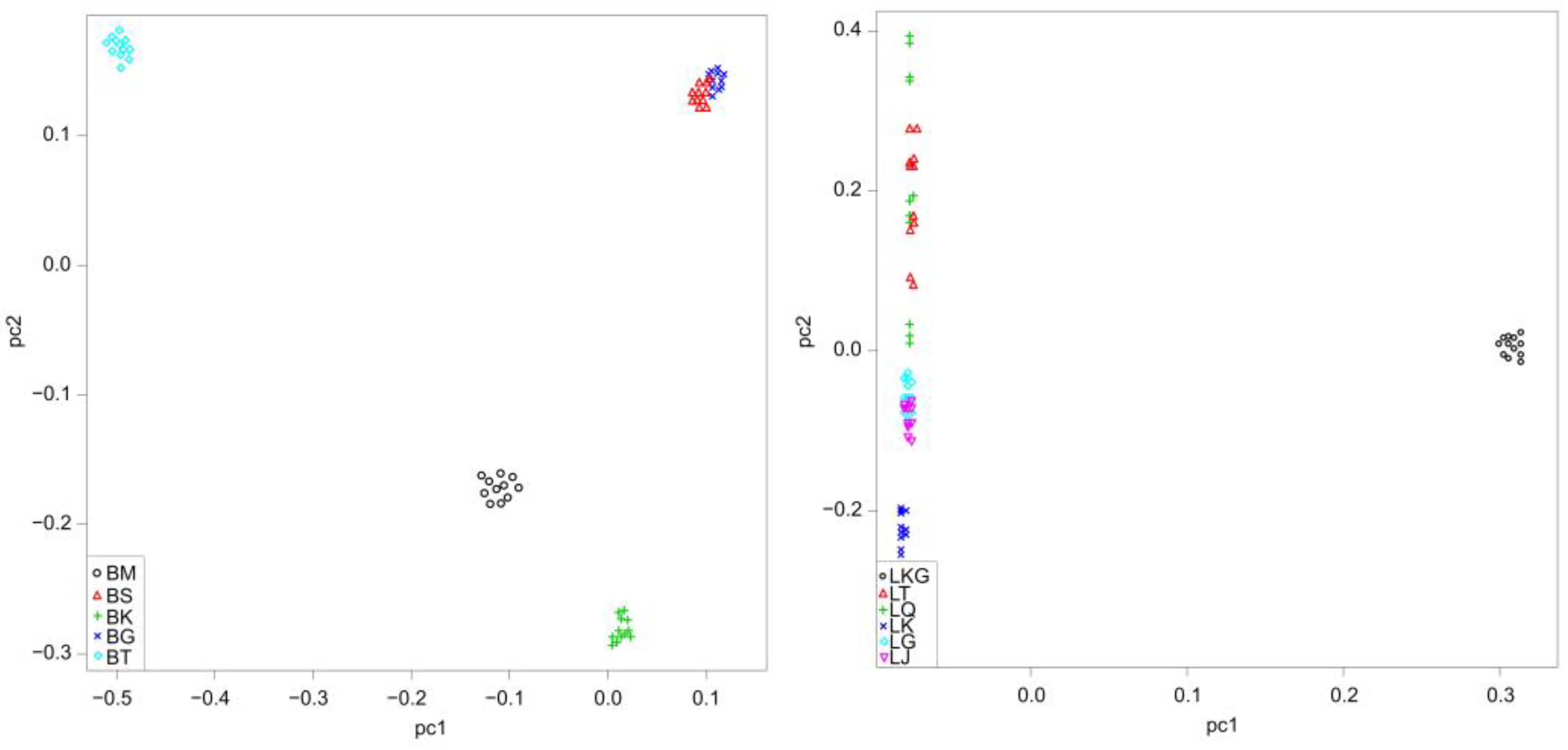
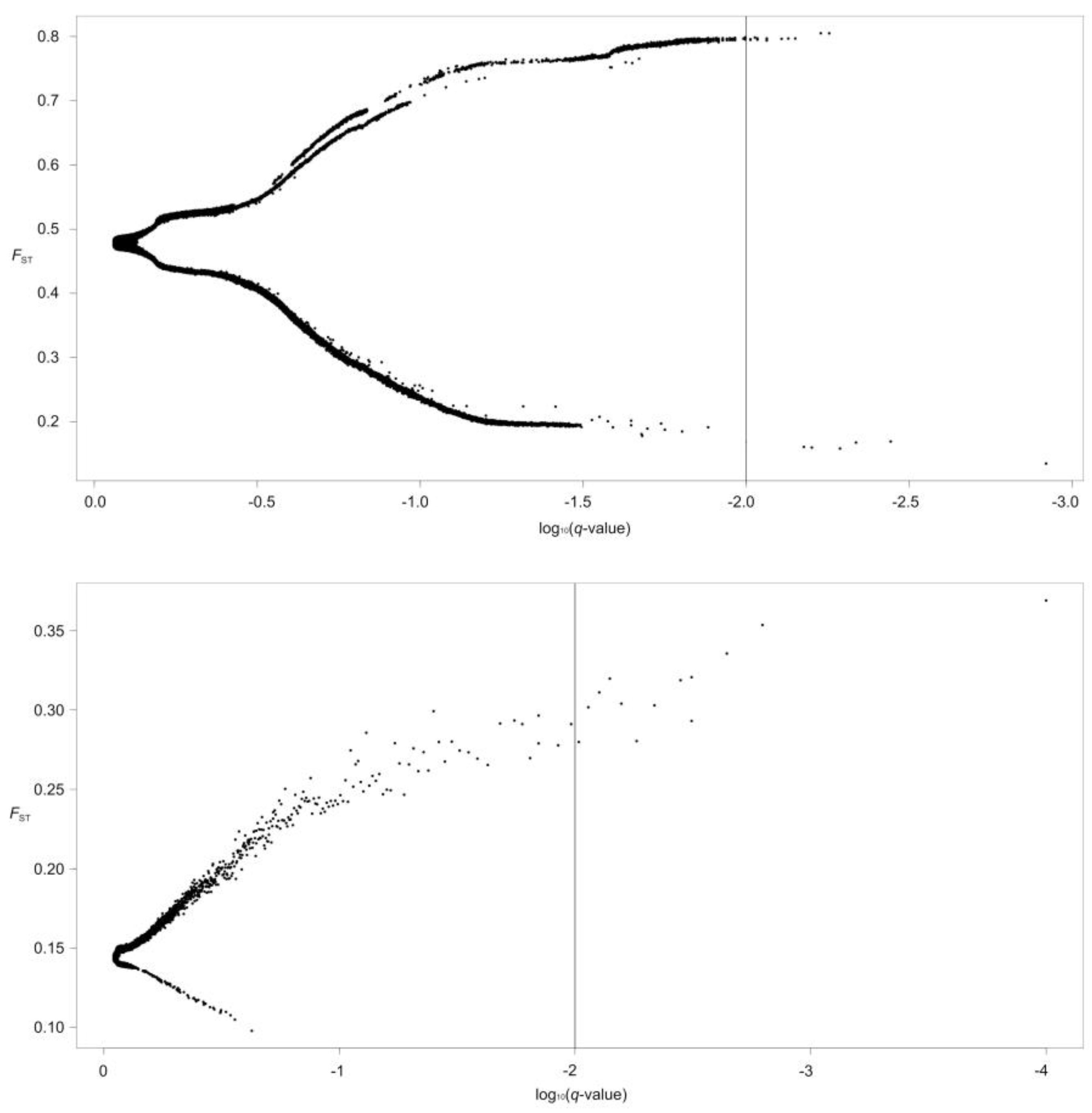
| Species | Population | River | River Basin | Sample Size | Altitude (m) | Temperature (°C) |
|---|---|---|---|---|---|---|
| Diptychus maculates | BM | Muzat River | Tarim River | 11 | 2935.28 | 5.03 |
| BK | Kumarik River | 12 | 2876.90 | 3.03 | ||
| BT | Tashkurgan River | 12 | 3882.09 | −2.84 | ||
| BG | Kunes River | Yili River | 11 | 1415.33 | 1.67 | |
| BS | Kashi River | 11 | 1562.87 | 2.2 | ||
| Gymnodiptychus dybowskii | LKG | Kaidu River | Tarim River | 12 | 1673.75 | 3.49 |
| LQ | Qiapugihai Reservoir | Yili River | 11 | 2776.23 | 3.67 | |
| LK | Kashi River | 11 | 1562.87 | 2.2 | ||
| LG | Kunes River | 11 | 1415.33 | 1.67 | ||
| LJ | Yamadu | 11 | 1988.16 | 4.49 | ||
| LT | Tekes River | 11 | 2745.82 | 1.99 |
| Population | Obs Het | Exp Het | Pi | Fis |
|---|---|---|---|---|
| BM | 0.1549 | 0.0961 | 0.1282 | −0.0401 |
| BS | 0.2439 | 0.1750 | 0.1912 | −0.1017 |
| BK | 0.2948 | 0.2146 | 0.2313 | −0.1257 |
| BG | 0.2419 | 0.1735 | 0.1895 | −0.1007 |
| BT | 0.1789 | 0.1191 | 0.1429 | −0.0604 |
| LKG | 0.1323 | 0.1071 | 0.1146 | −0.0328 |
| LT | 0.2687 | 0.2115 | 0.2311 | −0.0742 |
| LQ | 0.2616 | 0.2097 | 0.2293 | −0.0635 |
| LK | 0.2805 | 0.2131 | 0.2327 | −0.0961 |
| LG | 0.2622 | 0.2069 | 0.2261 | −0.0712 |
| LJ | 0.2554 | 0.2059 | 0.2251 | −0.0584 |
| IBD | IBE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Altitude | Temperature | |||||||||
| Mantel Test | Partial Mantel Test | Mantel Test | Partial Mantel Test | |||||||
| rs | p | rs | p | rs | p | rs | p | rs | p | |
| D. maculates | 0.65 | 0.05 | 0.42 | 0.09 | 0.06 | 0.47 | 0.92 | 0.01 | 0.74 | 0.17 |
| G. dybowskii | 0.50 | 0.10 | −0.12 | 0.75 | −0.35 | 0.92 | −0.18 | 0.77 | 0.18 | 0.25 |
| BM | BS | BK | BG | BT | |
|---|---|---|---|---|---|
| BM | 0.28391 | 0.21963 | 0.28838 | 0.44715 | |
| BS | 0.28381 | 0.17747 | 0.05600 | 0.32412 | |
| BK | 0.21963 | 0.17737 | 0.18040 | 0.27498 | |
| BG | 0.28827 | 0.05599 | 0.18029 | 0.32785 | |
| BT | 0.44715 | 0.32404 | 0.27498 | 0.32776 |
| LKG | LT | LQ | LK | LG | LJ | |
|---|---|---|---|---|---|---|
| LKG | 0.32159 | 0.32137 | 0.32192 | 0.32457 | 0.32348 | |
| LT | 0.32158 | 0.04007 | 0.04102 | 0.04161 | 0.04117 | |
| LQ | 0.32137 | 0.04006 | 0.04078 | 0.04166 | 0.04085 | |
| LK | 0.32192 | 0.04101 | 0.04078 | 0.04136 | 0.04016 | |
| LG | 0.32457 | 0.04160 | 0.04165 | 0.04135 | 0.04192 | |
| LJ | 0.32347 | 0.04116 | 0.04085 | 0.04016 | 0.04192 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, T.; Meng, W.; Guo, B. Population Genomic Analysis of Two Endemic Schizothoracins Reveals Their Genetic Differences and Underlying Selection Associated with Altitude and Temperature. Animals 2020, 10, 447. https://doi.org/10.3390/ani10030447
Yang T, Meng W, Guo B. Population Genomic Analysis of Two Endemic Schizothoracins Reveals Their Genetic Differences and Underlying Selection Associated with Altitude and Temperature. Animals. 2020; 10(3):447. https://doi.org/10.3390/ani10030447
Chicago/Turabian StyleYang, Tianyan, Wei Meng, and Baocheng Guo. 2020. "Population Genomic Analysis of Two Endemic Schizothoracins Reveals Their Genetic Differences and Underlying Selection Associated with Altitude and Temperature" Animals 10, no. 3: 447. https://doi.org/10.3390/ani10030447
APA StyleYang, T., Meng, W., & Guo, B. (2020). Population Genomic Analysis of Two Endemic Schizothoracins Reveals Their Genetic Differences and Underlying Selection Associated with Altitude and Temperature. Animals, 10(3), 447. https://doi.org/10.3390/ani10030447





