Comparison of Methods for Measuring Protein Concentration in Venom Samples
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Pierce™ BCA Protein Assay Kit
2.2. 2-D Quant Kit
2.3. Bradford Assay
2.4. Qubit® Protein Assay Kit
2.5. NanoDrop
2.6. Analysis of Results
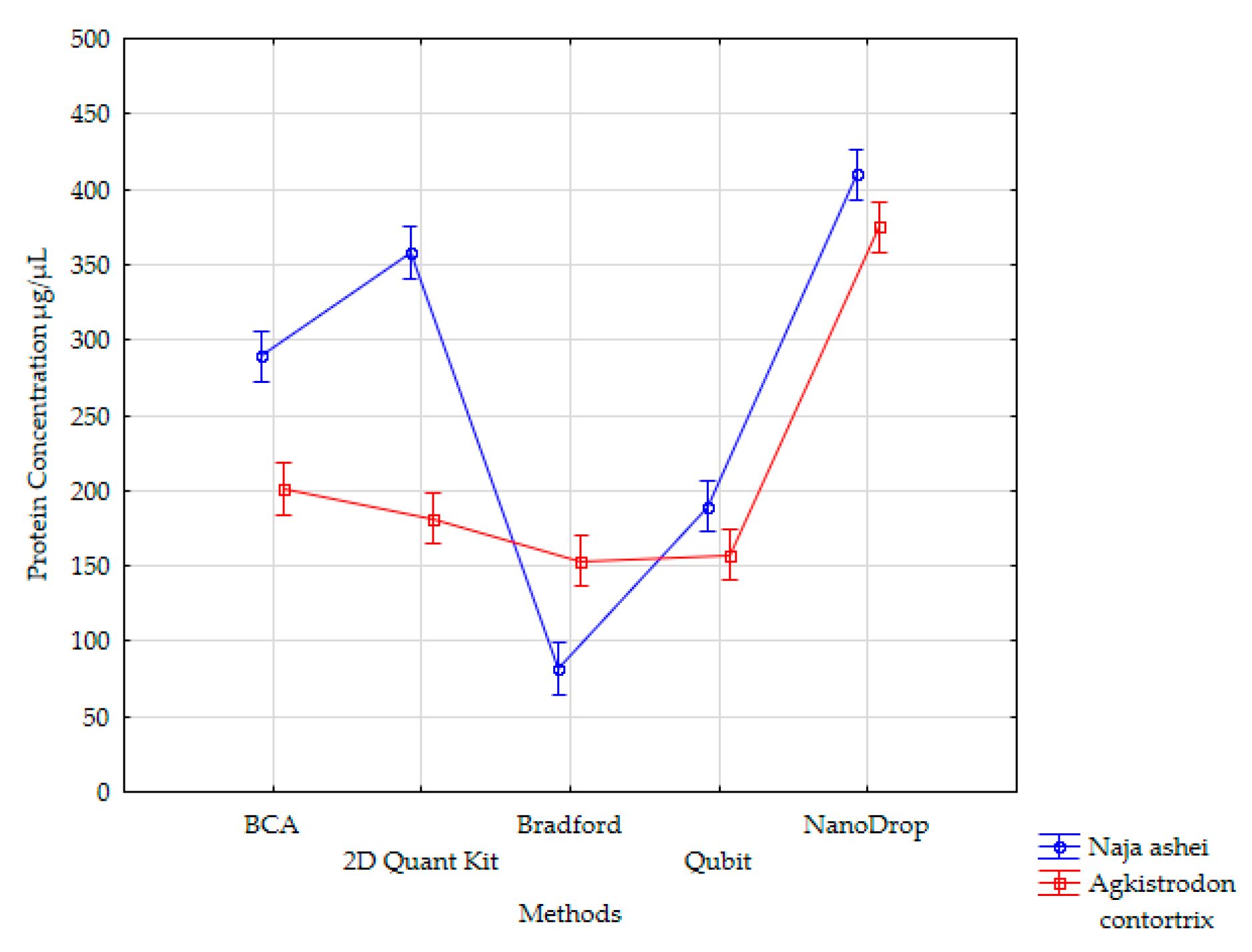
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Chippaux, J.P.; Williams, V.; White, J. Snake venom variability: methods of study, results and interpretation. Toxicon 1991, 29, 1279–1303. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Bieber, A.L. Metal and Nonprotein Constituents in Snake Venoms. In Snake Venoms Handbook of Experimental Pharmacology (Continuation of Handbuch der experimentellenPharmakologie); Lee, C.Y., Ed.; Springer: Berlin/Heidelberg, Germany, 1979; Volume 52, pp. 295–306. [Google Scholar]
- Li, L.; Jianzhong, H.; Yao, L. Snake Venoms in Cancer Therapy: Past, Present and Future. Toxins 2018, 10, 346. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Baron, A.; Salinas, M.; Douguet, D.; Scarzello, S.; Dabert-Gay, A.S.; Debayle, D.; Friend, V.; Alloui, A.; Lazdunski, M.; et al. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 2012, 490, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Bocian, A.; Hus, K.K. Antibacterial properties of snake venom components. Chem. Pap. 2020, 74, 407–419. [Google Scholar] [CrossRef]
- Koh, C.Y.; Kini, R.M. From snake venom toxins to therapeutics--cardiovascular examples. Toxicon 2012, 59, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Vejayan, J.; Too Lay Khoon, T.L.; Ibrahim, H. Comparative analysis of the venom proteome of four important Malaysian snake species. J. Venom. Anim. Toxins incl. Trop. Dis. 2014, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.L.H.; Tan, N.H.; Fung, S.Y.; Tan, C.H. Comparative proteomes, immunoreactivities and neutralization of procoagulant activities of Calloselasma rhodostoma (Malayan pit viper) venoms from four regions in Southeast Asia. Toxicon 2019, 169, 91–102. [Google Scholar] [CrossRef]
- Bocian, A.; Urbanik, M.; Hus, K.; Łyskowski, A.; Petrilla, V.; Andrejčáková, Z.; Petrillová, M.; Legáth, J. Proteome and peptidome of Vipera berus berus venom. Molecules 2016, 21, 1398. [Google Scholar] [CrossRef]
- Latinovic, Z.; Leonardi, A.; Šribar, J.; Sajevic, T.; Žužek, M.C.; Frangež, R.; Halassy, B.; Trampuš-Bakija, A.; Pungerčar, J.; Križaj, I. Venomics of Vipera berus berus to explain differences in pathology elicited by Vipera ammodytes ammodytes envenomation: Therapeutic implications. J. Proteom. 2016, 146, 34–47. [Google Scholar] [CrossRef]
- Lipps, B.V.; Khan, A.A. Antigenic cross reactivity among the venoms and toxins from unrelated diverse sources. Toxicon 2000, 38, 973–980. [Google Scholar] [CrossRef]
- Casewell, N.R.; Al-Abdulla, I.; Smith, D.; Coxon, R.; Landon, J. Immunological cross-reactivity and neutralisation of European viper venoms with the monospecific Vipera berus antivenom ViperaTAb. Toxins 2014, 6, 2471–2482. [Google Scholar] [CrossRef] [PubMed]
- Ledsgaard, L.; Jenkins, T.P.; Davidsen, K.; Krause, K.E.; Martos-Esteban, A.; Engmark, M.; Rørdam Andersen, M.; Lund, O.; Laustsen, A.H. Antibody Cross-Reactivity in Antivenom Research. Toxins 2018, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Slagboom, J.; Alomran, N.; Pla, D.; Alhamdi, Y.; King, S.I.; Bolton, F.M.S.; Gutiérrez, J.M.; Vonk, F.J.; Toh, C.H.; et al. The paraspecific neutralisation of snake venom induced coagulopathy by antivenoms. Commun. Biol. 2018, 1, 34. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Kalita, B.; Mackessy, S.P. A proteomic analysis of Pakistan Daboia russelii russelii venom and assessment of potency of Indian polyvalent and monovalent antivenom. J. Proteom. 2016, 144, 73–86. [Google Scholar] [CrossRef]
- Kalita, B.; Patra, A.; Mukherjee, A.K. Unraveling the Proteome Composition and Immuno-profiling of Western India Russell’s Viper Venom for In-Depth Understanding of Its Pharmacological Properties, Clinical Manifestations, and Effective Antivenom Treatment. J. Proteome. Res. 2017, 16, 583–598. [Google Scholar] [CrossRef]
- Faisal, T.; Tan, K.Y.; Sim, S.M.; Quraishi, N.; Tan, N.H.; Tan, C.H. Proteomics, functional characterization and antivenom neutralization of the venom of Pakistani Russell’s viper (Daboia russelii) from the wild. J. Proteom. 2018, 183, 1–13. [Google Scholar] [CrossRef]
- Deka, A.; Gogoi, A.; Das, D.; Purkayastha, J.; Doley, R. Proteomics of Naja kaouthia venom from North East India and assessment of Indian polyvalent antivenom by third generation antivenomics. J. Proteom. 2019, 207, 103463. [Google Scholar] [CrossRef]
- Aird, S.D.; Watanabe, Y.; Villar-Briones, A.; Roy, M.C.; Terada, K.; Mikheyev, A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genom. 2013, 14, 1–62. [Google Scholar] [CrossRef]
- Ziganshin, R.H.; Kovalchuk, S.I.; Arapidi, G.P.; Starkov, V.G.; Hoang, A.N.; Thi Nguyen, T.T.; Nguyen, K.C.; Shoibonov, B.B.; Tsetlin, V.I.; Utkin, Y.N. Quantitative proteomic analysis of Vietnamese krait venoms: Neurotoxins are the major components in Bungarus multicinctus and phospholipases A2 in Bungarusfasciatus. Toxicon 2015, 107, 197–209. [Google Scholar] [CrossRef]
- Kovalchuk, S.I.; Ziganshin, R.H.; Starkov, V.G.; Tsetlin, V.I.; Utkin, Y.N. Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components. Toxins 2016, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Tan, N.H. Revisiting Notechisscutatus venom: On shotgun proteomics and neutralization by the "bivalent" Sea Snake Antivenom. J. Proteom. 2016, 144, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.L.H.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Venomics of Calloselasma rhodostoma, the Malayan pit viper: A complex toxin arsenal unraveled. J. Proteom. 2016, 148, 44–56. [Google Scholar] [CrossRef]
- Lauridsen, L.P.; Laustsen, A.H.; Lomonte, B.; Gutiérrez, J.M. Toxicovenomics and antivenom profiling of the Eastern green mamba snake (Dendroaspis angusticeps). J. Proteom. 2016, 136, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y.; Tan, C.H.; Tan, K.Y.; Quraishi, N.H.; Tan, N.H. Elucidating the biogeographical variation of the venom of Naja naja (spectacled cobra) from Pakistan through a venom-decomplexing proteomic study. J. Proteom. 2018, 175, 156–173. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.K.; Saviola, A.J.; Reilly, S.B.; Stubbs, A.L.; Arida, E.; Iskandar, D.T.; McGuire, J.A.; Yates, J.R.; Mackessy, S.P. Venom Composition in a Phenotypically Variable Pit Viper (Trimeresurus insularis) across the Lesser Sunda Archipelago. J. Proteome Res. 2018, 18, 2206–2220. [Google Scholar] [CrossRef]
- Nicolau, C.A.; Carvalho, P.C.; Junqueira-de-Azevedo, I.L.; Teixeira-Ferreira, A.; Junqueira, M.; Perales, J.; Neves-Ferreira, A.G.; Valente, R.H. An in-depth snake venom proteopeptidome characterization: Benchmarking Bothrops jararaca. J. Proteom. 2017, 151, 214–231. [Google Scholar] [CrossRef]
- Moridikia, A.; Zargan, J.; Sobati, H.; Goodarzi, H.-R. Anticancer and antibacterical Effects of Iranian Viper (Vipera latifii) Venom; an in-vitro study. J. Cell. Physiol. 2018, 233, 6790–6797. [Google Scholar] [CrossRef]
- Román-Domínguez, L.; Neri-Castro, E.; VázquezLópez, H.; García-Osorio, B.; Archundia, I.G.; Ortiz-Medina, J.A.; Petricevich, V.L.; Alagón, A.; Bénard-Valle, M. Biochemical and immunochemical characterization of venoms from snakes of the genus Agkistrodon. Toxicon 2019, 4, 100013. [Google Scholar] [CrossRef]
- Noble, J.E.; Knight, A.E.; Reason, A.J.; Di Matola, A.; Bailey, M.J.A. A Comparison of Protein Quantitation Assays for Biopharmaceutical Applications. Mol. Biotechno. 2007, 37, 99–111. [Google Scholar] [CrossRef]
- Calderón-Celis, F.; Diez-Fernández, S.; Costa-Fernández, J.M.; Encinar, J.R.; Calvete, J.J.; Sanz-Medel, A. Elemental Mass Spectrometry for Absolute Intact Protein Quantification without Protein-Specific Standards: Application to Snake Venomics. Anal. Chem. 2016, 88, 9699–9706. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Bocian, A.; Urbanik, M.; Hus, K.; Łyskowski, A.; Petrilla, V.; Andrejčáková, Z.; Petrillová, M.; Legáth, J. Proteomic Analyses of Agkistrodon contortrix contortrix Venom Using 2D Electrophoresis and MS Techniques. Toxins 2016, 8, 372. [Google Scholar] [CrossRef] [PubMed]
- Hus, K.; Buczkowicz, J.; Petrilla, V.; Petrillová, M.; Ł yskowski, A.; Legáth, J.; Bocian, A. First Look at the venom of Naja ashei. Molecules 2018, 23, 609. [Google Scholar] [CrossRef] [PubMed]
- Freeman, H.C.; Smith, J.E.W.L.; Taylor, J.C. Crystallographic studies of the biuret reaction. I. Potassium bis-biuret cuprate (II) tetrahydrate. Acta. Cryst. 1961, 14, 407–418. [Google Scholar] [CrossRef]
- Huang, H.W.; Liu, B.S.; Chien, K.Y.; Chiang, L.C.; Huang, S.Y.; Sung, W.C.; Wu, W.G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015, 128, 92–104. [Google Scholar] [CrossRef]
- Samel, M.; Vija, H.; Kurvet, I.; Kunnis-Beres, K.; Trummal, K.; Subbi, J.; Kahru, A.; Sugur, J. Interactions of PLA2-s from Vipera lebetina, Vipera berus berus and Naja naja oxiana Venom with Platelets, Bacterial and Cancer Cells. Toxins 2013, 5, 203–223. [Google Scholar] [CrossRef]
- Syed, A.; Baumann, K.; Jackson, T.N.W.; Wood, K.; Mason, S.; Undheim, E.A.B.; Nouwend, A.; Koludarov, I.; Hendrikx, I.; Jones, A.; et al. Proteomic comparison of Hypnale hypnale (Hump-Nosed Pit-Viper) and Calloselasma rhodostoma (Malayan Pit-Viper) venoms. J. Protemics 2013, 91, 338–343. [Google Scholar]
- Lewinska, A.; Bocian, A.; Petrilla, V.; Adamczyk-Grochala, J.; Szymura, K.; Hendzel, W.; Kaleniuk, E.; Hus, K.K.; Petrillova, M.; Wnuk, M. Snake venoms promote stress-induced senescence in human fibroblasts. J. Cell Physiol. 2019, 234, 6147–6160. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Göçmen, B.; Heiss, P.; Petras, D.; Nalbantsoy, A.; Sussmuth, D.R. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon 2015, 107, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Tashima, A.K.; Pinto, A.F.M.; Leme, A.F.; Stuginski, D.R.; Furtado, M.F.; Sherman, N.E.; Ho, P.L.; Fox, J.W.; Serrano, S.M.T. Bothrops jararaca venom proteome rearrangement upon neonate to adult transition. Proteomics 2011, 11, 4218–4228. [Google Scholar] [CrossRef] [PubMed]
- Menezes, M.C.; Fustado, M.F.; Travaglia-Cardoso, S.R.; Camargo, A.C.M.; Serrano, S.M.T. Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 2006, 47, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Binh, D.V.; Thanh, T.T.; Chi, P.V. Proteomic characterization of the thermostable toxins from Naja venom. J. Venom. Anim. Toxins incl. Trop. Dis. 2010, 16, 631–638. [Google Scholar] [CrossRef]
- Camacho, E.; Sanz, L.; Escalante, T.; Pérez, A.; Villalta, F.; Lomonte, B.; Neves-Ferreira, A.G.C.; Feoli, A.; Calvete, J.J.; Gutiérrez, J.M.; et al. Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif. Toxins 2016, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Oulion, B.; Dobson, J.S.; Zdenek, C.N.; Arbuckle, K.; Lister, C.; Coimbra, F.C.P.; Op den Brouw, B.; Debono, J.; Rogalski, A.; Violette, A.; et al. Factor X activating Atractaspis snake venoms and the relative coagulotoxicity neutralizing efficacy of African antivenoms. Toxicol. Lett. 2018, 288, 119–128. [Google Scholar] [PubMed]
- Chaisakul, J.; Alsolaiss, J.; Charoenpitakchai, M.; Wiwatwarayos, K.; Sookprasert, N.; Harrison, R.A.; Chaiyabutr, N.; Chanhome, L.; Tan, C.H.; Casewell, N.R. Evaluation of the geographical utility of Eastern Russel’s viper (Daboia siamensis) antivenom from Thailand and an assessment of its protective effects against venom-induced nephrotoxicity. PLoS Negl. Trop. Dis. 2019, 13, e0007338. [Google Scholar]
- Bell, P.J.L.; Karuso, P. Epicocconone, A Novel Fluorescent Compound from the Fungus Epicoccum nigrum. J. Am. Chem. Soc. 2003, 125, 9304–9305. [Google Scholar] [CrossRef]
- Lomonte, B.; Tsai, W.-C.; Ureña-Diaz, J.M.; Sanz, L.; Mora-Obando, D.; Sánchez, E.E.; Fry, B.G.; Gutiérrez, J.M.; Gibbs, H.L.; Sovic, M.G.; et al. Venomics of New World pit vipers: Genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteom. 2014, 96, 103–116. [Google Scholar] [CrossRef]
- Bocian, A.; Ciszkowicz, E.; Hus, K.K.; Buczkowicz, J.; Lecka-Szlachta, K.; Pietrowska, M.; Petrílla, V.; Petrillova, M.; Legáth, L.; Legáth, J. Antimicrobial Activity of Protein Fraction from Naja ashei Venom Against Staphylococcus epidermidis. Molecules 2020, 25, 293. [Google Scholar] [CrossRef]
| Venom | Protein Concentration (µg/ µL) | |||
|---|---|---|---|---|
| Ist Replicate | IInd Replicate | IIIrd Replicate | Mean | |
| Pierce™ BCA Protein Assay Kit | ||||
| Naja ashei | 258.96 | 291.16 | 317.68 | 289.269 |
| Agkistrodon contortrix | 211.27 | 190.86 | 202.01 | 201.380 a |
| 2-D Quant Kit | ||||
| Naja ashei | 361.38 | 341.17 | 370.96 | 357.837 |
| Agkistrodon contortrix | 180.69 | 162.61 | 201.44 | 181.578 a,b |
| Bradford Assay | ||||
| Naja ashei | 81.88 | 90.73 | 73.02 | 81.875 |
| Agkistrodon contortrix | 151.15 | 158.96 | 149.58 | 153.229 b |
| Qubit® Protein Assay Kit | ||||
| Naja ashei | 189.50 | 189.18 | 190.23 | 189.637 |
| Agkistrodon contortrix | 156.83 | 157.10 | 157.83 | 157.254 b |
| NanoDrop™ | ||||
| Naja ashei | 405.05 | 407.14 | 417.11 | 409.77 |
| Agkistrodon contortrix | 391.52 | 374.86 | 358.54 | 374.97 c |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bocian, A.; Sławek, S.; Jaromin, M.; Hus, K.K.; Buczkowicz, J.; Łysiak, D.; Petrílla, V.; Petrillova, M.; Legáth, J. Comparison of Methods for Measuring Protein Concentration in Venom Samples. Animals 2020, 10, 448. https://doi.org/10.3390/ani10030448
Bocian A, Sławek S, Jaromin M, Hus KK, Buczkowicz J, Łysiak D, Petrílla V, Petrillova M, Legáth J. Comparison of Methods for Measuring Protein Concentration in Venom Samples. Animals. 2020; 10(3):448. https://doi.org/10.3390/ani10030448
Chicago/Turabian StyleBocian, Aleksandra, Sonja Sławek, Marcin Jaromin, Konrad K. Hus, Justyna Buczkowicz, Dawid Łysiak, Vladimir Petrílla, Monika Petrillova, and Jaroslav Legáth. 2020. "Comparison of Methods for Measuring Protein Concentration in Venom Samples" Animals 10, no. 3: 448. https://doi.org/10.3390/ani10030448
APA StyleBocian, A., Sławek, S., Jaromin, M., Hus, K. K., Buczkowicz, J., Łysiak, D., Petrílla, V., Petrillova, M., & Legáth, J. (2020). Comparison of Methods for Measuring Protein Concentration in Venom Samples. Animals, 10(3), 448. https://doi.org/10.3390/ani10030448






