High Diversity of Leptospira Species Infecting Bats Captured in the Urabá Region (Antioquia-Colombia)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Considerations
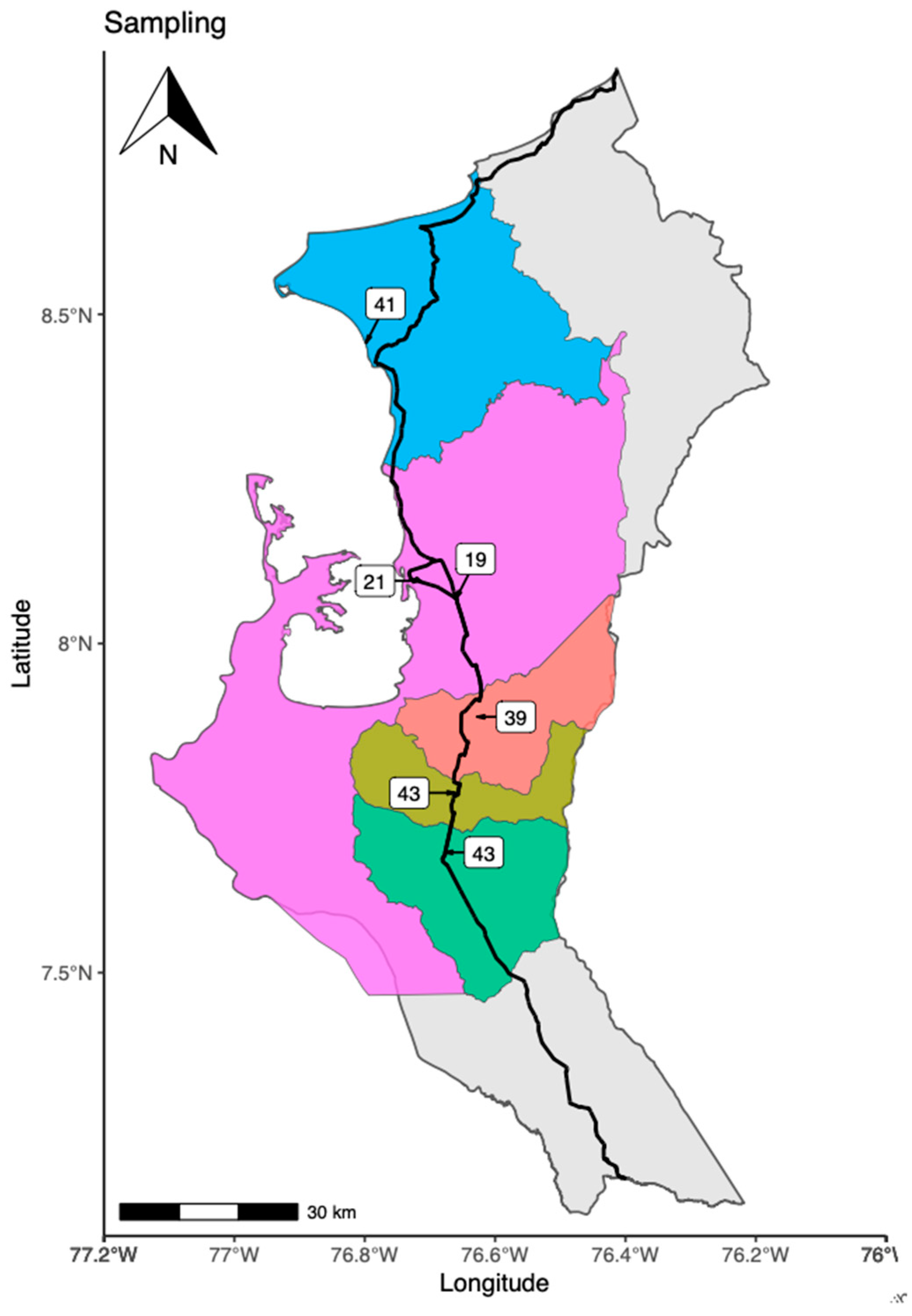
2.2. Characteristics of the Capture Area of Specimens
2.3. Capture of Bats
2.4. Euthanasia of Captured Bats
2.5. DNA Extraction
2.6. PCR-16S Ribosomal Gene Conditions
2.7. 16S Ribosomal Gene Sequencing from Kidneys of Bats
2.8. Phylogenetic Analysis 16S Ribosomal Gene
3. Results
3.1. Places of Bat’s Capture
3.2. Families, Genera and Species of Captured Bats
3.3. Detection of Leptospira spp. in Bats by Conventional PCR
3.4. Identification of Leptospira Species by Phylogenetic Analysis
3.5. Host-Pathogen Relationship between Bats and Leptospira
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Hagan, J.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [Green Version]
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neel, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [Green Version]
- Cerqueira, G.M.; Picardeau, M. A century of Leptospira strain typing. Infect. Genet. Evol. 2009, 9, 760–768. [Google Scholar] [CrossRef]
- Bello, S.; Rodríguez, M.; Paredes, A.; Mendivelso, F.; Walteros, D.; Rodríguez, F.; Realpe, M.E. Comportamiento de la vigilancia epidemiológica de la leptospirosis humana en Colombia, 2007–2011. Biomédica 2013, 33 (Suppl. 1), 153–160. [Google Scholar] [CrossRef] [Green Version]
- Carreño, L.A.; Salas, D.; Beltrán, K.B. Prevalencia de leptospirosis en Colombia: Revisión sistemática de literatura. Prevalence of leptospirosis in Colombia: Systematic literature review. Rev. Salud Pública 2017, 19, 204–209. [Google Scholar] [CrossRef] [Green Version]
- Agudelo, F.P.; Restrepo-Jaramillo, B.N.; Arboleda-Naranjo, M. Situación de la leptospirosis en el Urabá antioqueño colombiano: Estudio seroepidemiológico y factores de riesgo en población general urbana. Cad. Saúde Pública 2007, 23, 2094–2102. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, R.G.P.; Lopez, J.; Pereira, M.M.; Naranjo, M.A.; Agudelo-Flórez, P. Genetic diversity of Leptospira in northwestern Colombia: First report of Leptospira santarosai as a recognised leptospirosis agent. Mem. Inst. Oswaldo Cruz. 2016, 111, 737–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mühldorfer, K. Bats and bacterial pathogens: A review. Zoonoses Public Health 2013, 60, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Juliane Schaer, J.; Perkins, S.L.; Decher, J.; Leendertz, F.H.; Fahr, J.; Weber, N.; Kai Matuschewski, K. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc. Natl. Acad. Sci. USA 2013, 110, 17415–17419. [Google Scholar] [CrossRef] [Green Version]
- Mok, W.Y.; Luizão, R.C.; Barreto da Silva, M.S. Isolation of fungi from bats of the Amazon basin. Appl. Environ. Microbiol. 1982, 44, 570–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, D.R. Migrations and homing in bats. In Biology of Bats, 2nd ed.; Wimsatt, W.A., Ed.; Academic Press: New York, NY, USA, 1970; pp. 233–264. [Google Scholar]
- Amador, L.I.; Moyers-Arévalo, R.L.; Almeida, F.C.; Catalano, S.A.; Giannini, N.P. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J. Mammal. Evol. 2018, 25, 37–70. [Google Scholar] [CrossRef]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef]
- Holland, R.A.; Waters, D.A.; Rayner, J.M. Echolocation signal structure in the megachiropteran bat Rousettus aegyptiacus Geoffroy 1810. J. Exp. Biol. 2004, 207, 4361–4369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyman, C.P. Thermoregulation and metabolism in bats. In Biology of Bats, 2nd ed.; Wimsatt, W.A., Ed.; Academic Press: New York, NY, USA, 1970; pp. 301–330. [Google Scholar]
- Constantine, D.G. Activity Patterns of the Mexican Free-Tailed Bat; University of New Mexico Press: Albuquerque, NM, USA, 1967; pp. 1–79. [Google Scholar]
- Cockrum, E.L. Migration in the Guano Bat, Tadarida Brasiliensis; University of Kansas Museum of Natural History: Lawrence, KS, USA, 1969; pp. 303–336. [Google Scholar]
- Eisenberg, J.F. The density and biomass of tropical mammals. In Conservation Biology; Soule, M., Wilcox, B.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1980; pp. 35–56. [Google Scholar]
- Dietrich, M.; Mühldorfer, K.; Tortosa, P.; Markotter, W. Leptospira and bats: Story of an emerging friendship. PLOS Pathog. 2015, 11, e1005176. [Google Scholar] [CrossRef]
- Bunnell, J.E.; Hice, C.L.; Watts, D.M.; Montrueil, V.; Tesh, R.B.; Vinetz, J.M. Detection of pathogenic Leptospira spp. infections among mammals captured in the Peruvian Amazon basin region. Am. J. Trop. Med. Hyg. 2000, 63, 255–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetun, C.; Hoffmann, J.; Silva, R.; Souza, L.; Langoni, H. Leptospira spp. and Toxoplasma gondii antibodies in vampire bats (Desmodus rotundus) in Botucatu region, Brazil. J. Venom. Anim. Toxins Incl. Trop. Dis. 2009, 15, 546–552. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, N.N.; Alegre, E.A.; De Biasio, M.B.; Bastiani, C.E. Detecciósn de leptospiras patógenas en tejido renal de murciélagos de Corrientes, Argentina. Rev. Vet. 2014, 25, 16–20. [Google Scholar] [CrossRef]
- Smythe, L.D.; Field, H.E.; Barnett, L.; Smith, C.S.; Sohnt, M.F.; Moore, M.R.; Rolfe, P.F. Leptospiral antibodies in flying foxes in Australia. J. Wildl. Dis. 2002, 38, 182–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagadec, E.; Gomard, Y.; Guernier, V.; Dietrich, M.; Pascalis, H.; Symonds, M.L.; Moore, M.R.; Rolfe, P.F. Pathogenic Leptospira spp. in bats, Madagascar and Union of the Comoros. Emerg. Infect. Dis. 2021, 18, 1696–1698. [Google Scholar] [CrossRef]
- Desvars, A.; Naze, F.; Benneveau, A.; Cardinale, E.; Michault, A. Endemicity of leptospirosis in domestic and wild animal species from Reunion Island (Indian Ocean). Epidemiol. Infect. 2013, 141, 1154–1165. [Google Scholar] [CrossRef] [Green Version]
- Desvars, A.; Naze, F.; Vourc’h, G.; Cardinale, E.; Picardeau, M.; Bourhy, P. Similarities in Leptospira serogroup and species distribution in animals and humans in the Indian ocean island of Mayotte. Am. J. Trop. Med. Hyg. 2021, 87, 134–140. [Google Scholar] [CrossRef]
- Van Peenen, P.F.D.; Light, R.H.; Sulianti-Saroso, J. Leptospirosis in wild mammals of Indonesia-Recent surveys. Southeast Asian J. Trop. Med. Public Health 1971, 2, 496–502. [Google Scholar] [PubMed]
- Thayaparan, S.; Robertson, I.A.N.; Amraan, F.; Ut, L.S.U.; Abdullah, M.T. Serological prevalence of leptospiral Infection in wildlife in Sarawak, Malaysia. Borneo J. Res. Sci. Technol. 2013, 2, 71–74. [Google Scholar] [CrossRef] [Green Version]
- Mgode, G.F.; Mbugi, H.A.; Mhamphi, G.G.; Ndanga, D.; Nkwama, E.L. Seroprevalence of Leptospira infection in bats roosting in human settlements in Morogoro municipality in Tanzania. Tanzan. J. Health Res. 2014, 16, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, C.R.; Fraser-Chanpong, G.M.; Bhagwandin, L.J.; Race, M.W.; James, A.C. Leptospires in wildlife from Trinidad and Grenada. J. Wildl. Dis. 1983, 19, 192–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebek, Z.; Sixl, W.; Reinthaler, F.; Valova, M.; Scneeweiss, W.; Stünzner, D.; Mascher, F. Results of serological examination for leptospirosis of domestic and wild animals in the Upper Nile province (Sudan). J. Hyg. Epidemiol. Microb. Immunol. 1989, 33, 337–345. [Google Scholar]
- Ogawa, H.; Koizumi, N.; Ohnuma, A.; Mutemwa, A.; Hang, B.M.; Mweene, A.S.; Takada, A.; Sugimoto, C.; Suzuki, Y.; Kida, H.; et al. Molecular epidemiology of pathogenic Leptospira spp. in the straw-colored fruit bat (Eidolon helvum) migrating to Zambia from the Democratic Republic of Congo. Infect. Genet. Evol. 2015, 32, 143–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobbins, S.E.; Alexander, K.A. Evidence of Leptospira sp. infection among a diversity of African wildlife species: Beyond the usual suspects. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Tagi-Zade, T.A.; Mardanly, A.S.; Akhmedov, I.B.; Alekperov, F.P.; Gasanov, S.N. Examination of bats for leptospirosis in the territory of Azerbaijan SSR. Zhurnal Mikrobiol. Epidemiol. I Immunobiol. 1970, 9, 118–121. [Google Scholar]
- Victoria, R.J.; Iriarte, L.J.; Sampedro, A.C. Presence of Leptospira spp. in urban bats from Sincelejo, Sucre, Colombia. Int. J. Pharm. Tech. Res. 2018, 11, 218–225. [Google Scholar]
- Mateus, J.; Gómez, N.; Herrera-Sepúlveda, M.T.; Hidalgo, M.; Pérez-Torres, J.; Cuervo, C. Bats are a potential reservoir of pathogenic Leptospira species in Colombia. J. Infect. Dev. Ctries. 2019, 13, 278–283. [Google Scholar] [CrossRef] [Green Version]
- Díaz, M.M.; Solari, S.; Aguirre, L.F.; Aguiar, L.M.S.; Barquez, R.M. Clave de identificación de los murciélagos de Sudamérica/Chave de identificação dos morcegos da América do Sul. Publ. Espec. Nro 2016, 2, 160. [Google Scholar]
- Peláez Sánchez, R.G.; Quintero, J.Á.L.; Pereira, M.M.; Agudelo-Flórez, P. High-resolution melting curve analysis of the 16S ribosomal gene to detect and identify pathogenic and saprophytic Leptospira species in colombian isolates. Am. J. Trop. Med. Hyg. 2017, 96, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Boey, K.; Shiokawa, K.; Rajeev, S. Leptospira infection in rats: A literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 2019, 13, e0007499. [Google Scholar] [CrossRef]
- Talpada, M.D.; Garvey, N.; Sprowls, R.; Eugster, A.K.; Vinetz, J.M. Prevalence of leptospiral infection in Texas Cattle: Implications for transmission to humans. Vector Borne Zoonotic Dis. 2003, 3, 141–147. [Google Scholar] [CrossRef]
- Calderón, A.; Rodríguez, V.; Máttar, S.; Arrieta, G. Leptospirosis in pigs, dogs, rodents, humans, and water in an area of the Colombian tropics. Trop. Anim. Health Prod. 2014, 46, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Alton, G.D.; Berke, O.; Reid-Smith, R.; Ojkic, D.; Prescott, J.F. Increase in seroprevalence of canine leptospirosis and its risk factors, Ontario 1998–2006. Can. J. Vet. Res. 2009, 73, 167–175. [Google Scholar]
- Jorge, S.; Monte, L.G.; Coimbra, M.A.; Albano, A.P.; Hartwig, D.D.; Lucas, C.; Seixas, F.K.; Dellagostin, O.A.; Hartleben, C.P. Detection of virulence factors and molecular typing of pathogenic Leptospira from capybara (Hydrochaeris hydrochaeris). Curr. Microbiol. 2012, 65, 461–464. [Google Scholar] [CrossRef]
- Szonyi, B.; Agudelo-Flórez, P.; Ramírez, M.; Moreno, N.; Ko, A.I. An outbreak of severe leptospirosis in capuchin (Cebus) monkeys. Vet. J. 2011, 188, 237–239. [Google Scholar] [CrossRef] [Green Version]
- Dezzutto, D.; Barbero, R.; Canale, G.; Acutis, P.L.; Biolatti, C.; Dogliero, A.; Mitzy, M.D.; Francone, P.; Colzani, A.; Bergagna, S.; et al. Detection of Leptospira spp. in water turtle (trachemys scripta) living in ponds of Urban Parks. Vet. Sci. 2017, 4, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colagross-Schouten, A.M.; Mazet, J.A.; Gulland, F.M.; Miller, M.A.; Hietala, S. Diagnosis and seroprevalence of leptospirosis in California sea lions from coastal California. J. Wildl. Dis. 2002, 38, 7–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindtner-Knific, R.; Vergles-Rataj, A.; Vlahović, K.; Zrimšek, P.; Dovč, A. Prevalence of antibodies against Leptospira sp. in snakes, lizards and turtles in Slovenia. Acta Vet. Scand. 2013, 55, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bessa, T.A.F.; Spichler, A.; Chapola, E.G.B.; Husch, A.C.; de Almeida, M.F.; Husch, A.C. The contribution of bats to leptospirosis transmission in Sao Paulo City, Brazil. Am. J. Trop. Med. Hyg. 2010, 82, 315–317. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Mejía, A.F.; Urbina-Cardona, J.N.; Sánchez, F. Functional diversity of phyllostomid bats in an urban–rural landscape: A scale-dependent analysis. Biotropica 2020, 52, 1168–1182. [Google Scholar] [CrossRef]
- Jung, K.; Kalko, E.K. Adaptability and vulnerability of high flying Neotropical aerial insectivorous bats to urbanization. Divers. Distrib. 2011, 17, 262–274. [Google Scholar] [CrossRef]
- Russo, D.; Ancillotto, L. Sensitivity of bats to urbanization: A review. Mamm. Biol. 2015, 80, 205–212. [Google Scholar] [CrossRef]
- Kalko, E.K.; Handley, C.O.; Handley, D. Organization, diversity, and long-term dynamics of a Neotropical bat community. In Long-Term Studies of Vertebrate Communities; Cody, M.L., Smallwood, J.A., Eds.; Academic Press: New York, NY, USA, 1996; pp. 503–553. [Google Scholar]
- Meyer, C.F.J.; Struebig, M.J.; Willig, M.R. Responses of tropical bats to habitat fragmentation, logging, and deforestation. In Bats in the Anthropocene: Conservation of Bats in a Changing World; Voigt, C., Kingston, T., Eds.; Springer: Cham, Switzerland, 2016; pp. 63–103. [Google Scholar]
- Tamsitt, J.R.; Valdivieso, D. Los murciélagos y la salud pública: Estudio con especial referencia a Puerto Rico. Boletín Oficina Sanit. Panam. 1970, 69, 122–140. [Google Scholar]
- Drews, C. Convivencia con murciélagos de Costa Rica. Ambientico 2002, 103, 12–13. [Google Scholar]



| Species | Number | Percentage (%) | Frequency | Feeding Habits |
|---|---|---|---|---|
| Artibeus jamaicensis | 1 | 0.49% | 0.005 | frugivore |
| Carollia brevicauda | 13 | 6.31% | 0.063 | frugivore |
| Carollia castanea | 1 | 0.49% | 0.005 | frugivore |
| Carollia perspicillata | 13 | 6.31% | 0.063 | frugivore |
| Dermanura rava | 4 | 1.94% | 0.019 | frugivore |
| Glossophaga soricina | 43 | 20.87% | 0.209 | nectarivore |
| Sturnira bakeri | 17 | 8.25% | 0.083 | frugivore |
| Molossus molossus | 26 | 12.62% | 0.126 | insectivore |
| Artibeus lituratus | 5 | 2.43% | 0.024 | frugivore |
| Myotis caucensis | 9 | 4.37% | 0.044 | insectivore |
| Artibeus planirostris | 55 | 26.70% | 0.267 | frugivore |
| Uroderma convexum | 15 | 7.28% | 0.073 | frugivore |
| Rhogeessa sp. | 1 | 0.49% | 0.005 | insectivore |
| Phyllostomus hastatus | 2 | 0.97% | 0.010 | omnivore |
| Phyllostomus discolor | 1 | 0.49% | 0.005 | omnivore |
| TOTAL | 206 | 100% | 1 |
| Code | Phylogenetic Identification (16S Ribosomal Gene) | Infected Species | Feeding Habits | Gender | Municipality |
|---|---|---|---|---|---|
| ZM-022 | Leptospira kirschneri | Carollia perspicillata | Frugivore | Female | Carepa |
| ZM-025 | Leptospira kirschneri | Dermanura rava | Frugivore | Male | Carepa |
| ZM-047 | Leptospira interrogans | Glossophaga soricina | Nectarivore | Female | Apartadó |
| ZM-056 | Leptospira kirschneri | Glossophaga soricina | Nectarivore | Male | Apartadó |
| ZM-060 | Leptospira noguchii | Glossophaga soricina | Nectarivore | Female | Apartadó |
| ZN-083 | Leptospira noguchii | Uroderma convexum | Frugivore | Male | Chigorodó |
| ZN-087 | Leptospira noguchii | Uroderma convexum | Frugivore | Male | Chigorodó |
| ZN-107 | Leptospira noguchii | Uroderma convexum | Frugivore | Female | Chigorodó |
| ZN-125 | Leptospira noguchii | Molossus molossus | Insectivore | Female | Turbo |
| ZN-126 | Leptospira kirschneri | Molossus molossus | Insectivore | Male | Turbo |
| ZN-129 | Leptospira kirschneri | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-136 | Leptospira borgpetersenii | Glossophaga soricina | Nectarivore | Female | Turbo |
| ZN-138 | Leptospira borgpetersenii | Glossophaga soricina | Nectarivore | Female | Turbo |
| ZN-139 | Leptospira interrogans | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-141 | Leptospira alexanderi | Uroderma convexum | Frugivore | Female | Turbo |
| ZN-149 | Leptospira alexanderi | Glossophaga soricina | Nectarivore | Male | Turbo |
| ZN-150 | Leptospira borgpetersenii | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-163 | Leptospira kirschneri | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-168 | Leptospira noguchii | Uroderma convexum | Frugivore | Male | Necoclí |
| ZN-169 | Leptospira interrogans | Uroderma convexum | Frugivore | Female | Necoclí |
| Leptospira Species | Infected Bat Species | Infected Bats |
|---|---|---|
| Leptospira borgpetersenii | Glossophaga soricina (2) Artibeus planirostris (1) | 3 |
| Leptospira alexanderi | Uroderma convexum (1) Glossophaga soricina (1) | 2 |
| Leptospira noguchii | Glossophaga soricina (1) Uroderma convexum (4) Molossus molossus (1) | 6 |
| Leptospira interrogans | Glossophaga soricina (1) Artibeus planirostris (1) Uroderma convexum (1) | 3 |
| Leptospira kirschneri | Carollia perspicillata (1) Dermanura rava (1) Glossophaga soricina (1) Molossus molossus (1) Artibeus planirostris (2) | 6 |
| Total: 20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monroy, F.P.; Solari, S.; Lopez, J.Á.; Agudelo-Flórez, P.; Peláez Sánchez, R.G. High Diversity of Leptospira Species Infecting Bats Captured in the Urabá Region (Antioquia-Colombia). Microorganisms 2021, 9, 1897. https://doi.org/10.3390/microorganisms9091897
Monroy FP, Solari S, Lopez JÁ, Agudelo-Flórez P, Peláez Sánchez RG. High Diversity of Leptospira Species Infecting Bats Captured in the Urabá Region (Antioquia-Colombia). Microorganisms. 2021; 9(9):1897. https://doi.org/10.3390/microorganisms9091897
Chicago/Turabian StyleMonroy, Fernando P., Sergio Solari, Juan Álvaro Lopez, Piedad Agudelo-Flórez, and Ronald Guillermo Peláez Sánchez. 2021. "High Diversity of Leptospira Species Infecting Bats Captured in the Urabá Region (Antioquia-Colombia)" Microorganisms 9, no. 9: 1897. https://doi.org/10.3390/microorganisms9091897







