Diverse Bacteriophages Infecting the Bacterial Striped Catfish Pathogen Edwardsiella ictaluri
Abstract
1. Introduction
2. Materials and Methods
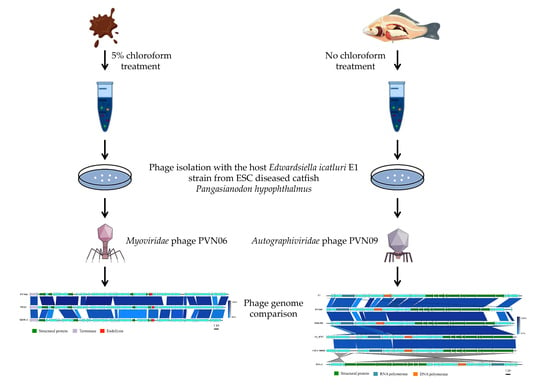
2.1. Phage Isolation and Purification
2.2. Host Range Analysis
2.3. Transmission Electron Microscopy (TEM)
2.4. One-Step Growth Curve
2.5. Bacteriophage Stability at Acidic and Alkaline pH, in Solvent, and at Different Temperatures
2.6. Phage Genomic DNA Extraction, Sequencing and Bioinformatics Analysis
3. Results
3.1. Two Novel Phages Infecting Edwardsiella Ictaluri Isolated in Vietnam Displayed Various Biological Features
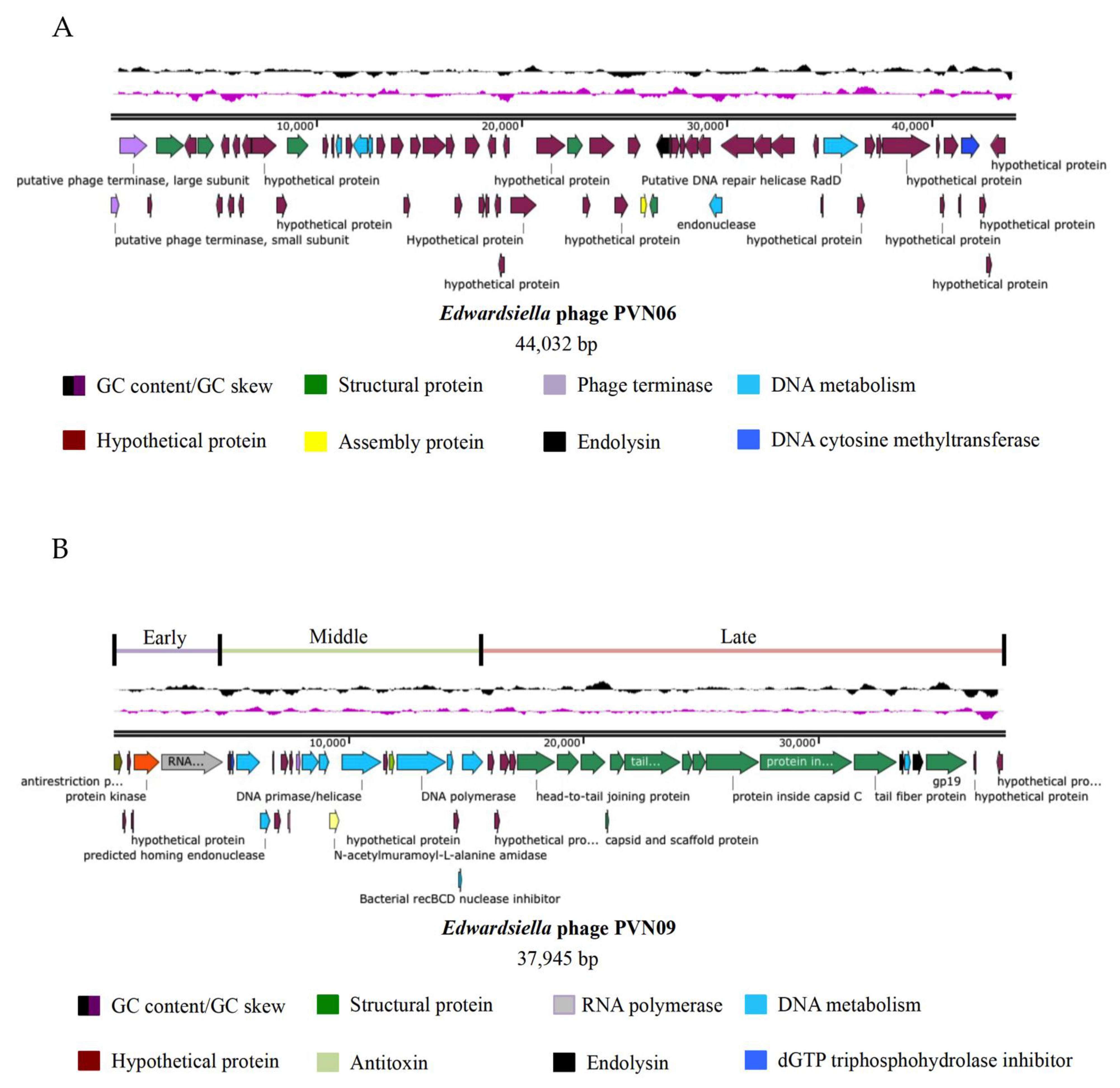
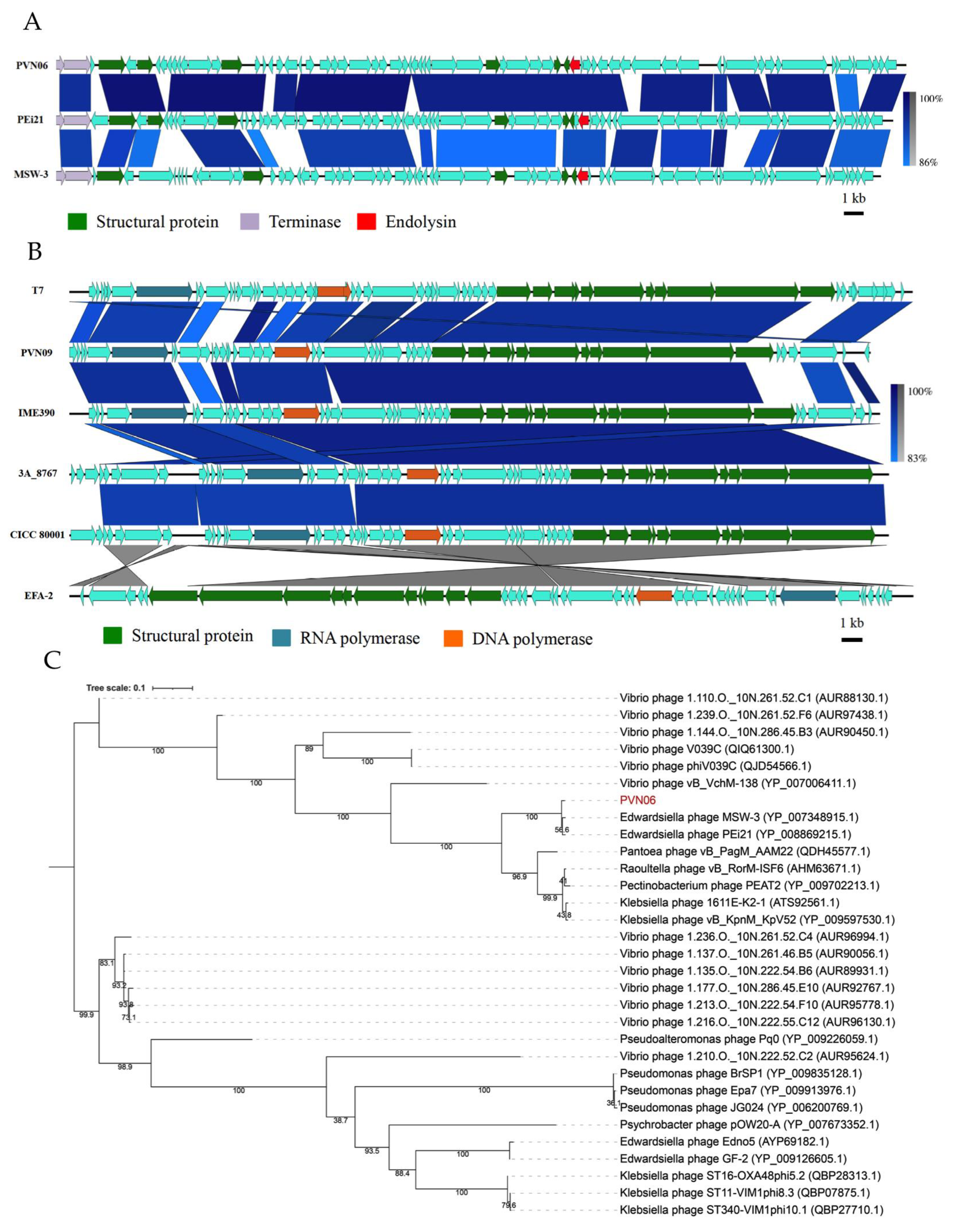
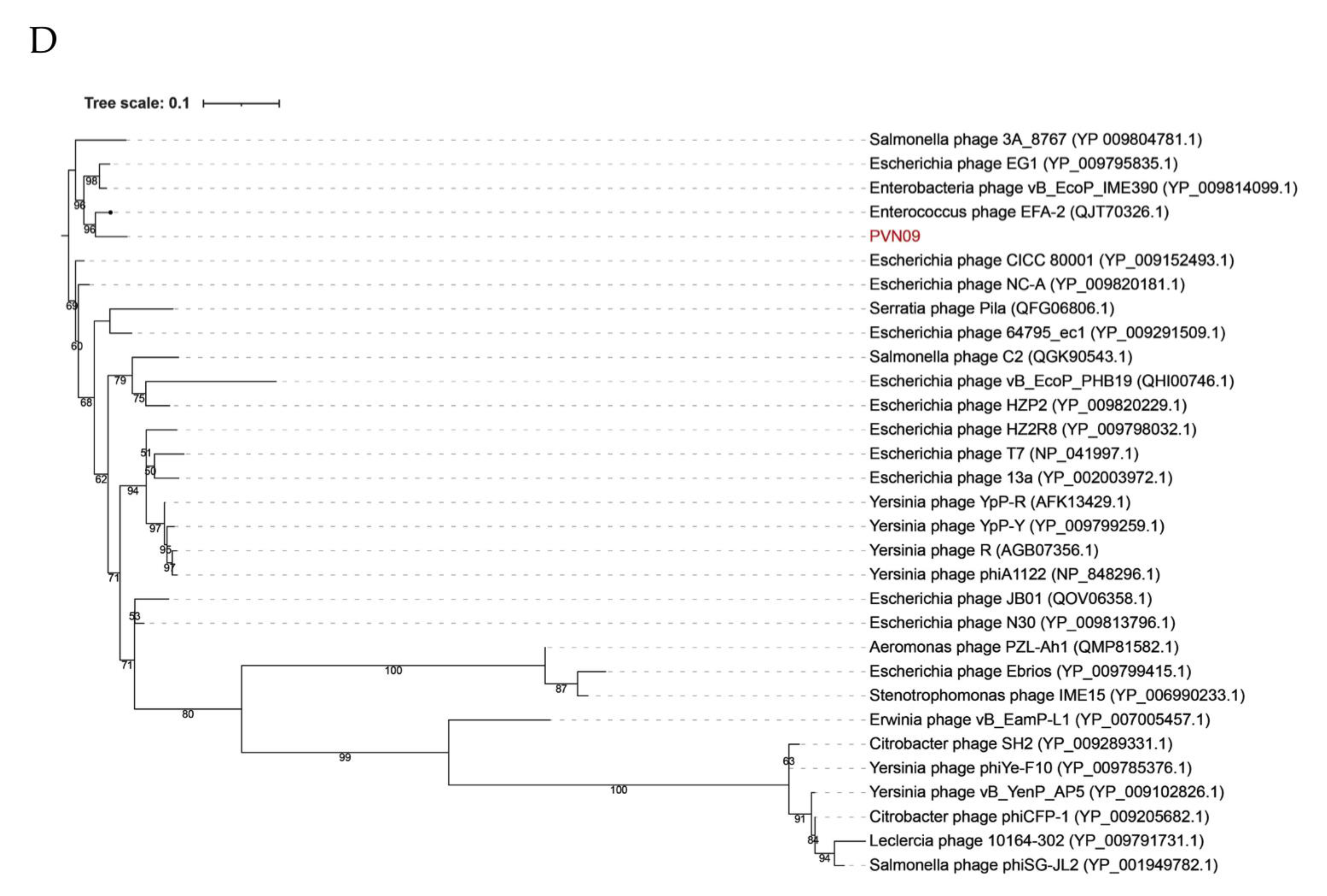
3.2. Genomic Variations of Two Novel E. ictaluri Phages
4. Discussion
4.1. Various Approaches in Phage Isolation Revealed the Diversity of Edwardsiella Phages in Natural Habitat
4.2. A Probable Adaptation of Phage vB_EiA_PVN09 to the Host E. ictaluri
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plumb, J.A.; Sanchez, D.J. Susceptibility of five species of fish to Edwardsiella ictaluri. J. Fish. Dis. 1983, 6, 261–266. [Google Scholar] [CrossRef]
- Crumlish, M.; Dung, T.T.; Turnbull, J.F.; Ngoc, N.T.N.; Ferguson, H.W. Identification of Edwardsiella ictaluri from diseased freshwater catfish, Pangasius hypophthalmus (Sauvage), cultured in the Mekong Delta, Vietnam. J. Fish. Dis. 2002, 25, 733–736. [Google Scholar] [CrossRef]
- Yuasa, K.; Kholidin, E.B.; Panigoro, N.; Hatai, K. First isolation of Edwardsiella ictaluri from cultured striped catfish Pangasius hypophthalmus in Indonesia. Fish. Pathol. 2003, 38, 181–183. [Google Scholar] [CrossRef][Green Version]
- Sakai, T.; Yuasa, K.; Ozaki, A.; Sano, M.; Okuda, R.; Nakai, T.; Iida, T. Genotyping of Edwardsiella ictaluri isolates in Japan using amplified-fragment length polymorphism analysis. Lett. Appl. Microbiol. 2009, 49, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Hawke, J.P.; Kent, M.; Rogge, M.; Baumgartner, W.; Wiles, J.; Shelley, J.; Savolainen, L.C.; Wagner, R.; Murray, K.; Peterson, T.S. Edwardsiellosis caused by Edwardsiella ictaluri in laboratory populations of Zebrafish Danio rerio. J. Aquat. Anim. Health 2013, 25, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Oanh, D.T.H.; Phuong, N.T. Detection of Edwardsiella ictaluri causing white spots in the internal organs of striped catfish Pangasianodon hypophthalmus by using polymerase chain reaction technique. J. Sci. Can Tho Univ. 2010, 13, 151–159. (In Vietnamese) [Google Scholar]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 21. [Google Scholar] [CrossRef]
- Tu, T.D.; Haesebrouck, F.; Nguyen, A.T.; Sorgeloos, P.; Baele, M.; Decostere, A. Antimicrobial susceptibility pattern of Edwardsiella ictaluri isolates from natural outbreaks of bacillary necrosis of Pangasianodon hypophthalmus in Vietnam. Microb. Drug Resist. Larchmt. N 2008, 14, 311–316. [Google Scholar] [CrossRef]
- Salako, D.; Trang, P.N.; Ha, N.C.; Miyamoto, T.; Thi, A. Prevalence of antibiotic resistance Escherichia coli isolated from pangasius catfish (Pangasius hypophthalmus) fillet during freezing process at two factories in Mekong Delta Vietnam. Food Res. 2020, 4, 1785–1793. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Pratama, A.A.; van Elsas, J.D. The “neglected” soil virome-potential role and impact. Trends Microbiol. 2018, 26, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Rigano, L.A.; Fineran, P.C. Phage-based biocontrol strategies and their application in agriculture and aquaculture. Biochem. Soc. Trans. 2018, 46, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Carrias, A.; Welch, T.J.; Waldbieser, G.C.; Mead, D.A.; Terhune, J.S.; Liles, M.R. Comparative genomic analysis of bacteriophages specific to the channel catfish pathogen Edwardsiella ictaluri. Virol. J. 2011, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Yasuike, M.; Sugaya, E.; Nakamura, Y.; Shigenobu, Y.; Kawato, Y.; Kai, W.; Nagai, S.; Fujiwara, A.; Sano, M.; Kobayashi, T.; et al. Complete genome sequence of a novel myovirus which infects atypical strains of Edwardsiella tarda. Genome Announc. 2013, 1, e00248-12. [Google Scholar] [CrossRef]
- Yasuike, M.; Kai, W.; Nakamura, Y.; Fujiwara, A.; Kawato, Y.; Hassan, E.S.; Mahmoud, M.M.; Nagai, S.; Kobayashi, T.; Ototake, M.; et al. Complete genome sequence of the Edwardsiella ctaluri-specific bacteriophage PEi21, isolated from river water in Japan. Genome Announc. 2014, 2, e00228-14. [Google Scholar] [CrossRef] [PubMed]
- Booth, N.J.; Beekman, J.B.; Thune, R.L. Edwardsiella ictaluri encodes an acid-activated urease that is required for intracellular replication in channel catfish (Ictalurus punctatus) macrophages. Appl. Environ. Microbiol. 2009, 75, 6712–6720. [Google Scholar] [CrossRef]
- Rogge, M.L.; Dubytska, L.; Jung, T.S.; Wiles, J.; Elkamel, A.A.; Rennhoff, A.; Oanh, D.T.; Thune, R.L. Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis. Aquat. Org. 2013, 106, 17–29. [Google Scholar] [CrossRef]
- Walakira, J.K.; Carrias, A.A.; Hossain, M.J.; Jones, E.; Terhune, J.S.; Liles, M.R. Identification and characterization of bacteriophages specific to the catfish pathogen, Edwardsiella ictaluri. J. Appl. Microbiol. 2008, 105, 2133–2142. [Google Scholar] [CrossRef] [PubMed]
- Hoang, H.A.; Yen, M.H.; Ngoan, V.T.; Nga, L.P.; Oanh, D.T.H. Virulent bacteriophage of Edwardsiella ictaluri isolated from kidney and liver of striped catfish Pangasianodon hypophthalmus in Vietnam. Dis. Aquat. Org. 2018, 132, 49–56. [Google Scholar] [CrossRef]
- Ackermann, H.-W.; Heldal, M. Basic electron microscopy of aquatic viruses. Man. Aquat. Viral Ecol. 2010, 18, 182–192. [Google Scholar] [CrossRef]
- Verma, V.; Harjai, K.; Chhibber, S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: A potential therapeutic agent. Curr. Microbiol. 2009, 59, 274–281. [Google Scholar] [CrossRef]
- Hoang, A.H.; Tran, T.T.X.; Le, P.N.; Dang, T.H.O. Selection of phages to control Aeromonas hydrophila—An infectious agent in striped catfish. Biocontrol. Sci. 2019, 24, 23–28. [Google Scholar] [CrossRef]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage ΦYeO3-12, specific for Yersinia enterocolitica serotype O:3, Is related to coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef]
- Jun, J.W.; Kim, J.H.; Shin, S.P.; Han, J.E.; Chai, J.Y.; Park, S.C. Protective effects of the Aeromonas phages PAh1-C and PAh6-C against mass mortality of the cyprinid loach (Misgurnus anguillicaudatus) caused by Aeromonas hydrophila. Aquaculture 2013, 416-417, 289–295. [Google Scholar] [CrossRef]
- Yamaki, S.; Omachi, T.; Kawai, Y.; Yamazaki, K. Characterization of a novel morganella morganii bacteriophage FSP1 isolated from river water. FEMS Microbiol. Lett. 2014, 359, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinform. Oxf. Engl. 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinform. Oxf. Engl. 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Cuccuru, G.; Orsini, M.; Pinna, A.; Sbardellati, A.; Soranzo, N.; Travaglione, A.; Uva, P.; Zanetti, G.; Fotia, G. Orione, a Web-based framework for NGS analysis in microbiology. Bioinform. Oxf. Engl. 2014, 30, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-Likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H.; Park, B.H. An enzyme produced by a phage-host cell system: II. The properties of the polysaccharide depolymerase. Virology 1956, 2, 719–736. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. (Praha) 2011, 56, 191–200. [Google Scholar] [CrossRef]
- Tekedar, H.C.; Blom, J.; Kalindamar, S.; Nho, S.; Karsi, A.; Lawrence, M.L. Comparative genomics of the fish pathogens Edwardsiella ictaluri 93-146 and Edwardsiella piscicida C07-087. Microb. Genom. 2020, 6, e000322. [Google Scholar] [CrossRef]
- Grynberg, M.; Godzik, A. NERD: A DNA processing-related domain present in the anthrax virulence plasmid, PXO1. Trends Biochem. Sci. 2004, 29, 106–110. [Google Scholar] [CrossRef]
- Philipson, C.W.; Voegtly, L.J.; Lueder, M.R.; Long, K.A.; Rice, G.K.; Frey, K.G.; Biswas, B.; Cer, R.Z.; Hamilton, T.; Bishop-Lilly, K.A. Characterizing phage genomes for therapeutic applications. Viruses 2018, 10, 188. [Google Scholar] [CrossRef]
- Bao, Y.; Chetvernin, V.; Tatusova, T. Improvements to pairwise seauence comparison (PASC): A genome-based web tool for virus classification. Arch. Virol. 2014, 159, 3293–3304. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Rodney Brister, J. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Shotts, E.B.; Blazer, V.S.; Waltman, W.D. Pathogenesis of experimental Edwardsiella ictaluri infections in Channel Catfish (Icta Lurus punctatus). Can. J. Fish. Aquat. Sci. 1986, 43, 36–42. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.-T.; Xuan, T.T.T.; Ngoc, T.H.; Duyen, L.T.M.; Vinh, T.Q.; My, P.D.T.; Hoang, H.A.; Nga, L.P. Diverse Bacteriophages Infecting the Bacterial Striped Catfish Pathogen Edwardsiella ictaluri. Microorganisms 2021, 9, 1830. https://doi.org/10.3390/microorganisms9091830
Nguyen T-T, Xuan TTT, Ngoc TH, Duyen LTM, Vinh TQ, My PDT, Hoang HA, Nga LP. Diverse Bacteriophages Infecting the Bacterial Striped Catfish Pathogen Edwardsiella ictaluri. Microorganisms. 2021; 9(9):1830. https://doi.org/10.3390/microorganisms9091830
Chicago/Turabian StyleNguyen, Tan-Trung, Tran T. T. Xuan, To H. Ngoc, Le T. My Duyen, Tu Q. Vinh, Pham D. T. My, Hoang A. Hoang, and Le P. Nga. 2021. "Diverse Bacteriophages Infecting the Bacterial Striped Catfish Pathogen Edwardsiella ictaluri" Microorganisms 9, no. 9: 1830. https://doi.org/10.3390/microorganisms9091830
APA StyleNguyen, T.-T., Xuan, T. T. T., Ngoc, T. H., Duyen, L. T. M., Vinh, T. Q., My, P. D. T., Hoang, H. A., & Nga, L. P. (2021). Diverse Bacteriophages Infecting the Bacterial Striped Catfish Pathogen Edwardsiella ictaluri. Microorganisms, 9(9), 1830. https://doi.org/10.3390/microorganisms9091830