Novel Microdialysis Technique Reveals a Dramatic Shift in Metabolite Secretion during the Early Stages of the Interaction between the Ectomycorrhizal Fungus Pisolithus microcarpus and Its Host Eucalyptus grandis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials and Growth Conditions
2.2. Experimental Design
2.3. Metabolite Analysis
3. Results
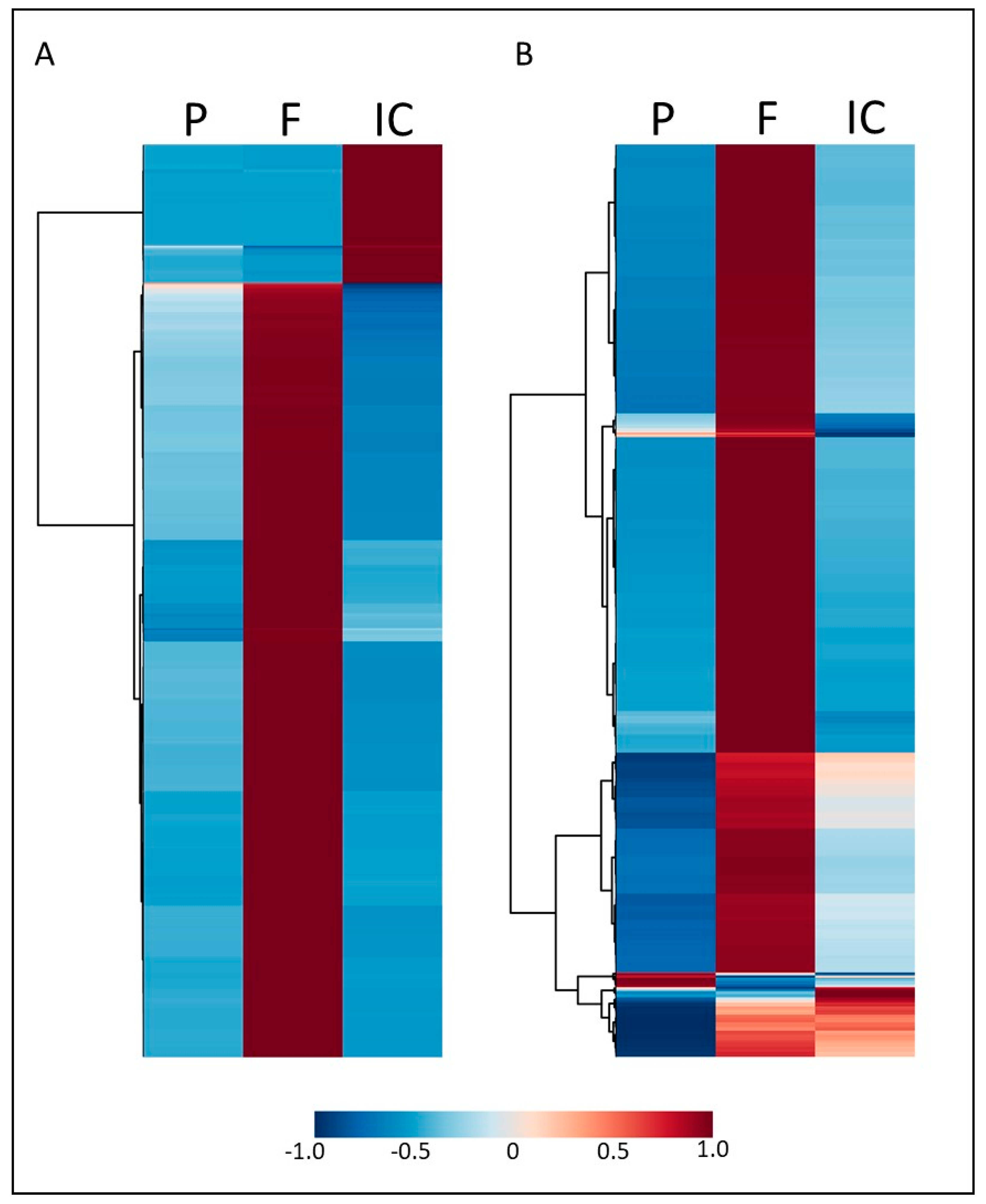
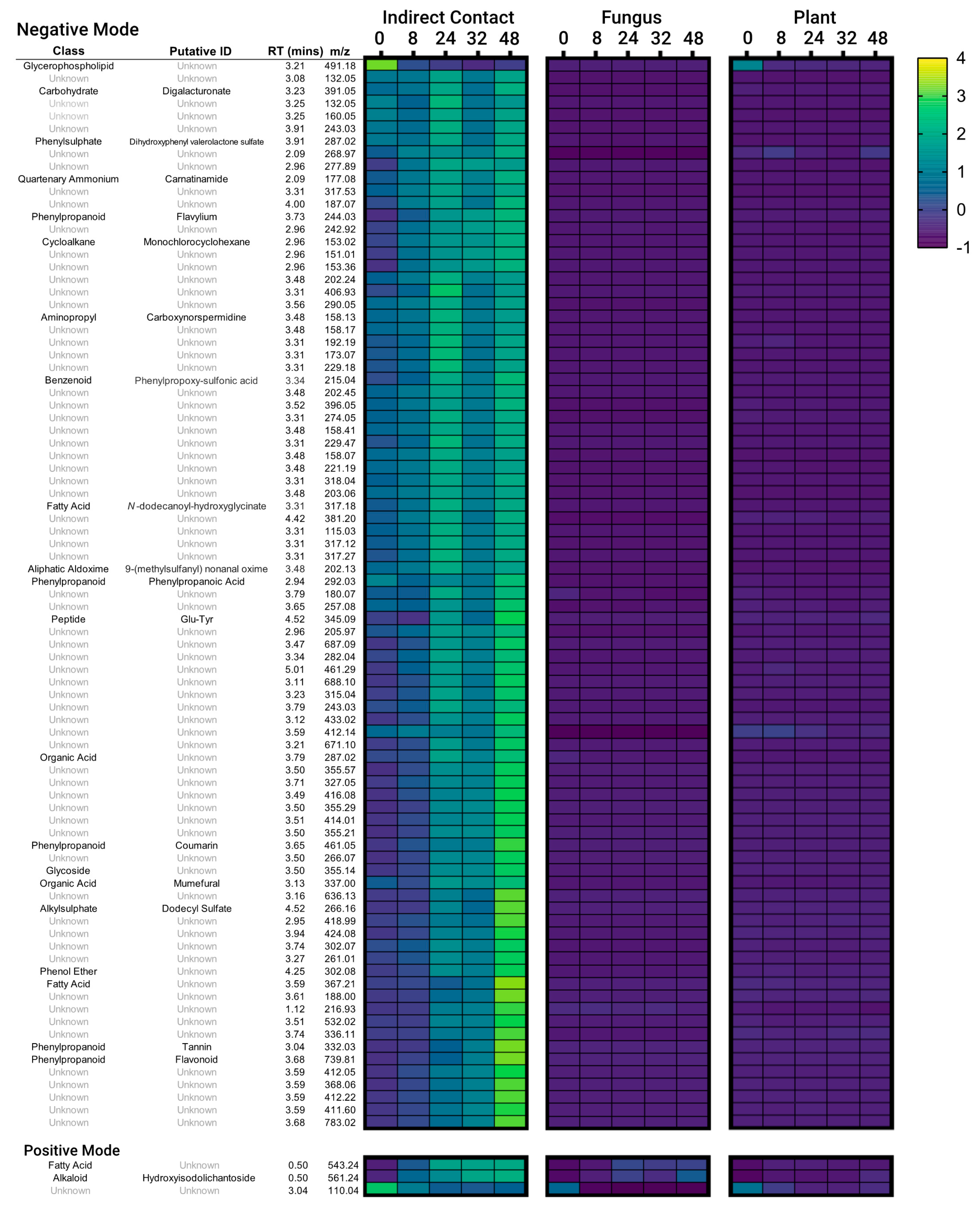
3.1. Indirect Contact between E. grandis and P. microcarpus Alters the Secretion of Secondary Metabolites in Both Organisms with the Induction of a New Set of Metabolites
3.2. Secondary Metabolites Induced by Indirect Contact between E. grandis and P. microcarpus Appear within Hours of Contact and Increase in Concentration over Time
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Steidinger, B.S.; Crowther, T.W.; Liang, J.; Van Nuland, M.E.; Werner, G.D.A.; Reich, P.B.; Nabuurs, G.J.; de-Miguel, S.; Zhou, M.; Picard, N.; et al. Climatic controls of decomposition drive the global biogeography of forest-tree symbioses. Nature 2019, 569, 404–408. [Google Scholar] [CrossRef] [Green Version]
- Becquer, A.; Guerrero-Galán, C.; Eibensteiner, J.L.; Houdinet, G.; Bucking, H.; Zimmermann, S.D.; Garcia, K. Chapter three—the ectomycorrhizal contribution to tree nutrition. Adv. Bot. Res. 2019, 89, 77–126. [Google Scholar]
- Garcia, K.; Delaux, P.-M.; Cope, K.R.; Ané, J.-M. Molecular signals required for the establishment and maintenance of ectomycorrhizal symbiosis. New Phytol. 2015, 208, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Martin, F.M. Know your enemy, embrace your friend: Using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 2018, 93, 729–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, P.E.; Sreedasyam, A.; Trivedi, G.; Desai, S.; Dai, Y.; Cseke, L.J.; Collart, F.R. Multi-omics approach identifies molecular mechanisms of plant-fungus mycorrhizal interaction. Front. Plant Sci. 2016, 6, 1061. [Google Scholar] [CrossRef]
- Plett, J.M.; Tisserant, E.; Brun, A.; Morin, E.; Grigoriev, I.V.; Kuo, A.; Martin, F.; Kohler, A. The mutualist Laccaria bicolor expresses a core gene regulon during the colonization of diverse host plants and a variable regulon to counteract host-specific defenses. MPMI 2015, 28, 261–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, J.W.H.; Lutz, A.; Natera, S.; Wang, M.; Ng, V.; Grigoriev, I.; Martin, F.; Roessner, U.; Anderson, I.C.; Plett, J.M. The influence of contrasting microbial lifestyles on the pre-symbiotic metabolite responses of Eucalyptus grandis roots. Front. Ecol. Evol. 2019, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Menotta, M.; Amicucci, A.; Sisti, D.; Gioacchini, A.M.; Stocchi, V. Differential gene expression during pre-symbiotic interaction between Tuber borchii Vittad. and Tilia americana L. Curr. Genet. 2004, 46, 158–165. [Google Scholar] [CrossRef]
- Felten, J.; Kohler, A.; Morin, E.; Bhalerao, R.P.; Palme, K.; Martin, F.; Ditengou, F.A.; Legué, V. The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signalling. Plant Physiol. 2009, 151, 1991–2005. [Google Scholar] [CrossRef] [Green Version]
- Ditengou, F.A.; Müller, A.; Rosenkranz, M.; Felten, J.; Lasok, H.; van Doorn, M.M.; Legué, V.; Palme, K.; Schnitzler, J.-P.; Polle, A. Volatile signalling by sesquiterpenes from ectomycorrhizal fungi reprogrammes root architecture. Nat. Commun. 2015, 6, 6279. [Google Scholar] [CrossRef]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legue, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tschaplinkski, T.J.; Plett, J.M.; Engle, N.L.; Deveau, A.; Cushman, K.C.; Martin, M.Z.; Doktycz, M.J.; Tuskan, G.A.; Brun, A.; Kohler, A.; et al. Populus trichocarpa and Populus deltoides exhibit different metabolomic responses to colonization by the symbiotic fungus Laccaria bicolor. Mol. Plant-Microbe Interact. 2014, 27, 546–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, K.; Matsushita, N.; Suzuki, K.; Hogetsu, T. Flavonoids induce germination of basidiospores of the ectomycorrhizal fungus Suillus bovinus. Mycorrhiza 2007, 17, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Martin, F. Poplar root exudates contain compounds that induce the expression of MiSSP7 in Laccaria bicolor. Plant Signal. Behav. 2012, 7, 12–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vayssières, A.; Pěnčík, A.; Felten, J.; Kohler, A.; Ljung, K.; Martin, F.; Legué, V. Development of the Poplar-Laccaria bicolor ectomycorrhiza modifies root auxin metabolism, signaling, and response. Plant Physiol. 2015, 169, 890–902. [Google Scholar] [CrossRef] [Green Version]
- Splivallo, R.; Fischer, U.; Gobel, C.; Feussner, I.; Karlovsky, P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 2009, 150, 2018–2029. [Google Scholar] [CrossRef] [Green Version]
- Cope, K.R.; Bascaules, A.; Irving, T.B.; Venkateshwaran, M.; Maeda, J.; Garcia, K.; Rush, T.A.; Ma, C.; Labbé, J.; Jawdy, S.; et al. The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize Populus roots. Plant Cell 2019, 31, 2386–2410. [Google Scholar] [CrossRef] [Green Version]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssières, A.; Deveau, D.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C.; et al. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollet, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Ann. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Plett, J.M.; Plett, K.L.; Wong-Bajracharya, J.; de Freitas Pereira, M.; Dutra Costa, M.; Kohler, A.; Martin, F.; Anderson, I.C. Mycorrhizal effector PaMiSSP10b alters polyamine biosynthesis in Eucalytpus root cells and promotes root colonization. New Phytol. 2020, 228, 712–727. [Google Scholar] [CrossRef] [PubMed]
- Pellegrin, C.; Daguerre, Y.; Ruytinx, J.; Guinet, F.; Kemppaine, M.; Frey, N.F.D.; Puech-Pagès, V.; Hecker, A.; Pardo, A.G.; Martin, F.M.; et al. Laccaria bicolor MiSSP8 is a small-secreted prtein decisive for the establishment of the ectomycorrhizal symbiosis. Environ. Microbiol. 2019, 21, 3765–3779. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Chen, X.; Kemppainen, M.; Pardo, A.G.; Veneault-Fourrey, C.; Kohler, A.; Martin, F.M. The small secreted effector protein MiSSP7.6 of Laccaria bicolor is required for the establishment of ectomycorrhizal symbiosis. Environ. Microbiol. 2020, 22, 1435–1446. [Google Scholar] [CrossRef]
- Wong, J.W.H.; Plett, K.L.; Natera, S.H.A.; Roessner, U.; Anderson, I.C.; Plett, J.M. Comparative metabolomics implicates threitol as a fungal signal supporting colonization of Armillaria luteobubalina on eucalypt roots. Plant Cell Environ. 2020, 43, 374–386. [Google Scholar] [CrossRef]
- Oburger, E.; Jones, D.L. Sampling root exudates—Mission impossible? Rhizosphere 2018, 6, 116–133. [Google Scholar] [CrossRef]
- Johansson, E.M.; Fransson, P.M.A.; Finlay, R.D.; van Hees, P.A.W. Quantitative analysis of root and ectomycorrhizal exudates as a response to Pb, Cd and As stress. Plant Soil 2008, 313, 39–54. [Google Scholar] [CrossRef]
- Johansson, E.M.; Fransson, P.M.A.; Finlay, R.D.; van Hees, P.A.W. Quantitative analysis of soluble exudates produced by ectomycorrhizal roots as a response to ambient and elevated CO2. Soil Biol. Biochem. 2009, 41, 1111–1116. [Google Scholar] [CrossRef]
- De Vries, F.T.; Williams, A.; Stringer, F.; Willcocks, R.; McEwing, R.; Langridge, H.; Straathof, A.L. Changes in root-exudate-induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. New Phytol. 2019, 224, 132–145. [Google Scholar] [CrossRef]
- Menotta, M.; Gioacchini, A.M.; Amicucci, A.; Buffalini, M.; Sisti, D.; Stocchi, V. Headspace solid-phase microextraction with gas chromatography and mass spectrometry in the investigation of volatile organic compounds in an ectomycorrhizae synthesis system. Rapid Commun. Mass Spectrom. 2004, 18, 206–210. [Google Scholar] [CrossRef]
- Buckley, S.; Allen, D.; Brackin, R.; Jämtgård, S.; Näsholm, T.; Schmidt, S. Microdialysis as an in situ technique for sampling soil enzymes. Soil Biol. Biochem. 2019, 135, 20–27. [Google Scholar] [CrossRef]
- Buckley, S.; Brackin, R.; Jämtgård, S.; Näsholm, T.; Schmidt, S. Microdialysis in soil environments: Current practice and future perspectives. Soil Biol. Biochem. 2020, 143, 107743. [Google Scholar] [CrossRef]
- Randewig, D.; Marchall, J.D.; Näsholm, T.; Jämtgård, S. Combining microdialysis with metabolomics to characterize the in situ composition of dissolved organic compounds in boreal forest soil. Soil Biol. Biochem. 2019, 136, 107530. [Google Scholar] [CrossRef]
- Duplessis, S.; Courty, P.-E.; Tagu, D.; Martin, F. Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol. 2005, 165, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Myberg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualist. Nature Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Plett, K.L.; Kohler, A.; Lebel, T.; Singan, V.R.; Bauer, D.; He, G.; Ng, V.; Grigoriev, I.V.; Martin, F.; Plett, J.M.; et al. Intra-species genetic variability drives carbon metabolism and symbiotic host interactions in the ectomycorrhizal fungus Pisolithus microcarpus. Environ. Microbiol. 2021, 23, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Kohler, A.; Khachane, A.; Keniry, K.; Plett, K.L.; Martin, F.; Anderson, I.C. The effect of elevated carbon dioxide on the interaction between Eucalyptus grandis and diverse isolates of Pisolithus sp. is associated with a complex shift in the root transcriptome. New Phytol. 2015, 206, 1423–1436. [Google Scholar] [CrossRef] [Green Version]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLD-DA and SIMCA classification. J. Chemom. 2007, 20, 341–351. [Google Scholar] [CrossRef]
- Wong-Bajracharya, J.; Castañeda-Gómez, L.; Plett, K.L.; Anderson, I.C.; Carrillo, Y.; Plett, J.M. Untangling the effect of roots and mutualistic ectomycorrhizal fungi on soil metabolite profiles under ambient and elevated carbon dioxide. Soil Biol. Biochem. 2020, 151, 108021. [Google Scholar] [CrossRef]
- Ghirardo, A.; Fochi, V.; Lange, B.; Witting, M.; Schnitzler, J.-P.; Perotto, S.; Balestrini, R. Metabolomic adjustments in the orchid mycorrhizal fungus Tulasnella calospora during symbiosis with Serapias vomeracea. New Phytol. 2020, 228, 1939–1952. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, C.J.; Broc, M.T.; van Diepen, L.T.A.; Maignien, L.; Ewers, B.E.; Weinig, C. The plant circadian clock influences rhizospheric community structure and function. ISME J. 2018, 12, 400–410. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizophere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, V. Phenylpropanoid biosynthesis. Molec Plant 2010, 3, 2–20. [Google Scholar]
- Weiss, M.; Mikolajewski, S.; Pieipp, H.; Schmitt, U.; Schmidt, J.; Wray, V.; Strack, D. Tissue-specific and development-dependent accumulation of phenylpropanoids in larch mycorrhizas. Plant Physiol. 1997, 114, 15–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, M.; Schmidt, J.; Neumann, D.; Wray, V.; Christ, R.; Strack, D. Phenylpropanoids in mycorrhizas of the Pinaceae. Planta 1999, 208, 491–502. [Google Scholar] [CrossRef]
- Feugey, L.; Strullu, D.-G.; Poupard, P.; Simoneau, P. Induced defence responses limit Hartig net formation in ectomycorrhizal birch roots. New Phytol. 2002, 144, 541–547. [Google Scholar] [CrossRef]
- Behr, M.; Baldacci-Cresp, F.; Kohler, A.; Morreel, K.; Goeminne, G.; Van Acker, R.; Veneault-Fourrey, C.; Mol, A.; Pilate, G.; Boerjan, W.; et al. Alterations in the phenylpropanoid pathway affect poplar ability for ectomycorrhizal colonisation and susceptibility to root-knot nemotodes. Mycorrhiza 2020, 30, 555–566. [Google Scholar] [CrossRef]
- Lagrange, H.; Jay-Allgmand, C.; Lapeyrie, F. Rutin, the phenolglycoside from eucalyptus root exudates, stimulates Pisolithus hyphal growth at picomolar concentrations. New Phytol. 2001, 149, 349–355. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plett, K.L.; Buckley, S.; Plett, J.M.; Anderson, I.C.; Lundberg-Felten, J.; Jämtgård, S. Novel Microdialysis Technique Reveals a Dramatic Shift in Metabolite Secretion during the Early Stages of the Interaction between the Ectomycorrhizal Fungus Pisolithus microcarpus and Its Host Eucalyptus grandis. Microorganisms 2021, 9, 1817. https://doi.org/10.3390/microorganisms9091817
Plett KL, Buckley S, Plett JM, Anderson IC, Lundberg-Felten J, Jämtgård S. Novel Microdialysis Technique Reveals a Dramatic Shift in Metabolite Secretion during the Early Stages of the Interaction between the Ectomycorrhizal Fungus Pisolithus microcarpus and Its Host Eucalyptus grandis. Microorganisms. 2021; 9(9):1817. https://doi.org/10.3390/microorganisms9091817
Chicago/Turabian StylePlett, Krista L., Scott Buckley, Jonathan M. Plett, Ian C. Anderson, Judith Lundberg-Felten, and Sandra Jämtgård. 2021. "Novel Microdialysis Technique Reveals a Dramatic Shift in Metabolite Secretion during the Early Stages of the Interaction between the Ectomycorrhizal Fungus Pisolithus microcarpus and Its Host Eucalyptus grandis" Microorganisms 9, no. 9: 1817. https://doi.org/10.3390/microorganisms9091817





