Genome-Wide Analysis of Nutrient Signaling Pathways Conserved in Arbuscular Mycorrhizal Fungi
Abstract
1. Introduction
2. Materials and Methods
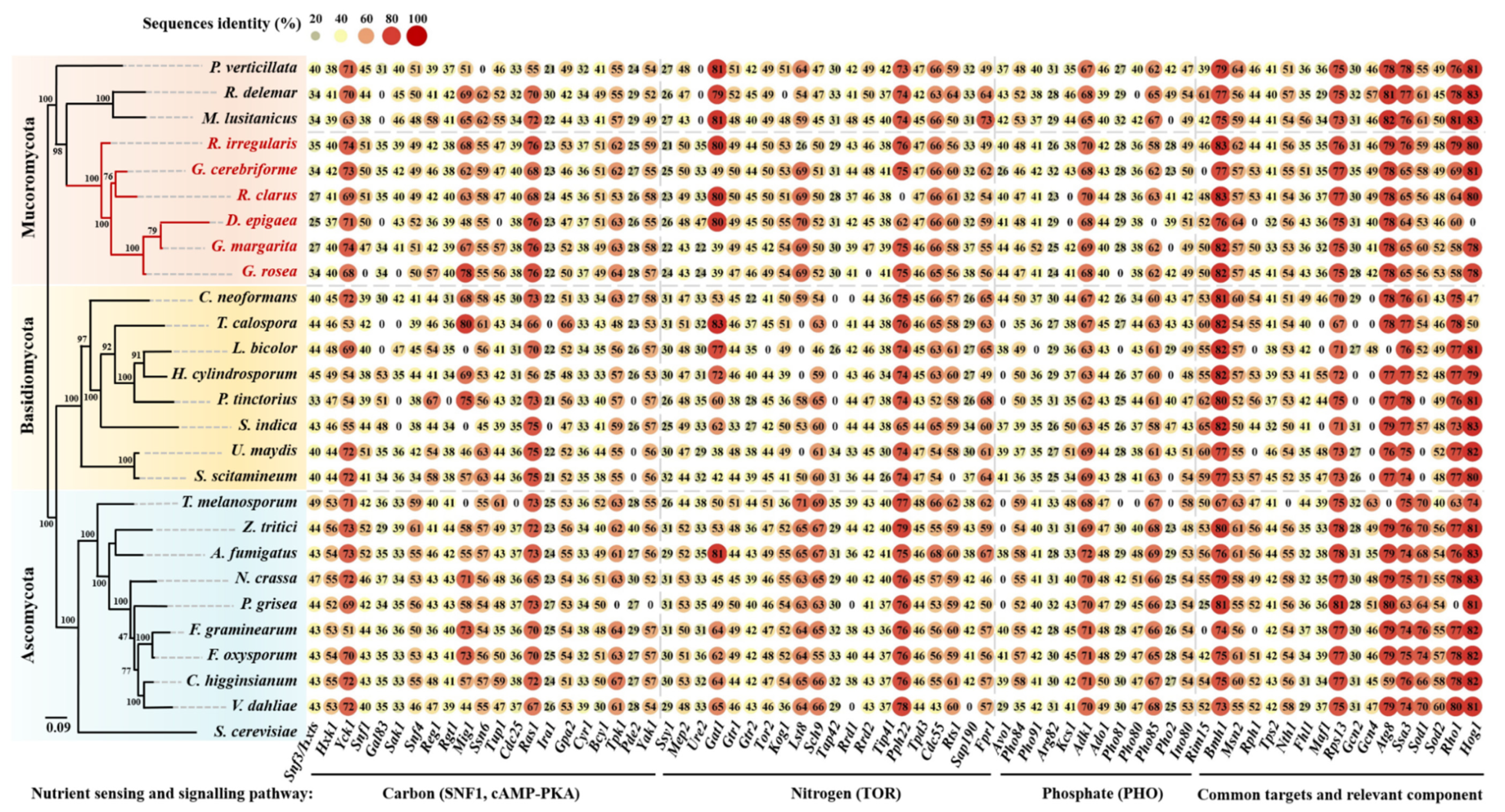
2.1. Genomic Sequences Searching and Analyses
2.2. Transcriptome Analysis
2.3. AM Fungal Materials and Mycorrhization
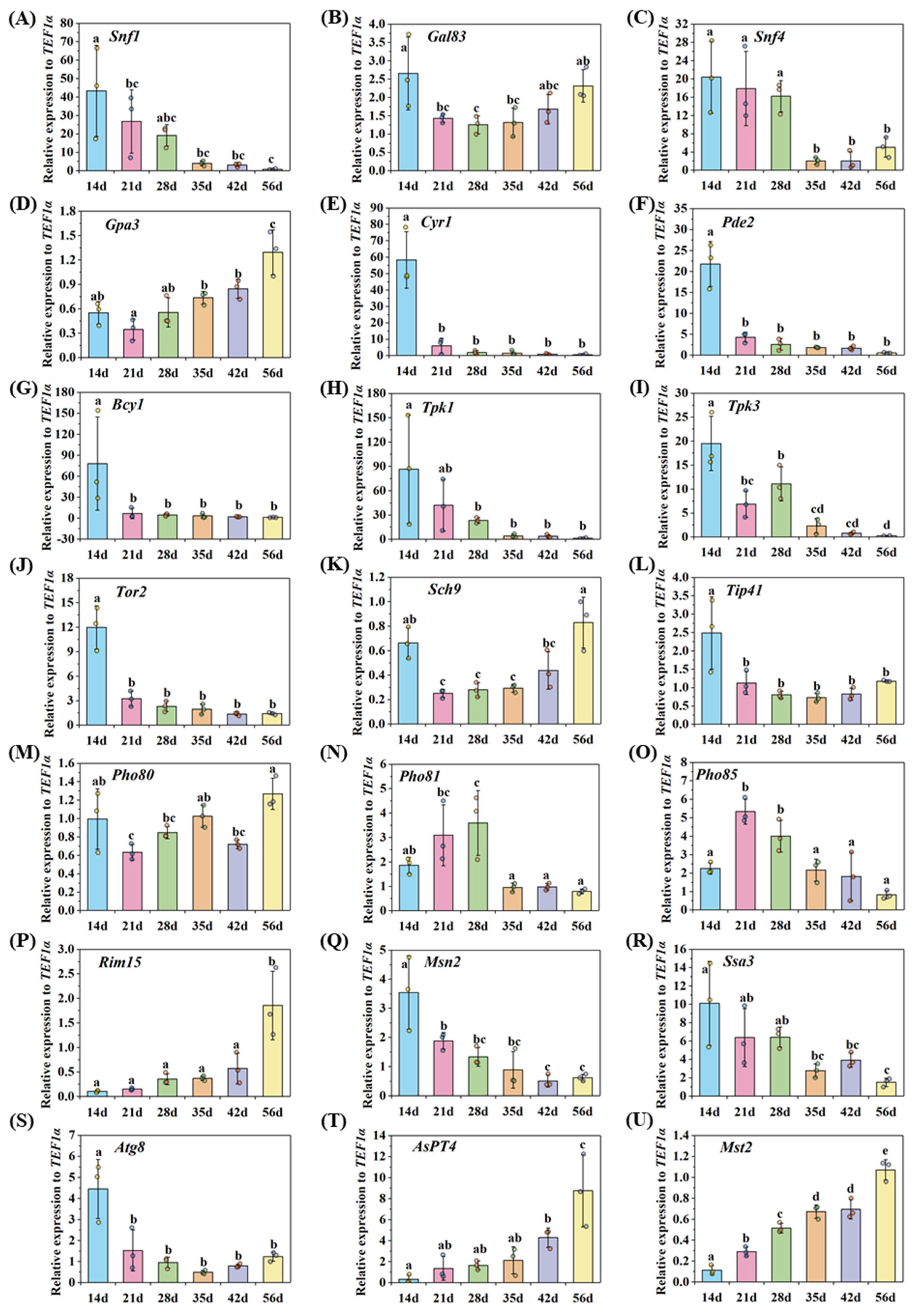
2.4. Gene Expression Detected by qRT-PCR
2.5. Staining Procedures
3. Results and Discussion
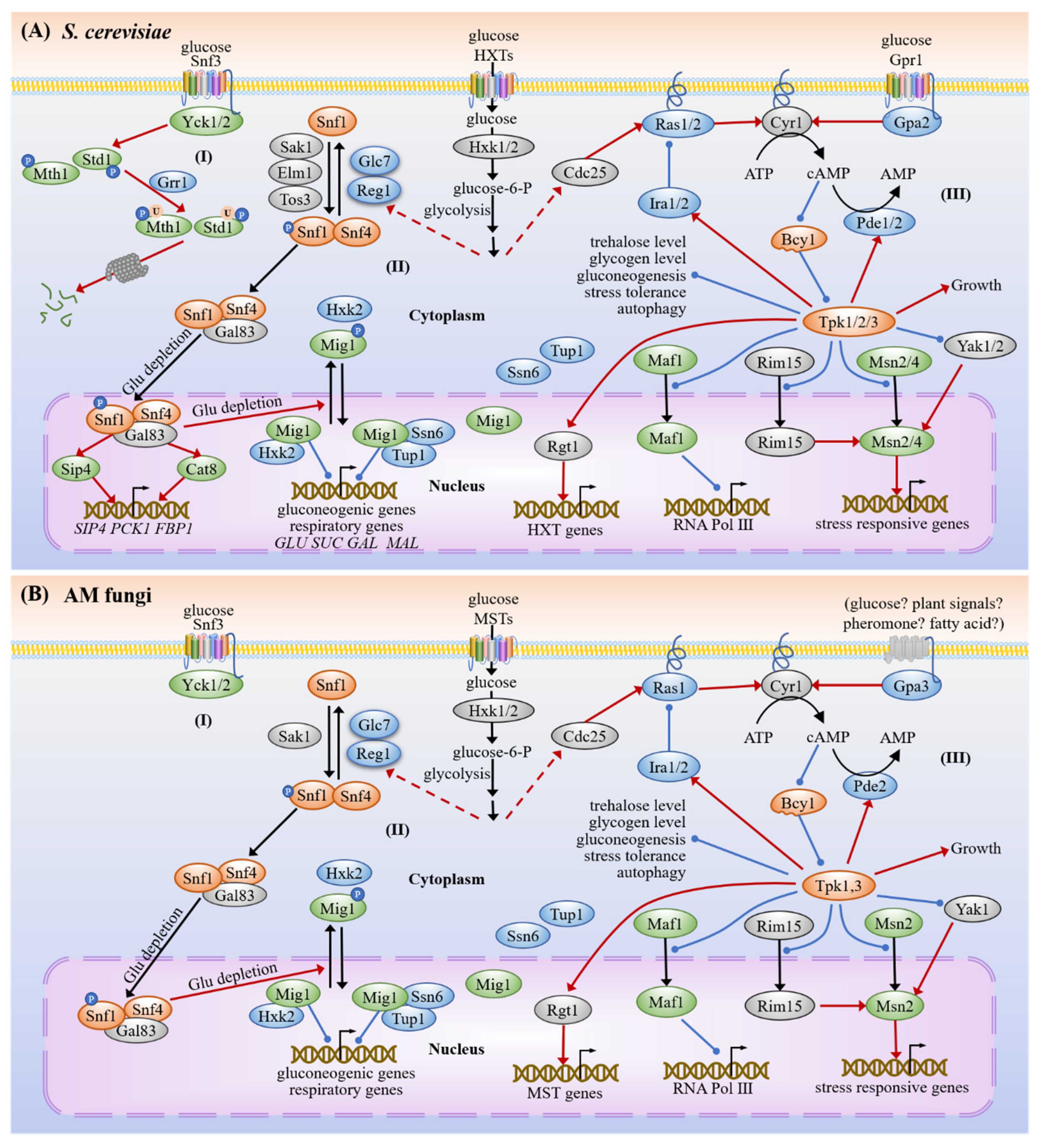
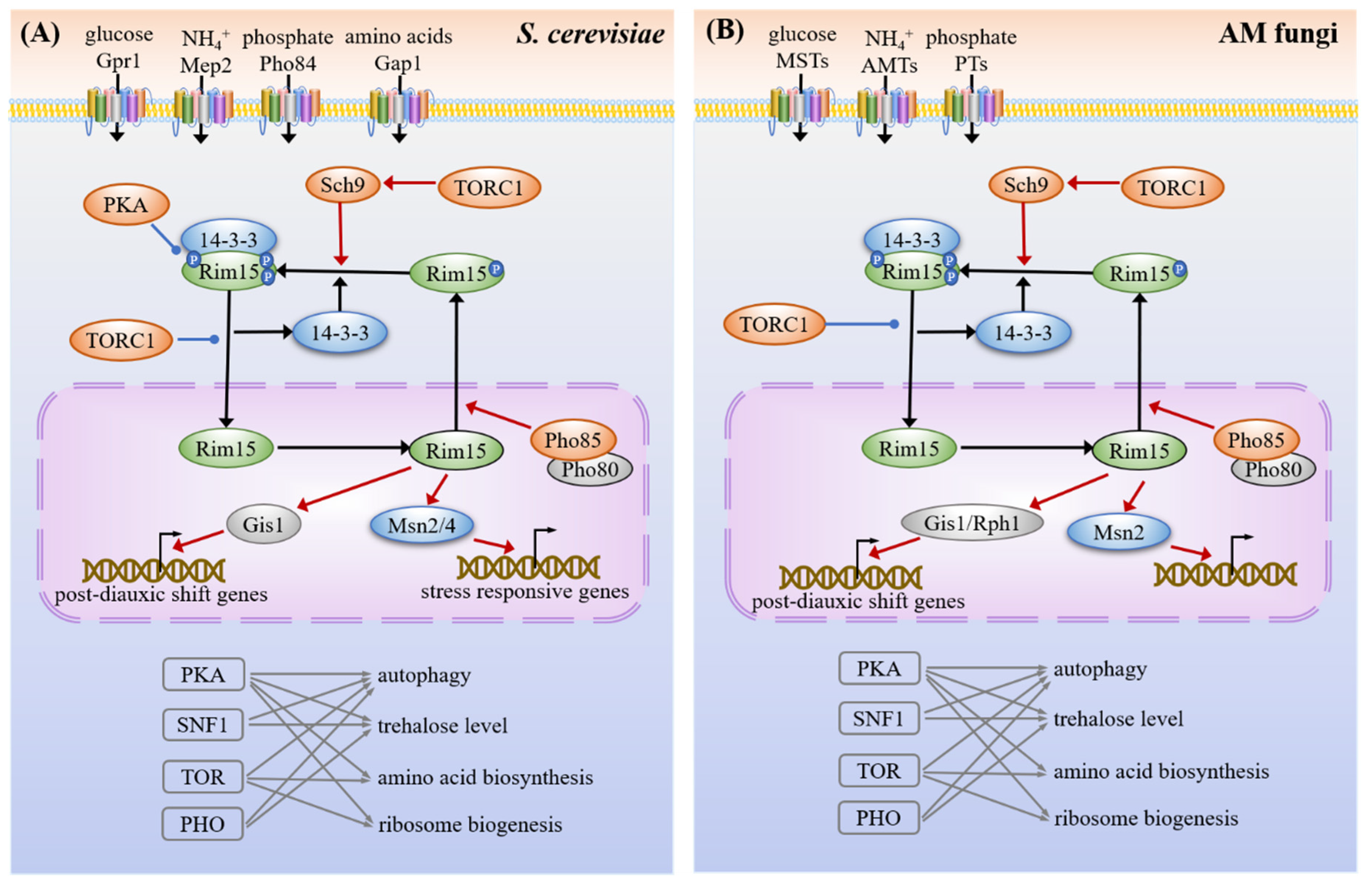
3.1. Carbon Sensing and Signaling
3.1.1. Snf3/Hxts
3.1.2. SNF1
3.1.3. cAMP-PKA
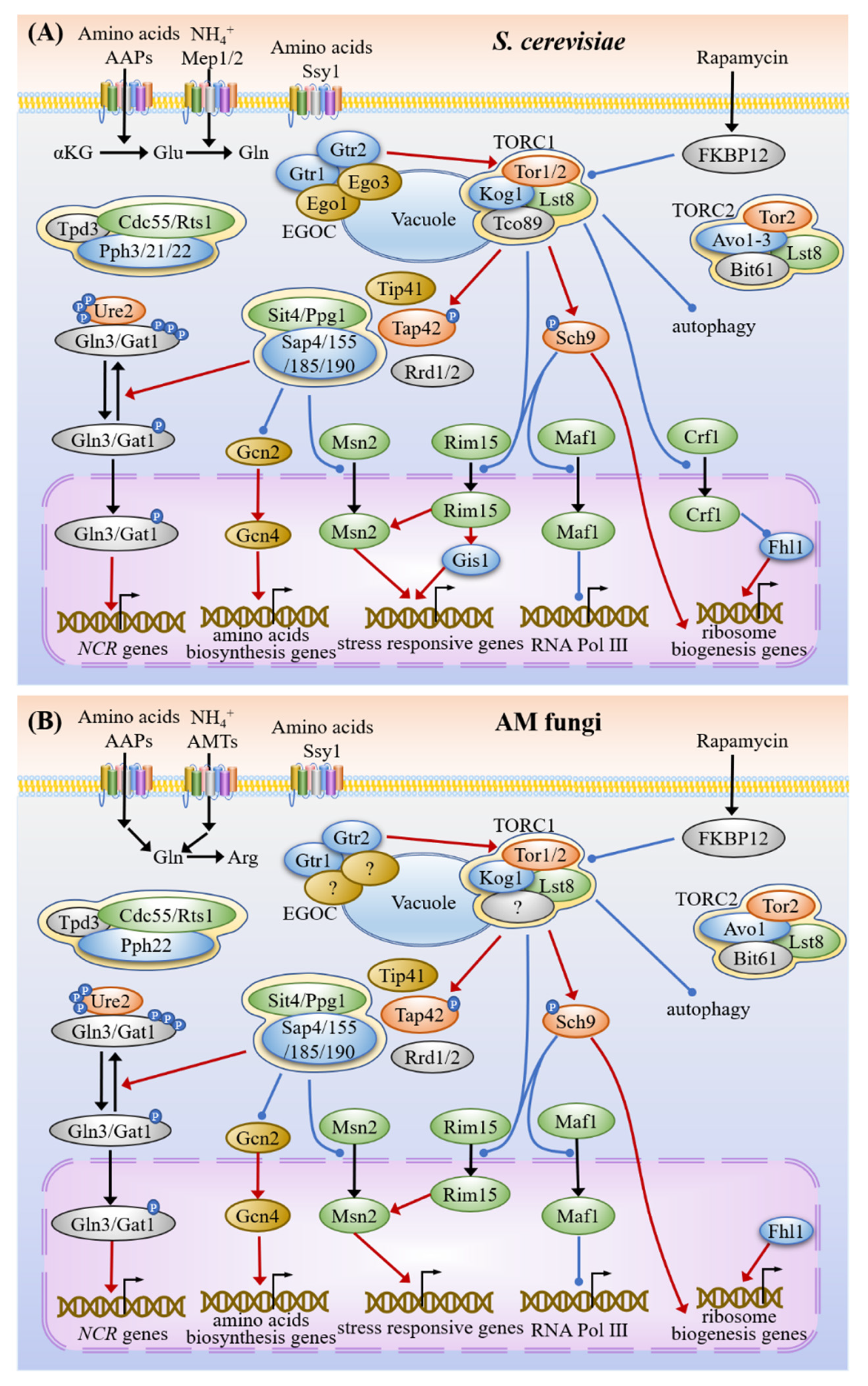
3.2. Nitrogen Sensing and Signaling
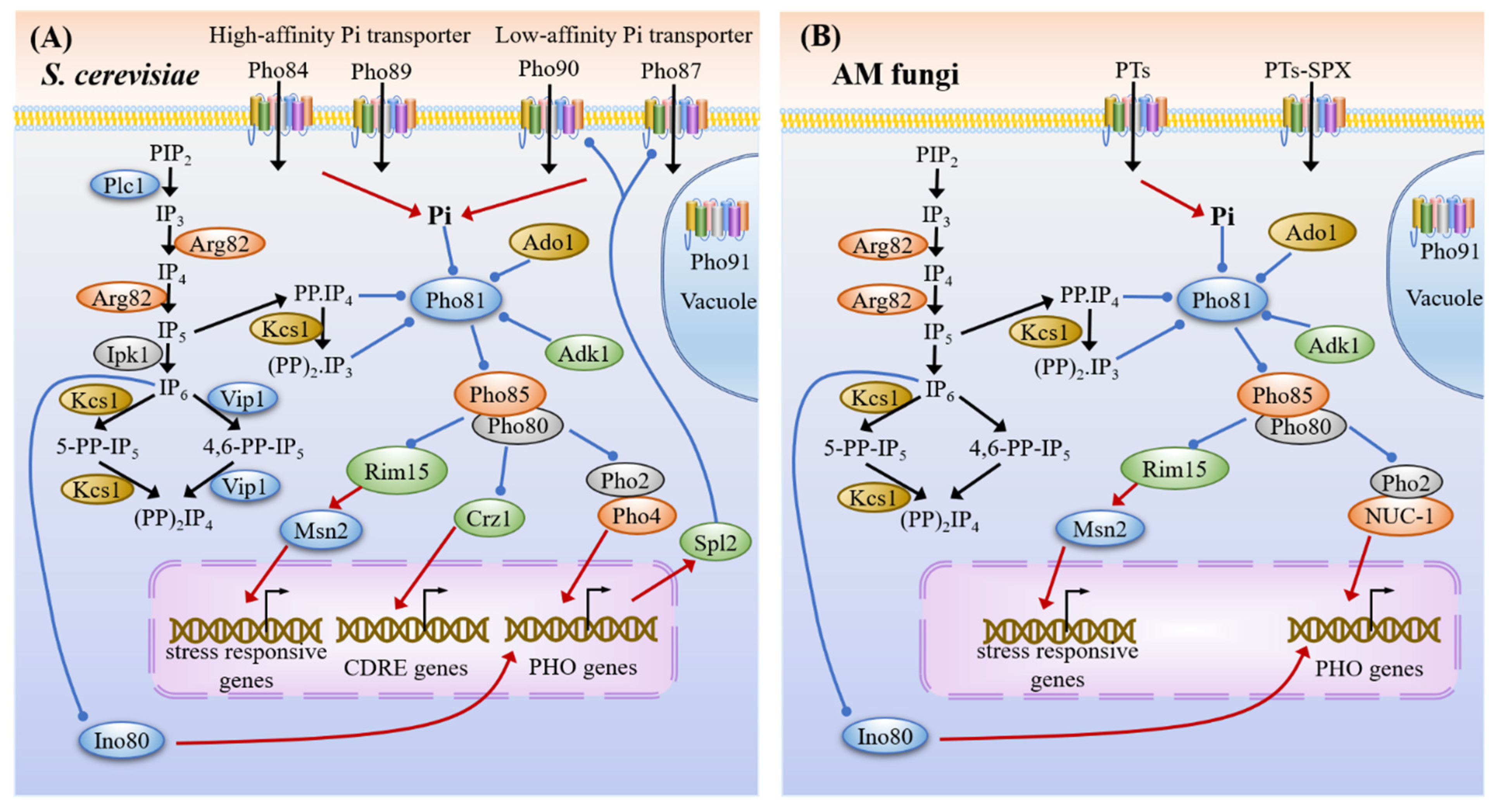
3.3. Phosphate Sensing and Signaling
3.4. Common Targets and Crosstalk
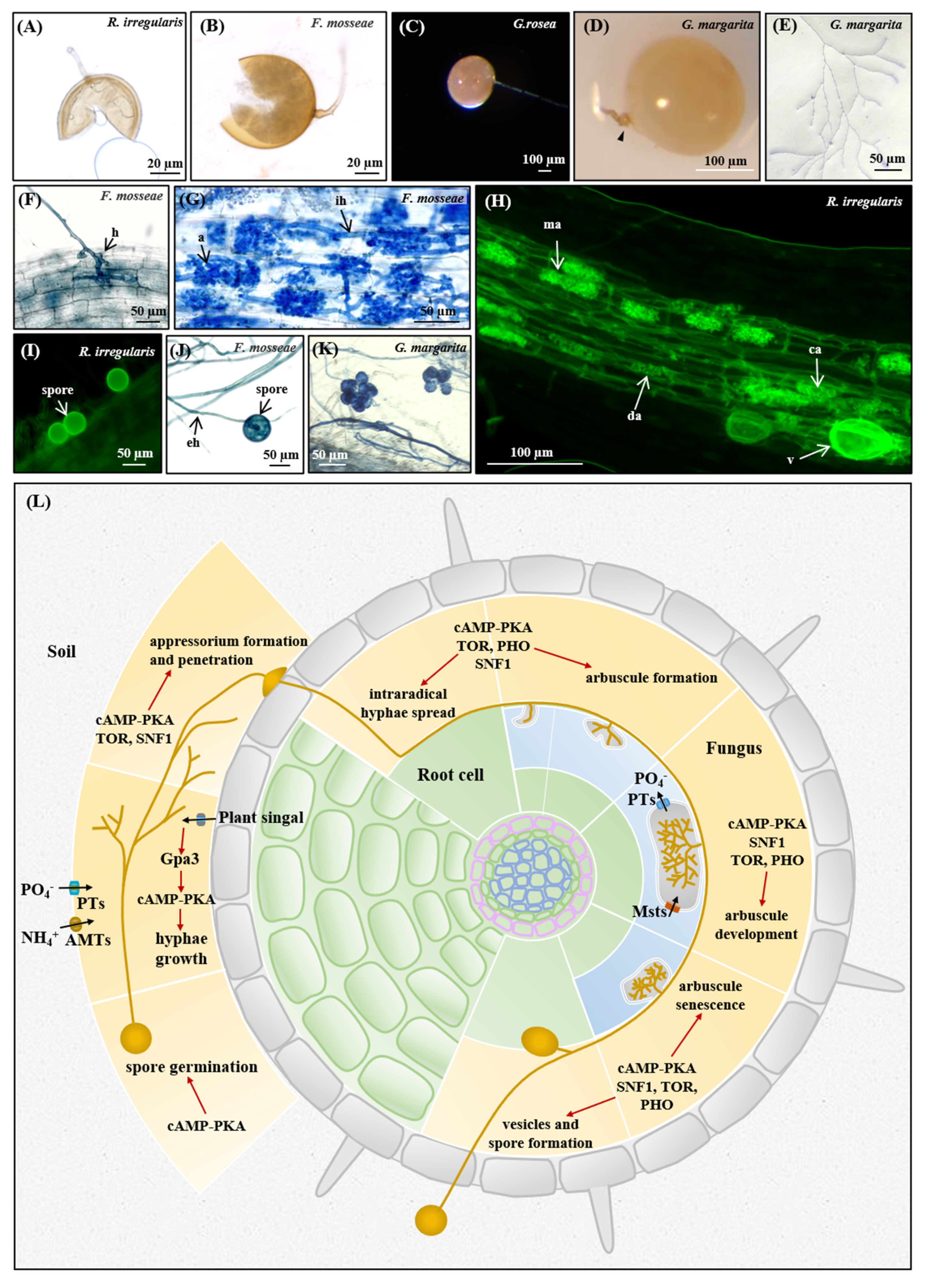
3.5. Nutrient Homeostasis in the Life Cycle of AM Fungi
3.5.1. Asymbiotic and Presymbiotic Stages
3.5.2. Appressorium Formation and Penetration
3.5.3. Arbuscule Formation and Development
- Carbon-phosphate trade in arbuscules
- Life span of arbuscule
3.5.4. Sporulation
4. Conclusions and Future Perspectives
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, P.E.; Douds Jr, D.D.; Becard, G.; Shachar-Hill, Y. Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol. 1999, 120, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, M.; Pfeffer, P.; Jin, H.; Abubaker, J.; Douds, D.; Allen, J.; Bucking, H.; Lammers, P.; Shachar-Hill, Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 2005, 435, 819–823. [Google Scholar] [CrossRef]
- Harrison, M.J.; van Buuren, M.L. A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 1995, 378, 626–629. [Google Scholar] [CrossRef]
- Jin, H.; Pfeffer, P.E.; Douds, D.D.; Piotrowski, E.; Lammers, P.J.; Shachar-Hill, Y. The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol. 2005, 168, 687–696. [Google Scholar] [CrossRef]
- Bago, B.; Pfeffer, P.E.; Shachar-Hill, Y. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 2000, 124, 949–957. [Google Scholar] [CrossRef]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 2017, 10, 1147–1158. [Google Scholar] [CrossRef]
- Ezawa, T.; Saito, K. How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol. 2018, 220, 1116–1121. [Google Scholar] [CrossRef]
- Perez-Tienda, J.; Testillano, P.S.; Balestrini, R.; Fiorilli, V.; Azcon-Aguilar, C.; Ferrol, N. GintAMT2, a new member of the ammonium transporter family in the arbuscular mycorrhizal fungus Glomus intraradices. Fungal Genet. Biol. 2011, 48, 1044–1055. [Google Scholar] [CrossRef]
- Calabrese, S.; Perez-Tienda, J.; Ellerbeck, M.; Arnould, C.; Chatagnier, O.; Boller, T.; Schussler, A.; Brachmann, A.; Wipf, D.; Ferrol, N.; et al. GintAMT3- a low-affinity ammonium transporter of the arbuscular mycorrhizal Rhizophagus irregularis. Front. Plant Sci. 2016, 7, 679. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasiborski, B.; Koul, R.; Lammers, P.J.; Bucking, H.; Shachar-Hill, Y. Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: Gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 2010, 153, 1175–1187. [Google Scholar] [CrossRef]
- Cappellazzo, G.; Lanfranco, L.; Fitz, M.; Wipf, D.; Bonfante, P. Characterization of an amino acid permease from the endomycorrhizal fungus Glomus mosseae. Plant Physiol. 2008, 147, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Lin, H.; Peng, X.; Xu, C.; Sun, Z.; Jiang, K.; Huang, A.; Wu, X.; Tang, N.; Salvioli, A.; et al. Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Mol. Plant Pathol. 2016, 9, 1583–1608. [Google Scholar] [CrossRef] [PubMed]
- Schuessler, A.; Martin, H.; Cohen, D.; Fitz, M.; Wipf, D. Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 2006, 444, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Helber, N.; Wippel, K.; Sauer, N.; Schaarschmidt, S.; Hause, B.; Requena, N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 2011, 23, 3812–3823. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hijikata, N.; Ohtomo, R.; Handa, Y.; Masayoshi, K.; Saito, K.; Masuta, C.; Ezawa, T. Aquaporin-mediated long-distance polyphosphate translocation directed towards the host in arbuscular mycorrhizal symbiosis: Application of virus-induced gene silencing. New Phytol. 2016, 211, 1202–1208. [Google Scholar] [CrossRef]
- Ezawa, T.; Smith, S.E.; Smith, F.A. P metabolism and transport in AM fungi. Plant Soil 2002, 244, 221–230. [Google Scholar] [CrossRef]
- Bago, B.; Pfeffer, P.; Shachar-Hill, Y. Could the urea cycle be translocating nitrogen in the arbuscular mycorrhizal symbiosis? New Phytol. 2001, 149, 4–8. [Google Scholar] [CrossRef]
- Johansen, A.; Finlay, R.D.; Olsson, P.A. Nitrogen metabolism of external hyphae of the arbuscular mycorrhizal fungus Glornus intraradices. New Phytol. 1996, 133, 705–712. [Google Scholar] [CrossRef]
- Guether, M.; Balestrini, R.; Hannah, M.; He, J.; Udvardi, M.K.; Bonfante, P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009, 182, 200–212. [Google Scholar] [CrossRef]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef]
- Mueller, L.M.; Harrison, M.J. Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 2019, 50, 132–139. [Google Scholar] [CrossRef]
- Chiu, C.H.; Paszkowski, U. Mechanisms and impact of symbiotic phosphate acquisition. Cold Spring Harb. Perspect. Biol. 2019, 11, a034603. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span-from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef]
- Smets, B.; Ghillebert, R.; De Snijder, P.; Binda, M.; Swinnen, E.; De Virgilio, C.; Winderickx, J. Life in the midst of scarcity: Adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 2010, 56, 1–32. [Google Scholar] [CrossRef]
- Lee, N.; D’Souza, C.A.; Kronstad, J.W. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 2003, 41, 399–427. [Google Scholar] [CrossRef]
- Yu, F.; Gu, Q.; Yun, Y.; Yin, Y.; Xu, J.R.; Shim, W.B.; Ma, Z. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014, 203, 219–232. [Google Scholar] [CrossRef]
- Mesquita, I.; Moreira, D.; Sampaio-Marques, B.; Laforge, M.; Cordeiro-da-Silva, A.; Ludovico, P.; Estaquier, J.; Silvestre, R. AMPK in pathogens. Exp. Suppl. 2016, 107, 287–323. [Google Scholar] [CrossRef] [PubMed]
- Ikeh, M.; Ahmed, Y.; Quinn, J. Phosphate acquisition and virulence in human fungal pathogens. Microorganisms 2017, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frei dit Frey, N.; Gianinazzi-Pearson, V.; et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, 20117–20122. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.C.H.; Morin, E.; Beaudet, D.; Noel, J.; Yildirir, G.; Ndikumana, S.; Charron, P.; St-Onge, C.; Giorgi, J.; Kruger, M.; et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018, 220, 1161–1171. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Maeda, T.; Yamaguchi, K.; Kameoka, H.; Tanaka, S.; Ezawa, T.; Shigenobu, S.; Kawaguchi, M. The genome of Rhizophagus clarus HR1 reveals a common genetic basis for auxotrophy among arbuscular mycorrhizal fungi. BMC Genom. 2018, 19, 465. [Google Scholar] [CrossRef]
- Morin, E.; Miyauchi, S.; San Clemente, H.; Chen, E.C.H.; Pelin, A.; de la Providencia, I.; Ndikumana, S.; Beaudet, D.; Hainaut, M.; Drula, E.; et al. Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. New Phytol. 2019, 222, 1584–1598. [Google Scholar] [CrossRef]
- Sun, X.; Chen, W.; Ivanov, S.; MacLean, A.M.; Wight, H.; Ramaraj, T.; Mudge, J.; Harrison, M.J.; Fei, Z. Genome and evolution of the arbuscular mycorrhizal fungus Diversispora epigaea (formerly Glomus versiforme) and its bacterial endosymbionts. New Phytol. 2019, 221, 1556–1573. [Google Scholar] [CrossRef]
- Venice, F.; Ghignone, S.; Salvioli di Fossalunga, A.; Amselem, J.; Novero, M.; Xianan, X.; Sedzielewska Toro, K.; Morin, E.; Lipzen, A.; Grigoriev, I.V.; et al. At the nexus of three kingdoms: The genome of the mycorrhizal fungus Gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ. Microbiol. 2020, 22, 122–141. [Google Scholar] [CrossRef] [PubMed]
- Malar, C.M.; Kruger, M.; Kruger, C.; Wang, Y.; Stajich, J.E.; Keller, J.; Chen, E.C.H.; Yildirir, G.; Villeneuve-Laroche, M.; Roux, C.; et al. The genome of Geosiphon pyriformis reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis. Curr. Biol. 2021, 31, 1578–1580. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Limpens, E.; Zhang, Z.; Ivanov, S.; Saunders, D.G.; Mu, D.; Pang, E.; Cao, H.; Cha, H.; Lin, T.; et al. Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 2014, 10, e1004078. [Google Scholar] [CrossRef]
- Navarro-Mendoza, M.I.; Perez-Arques, C.; Panchal, S.; Nicolas, F.E.; Mondo, S.J.; Ganguly, P.; Pangilinan, J.; Grigoriev, I.V.; Heitman, J.; Sanyal, K.; et al. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr. Biol. 2019, 29, 3791–3802.e6. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.J.; Ibrahim, A.S.; Skory, C.; Grabherr, M.G.; Burger, G.; Butler, M.; Elias, M.; Idnurm, A.; Lang, B.F.; Sone, T.; et al. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009, 5, e1000549. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canback, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Aerts, A.; Ahren, D.; Brun, A.; Danchin, E.G.; Duchaussoy, F.; Gibon, J.; Kohler, A.; Lindquist, E.; Pereda, V.; et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 2008, 452, 88–92. [Google Scholar] [CrossRef]
- Zuccaro, A.; Lahrmann, U.; Guldener, U.; Langen, G.; Pfiffi, S.; Biedenkopf, D.; Wong, P.; Samans, B.; Grimm, C.; Basiewicz, M.; et al. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 2011, 7, e1002290. [Google Scholar] [CrossRef] [PubMed]
- Dutheil, J.Y.; Mannhaupt, G.; Schweizer, G.; Sieber, C.M.; Munsterkotter, M.; Guldener, U.; Schirawski, J.; Kahmann, R. A tale of genome compartmentalization: The evolution of virulence clusters in smut fungi. Genome Biol. Evol. 2016, 8, 681–704. [Google Scholar] [CrossRef]
- Kamper, J.; Kahmann, R.; Bolker, M.; Ma, L.J.; Brefort, T.; Saville, B.J.; Banuett, F.; Kronstad, J.W.; Gold, S.E.; Muller, O.; et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 2006, 444, 97–101. [Google Scholar] [CrossRef]
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J., 3rd; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Martin, F.; Kohler, A.; Murat, C.; Balestrini, R.; Coutinho, P.M.; Jaillon, O.; Montanini, B.; Morin, E.; Noel, B.; Percudani, R.; et al. Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 2010, 464, 1033–1038. [Google Scholar] [CrossRef]
- Cuomo, C.A.; Guldener, U.; Xu, J.R.; Trail, F.; Turgeon, B.G.; Di Pietro, A.; Walton, J.D.; Ma, L.J.; Baker, S.E.; Rep, M.; et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 2007, 317, 1400–1402. [Google Scholar] [CrossRef]
- Ma, L.J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Gomez Luciano, L.B.; Tsai, I.J.; Chuma, I.; Tosa, Y.; Chen, Y.H.; Li, J.Y.; Li, M.Y.; Lu, M.J.; Nakayashiki, H.; Li, W.H. Blast fungal genomes show frequent chromosomal changes, gene gains and losses, and effector gene turnover. Mol. Biol. Evol. 2019, 36, 1148–1161. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Norvienyeku, J.; Chen, M.; Bao, J.; Lin, L.; Chen, L.; Lin, Y.; Wu, X.; Cai, Z.; Zhang, Q.; et al. Directional selection from host Plants is a major force driving host specificity in Magnaporthe species. Sci. Rep. 2016, 6, 25591. [Google Scholar] [CrossRef] [PubMed]
- Dallery, J.F.; Lapalu, N.; Zampounis, A.; Pigne, S.; Luyten, I.; Amselem, J.; Wittenberg, A.H.J.; Zhou, S.; de Queiroz, M.V.; Robin, G.P.; et al. Gapless genome assembly of Colletotrichum higginsianum reveals chromosome structure and association of transposable elements with secondary metabolite gene clusters. BMC Genom. 2017, 18, 667. [Google Scholar] [CrossRef]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N.; et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012, 44, 1060–1065. [Google Scholar] [CrossRef]
- Goodwin, S.B.; Ben M’Barek, S.; Dhillon, B.; Wittenberg, A.H.; Crane, C.F.; Hane, J.K.; Foster, A.J.; Van der Lee, T.A.; Grimwood, J.; Aerts, A.; et al. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011, 7, e1002070. [Google Scholar] [CrossRef] [PubMed]
- Amyotte, S.G.; Tan, X.; Pennerman, K.; Jimenez-Gasco, M.; Klosterman, S.J.; Ma, L.J.; Dobinson, K.F.; Veronese, P. Transposable elements in phytopathogenic Verticillium spp.: Insights into genome evolution and inter- and intra-specific diversification. BMC Genom. 2012, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef]
- Hewitt, E.J. Sand and water culture methods used in the study of plant nutrition. Technical Communication. Commonw. Agric. Bureaux. 1966, 22, 315–709. [Google Scholar]
- Xie, X.; Huang, W.; Liu, F.; Tang, N.; Liu, Y.; Lin, H.; Zhao, B. Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 2013, 198, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Song, J.; Xin, X.; Xie, X.; Zhao, B. Arbuscular mycorrhizal fungal 14-3-3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Front. Microbiol. 2018, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, S.; Dover, J.; Rosenwald, A.G.; Wölfl, S.; Johnston, M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 1996, 93, 12428–12432. [Google Scholar] [CrossRef]
- Palomino, A.; Herrero, P.; Moreno, F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 2006, 34, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Moriya, H.; Johnston, M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 2004, 10, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Jouandot II, D.; Roy, A.; Kim, J.H. Functional dissection of the glucose signaling pathways that regulate the yeast glucose transporter gene (HXT) repressor Rgt1. J. Cell. Biochem. 2011, 112, 3268–3275. [Google Scholar] [CrossRef]
- Wu, M.; Li, H.; Wei, S.; Wu, H.; Wu, X.; Bao, X.; Hou, J.; Liu, W.; Shen, Y. Simulating extracellular glucose signals enhances xylose metabolism in recombinant Saccharomyces cerevisiae. Microorganisms 2020, 8, 100. [Google Scholar] [CrossRef]
- Tomas-Cobos, L.; Viana, R.; Sanz, P. TOR kinase pathway and 14-3-3 proteins regulate glucose-induced expression of HXT1, a yeast low-affinity glucose transporter. Yeast 2005, 22, 471–479. [Google Scholar] [CrossRef]
- Tomás-Cobos, L.; Sanz, P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem. J. 2002, 368, 657–663. [Google Scholar] [CrossRef]
- Beullens, M.; Mbonyi, K.; Geerts, L.; Gladines, D.; Detremerie, K.; Jans, A.W.; Thevelein, J.M. Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 1988, 172, 227–231. [Google Scholar] [CrossRef]
- Momcilovic, M.; Iram, S.H.; Liu, Y.; Carlson, M. Roles of the glycogen-binding domain and Snf4 in glucose inhibition of SNF1 protein kinase. J. Biol. Chem. 2008, 283, 19521–19529. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Carlson, M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 1997, 17, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.V.; Heath, R.; Underwood, E.; Sanders, M.J.; Carmena, D.; McCartney, R.R.; Leiper, F.C.; Xiao, B.; Jing, C.; Walker, P.A.; et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011, 14, 707–714. [Google Scholar] [CrossRef]
- Hong, S.P.; Leiper, F.C.; Woods, A.; Carling, D.; Carlson, M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 2003, 100, 8839–8843. [Google Scholar] [CrossRef] [PubMed]
- Sanz, P.; Alms, G.R.; Haystead, T.A.; Carlson, M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 2000, 20, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Sanz, P.; Viana, R.; Garcia-Gimeno, M.A. AMPK in yeast: The SNF1 (Sucrose Non-fermenting 1) protein kinase complex. Exp. Suppl. 2016, 107, 353–374. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Gligoris, T.; Tzamarias, D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 2004, 5, 368–372. [Google Scholar] [CrossRef]
- Kaniak, A.; Xue, Z.; Macool, D.; Kim, J.H.; Johnston, M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell 2004, 3, 221–231. [Google Scholar] [CrossRef]
- Johnson, K.E.; Cameron, S.; Toda, T.; Wigler, M.; Zoller, M.J. Expression in Escherichia coli of BCY1, the regulatory subunit of cyclic AMP-dependent protein kinase from Saccharomyces cerevisiae. J. Biol. Chem. 1987, 262, 8636–8642. [Google Scholar] [CrossRef]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Wigler, M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 1987, 50, 277–287. [Google Scholar] [CrossRef]
- Ni, M.; Rierson, S.; Seo, J.A.; Yu, J.H. The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans. Eukaryot Cell 2005, 4, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, P.; Shen, Q.; Yang, F.; Naqvi, N.I. Cpk2, a catalytic subunit of cyclic AMP-PKA, regulates growth and pathogenesis in rice blast. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.; Kokkelink, L.; Huesmann, C.; Jimenez-Teja, D.; Collado, I.G.; Barakat, R.; Tudzynski, P.; Tudzynski, B. The cAMP-dependent signaling pathway and its role in conidial germination, growth, and virulence of the gray mold Botrytis cinerea. Mol. Plant-Microbe Interact. 2008, 21, 1443–1459. [Google Scholar] [CrossRef]
- Rolland, F.; de Winde, J.H.; Lemaire, K.; Boles, E.; Thevelein, J.M.; Winderickx, J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 2000, 38, 348–358. [Google Scholar] [CrossRef]
- Kataoka, T.; Broek, D.; Wigler, M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell 1985, 43, 493–505. [Google Scholar] [CrossRef]
- Regenfelder, E.; Spellig, T.; Hartmann, A.; Lauenstein, S.; Bölker, M.; Kahmann, R. G proteins in Ustilago maydis: Transmission of multiple signals? EMBO J. 1997, 16, 1934–1942. [Google Scholar] [CrossRef]
- Choi, J.; Jung, W.H.; Kronstad, J.W. The cAMP/protein kinase A signaling pathway in pathogenic basidiomycete fungi: Connections with iron homeostasis. J. Microbiol. 2015, 53, 579–587. [Google Scholar] [CrossRef]
- Krüger, J.; Loubradou, G.; Regenfelder, E.; Hartmann, A.; Kahmann, R. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 1998, 260, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, J.; Castermans, D.; Thevelein, J.M. Molecular mechanisms of feedback inhibition of protein kinase A on intracellular cAMP accumulation. Cell. Signal. 2012, 24, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Sass, P.; Field, J.; Nikawa, J.; Toda, T.; Wigler, M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1986, 83, 9303–9307. [Google Scholar] [CrossRef]
- Thines, E.; Weber, R.W.; Talbot, N.J. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 2000, 12, 1703–1718. [Google Scholar] [CrossRef]
- Jimenez-Jimenez, S.; Hashimoto, K.; Santana, O.; Aguirre, J.; Kuchitsu, K.; Cardenas, L. Emerging roles of tetraspanins in plant inter-cellular and inter-kingdom communication. Plant Signal Behav. 2019, 14, e1581559. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Hillmer, S.; Funaya, C.; Chiapello, M.; Schumacher, K.; Lo Presti, L.; Kahmann, R.; Paszkowski, U. Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat. Plants 2019, 5, 204–211. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, X.; Bian, Z.; Wei, J.; Xu, J.R. Coregulation of dimorphism and symbiosis by cyclic AMP signaling in the lichenized fungus Umbilicaria muhlenbergii. Proc. Natl. Acad. Sci. USA 2020. [Google Scholar] [CrossRef]
- Filteau, M.; Diss, G.; Torres-Quiroz, F.; Dube, A.K.; Schraffl, A.; Bachmann, V.A.; Gagnon-Arsenault, I.; Chretien, A.E.; Steunou, A.L.; Dionne, U.; et al. Systematic identification of signal integration by protein kinase A. Proc. Natl. Acad. Sci. USA 2015, 112, 4501–4506. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Cho, B.; Joo, H.; Hahn, J. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 2008, 70, 882–895. [Google Scholar] [CrossRef]
- Moir, R.D.; Lee, J.; Haeusler, R.A.; Desai, N.; Engelke, D.R.; Willis, I.M. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc. Natl. Acad. Sci. USA 2006, 103, 15044–15049. [Google Scholar] [CrossRef]
- Wei, M.; Fabrizio, P.; Hu, J.; Ge, H.; Cheng, C.; Li, L.; Longo, V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008, 4, e13. [Google Scholar] [CrossRef]
- Beck, T.; Hall, M.N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 1999, 402, 689–692. [Google Scholar] [CrossRef]
- Shertz, C.A.; Bastidas, R.J.; Li, W.; Heitman, J.; Cardenas, M.E. Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genom. 2010, 11, 510. [Google Scholar] [CrossRef]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Schreiber, S.L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 1991, 251, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Requena, N.; Mann, P.; Franken, P. A homologue of the cell cycle check point TOR2 from Saccharomyces cerevisiae exists in the arbuscular mycorrrhizal fungus Glomus mosseae. Protoplasma 2000, 212, 89–98. [Google Scholar] [CrossRef]
- Didion, T.; Regenberg, B.; Jørgensen, M.U.; Kielland-Brandt, M.C.; Andersen, H.A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 1998, 27, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Ljungdahl, P.O. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 2009, 37, 242–247. [Google Scholar] [CrossRef]
- Van Nuland, A.; Vandormael, P.; Donaton, M.; Alenquer, M.; Lourenço, A.; Quintino, E.; Versele, M.; Thevelein, J.M. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 2006, 59, 1485–1505. [Google Scholar] [CrossRef]
- Marini, A.M.; Soussi-Boudekou, S.; Vissers, S.; Andre, B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 4282–4293. [Google Scholar] [CrossRef]
- Zhang, T.; Péli-Gulli, M.P.; Yang, H.; De Virgilio, C.; Ding, J. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1. Structure 2012, 20, 2151–2160. [Google Scholar] [CrossRef]
- Jacinto, E.; Guo, B.; Arndt, K.T.; Schmelzle, T.; Hall, M.N. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 2001, 8, 1017–1026. [Google Scholar] [CrossRef]
- Liu, N.; Yun, Y.; Yin, Y.; Hahn, M.; Ma, Z.; Chen, Y. Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol. 2019, 223, 412–429. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Zhang, C.; Yu, F.; Yin, Y.; Shim, W.B.; Ma, Z. Protein kinase FgSch9 serves as a mediator of the target of rapamycin and high osmolarity glycerol pathways and regulates multiple stress responses and secondary metabolism in Fusarium graminearum. Environ. Microbiol. 2015, 17, 2661–2676. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Broach, J.R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999, 18, 2782–2792. [Google Scholar] [CrossRef] [PubMed]
- Di Como, C.J.; Arndt, K.T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996, 10, 1904–1916. [Google Scholar] [CrossRef]
- Huber, A.; Bodenmiller, B.; Uotila, A.; Stahl, M.; Wanka, S.; Gerrits, B.; Aebersold, R.; Loewith, R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009, 23, 1929–1943. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, V.A.; Hinnebusch, A.G. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003, 17, 859–872. [Google Scholar] [CrossRef]
- Secco, D.; Wang, C.; Shou, H.; Whelan, J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012, 586, 289–295. [Google Scholar] [CrossRef]
- Hurlimann, H.C.; Stadler-Waibel, M.; Werner, T.P.; Freimoser, F.M. Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4438–4445. [Google Scholar] [CrossRef]
- Ghillebert, R.; Swinnen, E.; De Snijder, P.; Smets, B.; Winderickx, J. Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem. J. 2011, 434, 243–251. [Google Scholar] [CrossRef]
- Nishizawa, M.; Komai, T.; Katou, Y.; Shirahige, K.; Ito, T.; Toh, E.A. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 2008, 6, 2817–2830. [Google Scholar] [CrossRef]
- Auesukaree, C.; Homma, T.; Tochio, H.; Shirakawa, M.; Kaneko, Y.; Harashima, S. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 17289–17294. [Google Scholar] [CrossRef]
- Auesukaree, C.; Tochio, H.; Shirakawa, M.; Kaneko, Y.; Harashima, S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 25127–25133. [Google Scholar] [CrossRef]
- Huang, S.; O’Shea, E.K. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics 2005, 169, 1859–1871. [Google Scholar] [CrossRef]
- Henry, T.C.; Power, J.E.; Kerwin, C.L.; Mohammed, A.; Weissman, J.S.; Cameron, D.M.; Wykoff, D.D. Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal transduction pathway in yeasts. Eukaryot Cell 2011, 10, 198–206. [Google Scholar] [CrossRef]
- Lee, Y.S.; Huang, K.; Quiocho, F.A.; O’Shea, E.K. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 2008, 4, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Magbanua, J.P.; Ogawa, N.; Harashima, S.; Oshima, Y. The transcriptional activators of the PHO regulon, Pho4p and Pho2p, interact directly with each other and with components of the basal transcription machinery in Saccharomyces cerevisiae. J. Biochem. 1997, 121, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Kaffman, A.; Herskowitz, I.; Tjian, R.; O’Shea, E.K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 1994, 263, 1153–1156. [Google Scholar] [CrossRef]
- O’Neill, E.M.; Kaffman, A.; Jolly, E.R.; O’Shea, E.K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 1996, 271, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Kaffman, A.; Rank, N.M.; O’Neill, E.M.; Huang, L.S.; O’Shea, E.K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 1998, 396, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Xiao, H.; Ranallo, R.; Wu, W.H.; Wu, C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 2003, 299, 112–114. [Google Scholar] [CrossRef]
- Steger, D.J.; Haswell, E.S.; Miller, A.L.; Wente, S.R.; O’Shea, E.K. Regulation of chromatin remodeling by inositol polyphosphates. Science 2003, 299, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Flanagan, P.R.; Zeng, J.; Jani, N.M.; Cardenas, M.E.; Moran, G.P.; Köhler, J.R. Phosphate is the third nutrient monitored by TOR in Candida albicans and provides a target for fungal-specific indirect TOR inhibition. Proc. Natl. Acad. Sci. USA 2017, 114, 6346–6351. [Google Scholar] [CrossRef] [PubMed]
- Steyfkens, F.; Zhang, Z.; Van Zeebroeck, G.; Thevelein, J.M. Multiple transceptors for macro- and micro-nutrients control diverse cellular properties through the PKA pathway in yeast: A paradigm for the rapidly expanding world of eukaryotic nutrient transceptors up to those in human cells. Front. Pharmacol. 2018, 9, 191. [Google Scholar] [CrossRef]
- Swinnen, E.; Wanke, V.; Roosen, J.; Smets, B.; Dubouloz, F.; Pedruzzi, I.; Cameroni, E.; De Virgilio, C.; Winderickx, J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006, 1. [Google Scholar] [CrossRef] [PubMed]
- Wanke, V.; Pedruzzi, I.; Cameroni, E.; Dubouloz, F.; De Virgilio, C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. Eur. Mol. Biol. Organ. J. 2005, 24, 4271–4278. [Google Scholar] [CrossRef] [PubMed]
- Pedruzzi, I.; Burckert, N.; Egger, P.; De Virgilio, C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. Eur. Mol. Biol. Organ. J. 2000, 19, 2569–2579. [Google Scholar] [CrossRef]
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 2009, 106, 17049–17054. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Geng, J.; Yen, W.; Wang, K.; Klionsky, D.J. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol. Cell 2010, 38, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Li, Y.; Wu, L.; Wu, C.; Zhang, Y.; Liu, J.; He, Z.; Wu, X.; Lu, C.; Wang, L.; et al. Atg11 is required for initiation of glucose starvation-induced autophagy. Autophagy 2020, 1–13. [Google Scholar] [CrossRef]
- Vlahakis, A.; Muniozguren, N.L.; Powers, T. Stress-response transcription factors Msn2 and Msn4 couple TORC2-Ypk1 signaling and mitochondrial respiration to ATG8 gene expression and autophagy. Autophagy 2017, 13, 1804–1812. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, E.; Rosseels, J.; Winderickx, J. The minimum domain of Pho81 is not sufficient to control the Pho85-Rim15 effector branch involved in phosphate starvation-induced stress responses. Curr. Genet. 2005, 48, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Zahringer, H.; Thevelein, J.M.; Nwaka, S. Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: Variations of PKA effect during stress and growth. Mol. Microbiol. 2000, 35, 397–406. [Google Scholar] [CrossRef] [PubMed]
- François, J.; Parrou, J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Winderickx, J.; De Winde, J.H.; Crauwels, M.; Hino, A.; Hohmann, S.; Van Dijck, P.; Thevelein, J.M. Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: Novel variations of STRE-mediated transcription control? Mol. Gen. Genet. 1996, 252, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Kankipati, H.N.; Kimpe, M.; Van Zeebroeck, G.; Zhang, Z.; Thevelein, J.M. The nutrient transceptor/PKA pathway functions independently of TOR and responds to leucine and Gcn2 in a TOR-independent manner. Fems Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR regulates ribosomal protein gene expression via PKA and the forkhead transcription factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Moir, R.D.; Willis, I.M. Regulation of RNA Polymerase III Transcription Involves SCH9-dependent and SCH9-independent Branches of the Target of Rapamycin (TOR) Pathway. J. Biol. Chem. 2009, 284, 12604–12608. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.; Orlova, M.; Maziarz, M.; Kuchin, S. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 2012, 11, 119–128. [Google Scholar] [CrossRef][Green Version]
- Orlova, M.; Kanter, E.; Krakovich, D.; Kuchin, S. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 2006, 5, 1831–1837. [Google Scholar] [CrossRef]
- Thevelein, J.M.; Gelade, R.; Holsbeeks, I.; Lagatie, O.; Popova, Y.; Rolland, F.; Stolz, F.; Van de Velde, S.; Van Dijck, P.; Vandormael, P.; et al. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 2005, 33, 253–256. [Google Scholar] [CrossRef]
- Nicastro, R.; Tripodi, F.; Gaggini, M.; Castoldi, A.; Reghellin, V.; Nonnis, S.; Tedeschi, G.; Coccetti, P. Snf1 phosphorylates adenylate cyclase and negatively regulates protein kinase A-dependent transcription in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 24715–24726. [Google Scholar] [CrossRef]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Soulard, A.; Cremonesi, A.; Moes, S.; Schütz, F.; Jenö, P.; Hall, M.N. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 2010, 21, 3475–3486. [Google Scholar] [CrossRef]
- Lu, J.Y.; Lin, Y.Y.; Sheu, J.C.; Wu, J.T.; Lee, F.J.; Chen, Y.; Lin, M.I.; Chiang, F.T.; Tai, T.Y.; Berger, S.L.; et al. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 2011, 146, 968–978. [Google Scholar] [CrossRef]
- Popova, Y.; Thayumanavan, P.; Lonati, E.; Agrochao, M.; Thevelein, J.M. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc. Natl. Acad. Sci. USA 2010, 107, 2890–2895. [Google Scholar] [CrossRef] [PubMed]
- Giots, F.; Donaton, M.C.V.; Thevelein, J.M. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 2003, 47, 1163–1181. [Google Scholar] [CrossRef]
- Li, L.; Kaplan, J.; Ward, D.M. The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J. Biol. Chem. 2017, 292, 15577–15586. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Lai, Y.; Jiang, Y. The TOR complex 1 is a direct target of Rho1 GTPase. Mol. Cell 2012, 45, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Tang, W.; Wang, J.; Liu, X.; Yang, L.; Gao, C.; Zhang, J.; Zhang, H.; Zheng, X.; Wang, P.; et al. Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2016, 17, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Liou, L.L.; Moy, V.N.; Diaspro, A.; SelverstoneValentine, J.; Gralla, E.B.; Longo, V.D. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 2003, 163, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, V.; Herman, P.K. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics 2011, 187, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Ocon, A.; Hampp, R.; Requena, N. Trehalose turnover during abiotic stress in arbuscular mycorrhizal fungi. New Phytol. 2007, 174, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Breuninger, M.; Requena, N. Recognition events in AM symbiosis: Analysis of fungal gene expression at the early appressorium stage. Fungal Genet. Biol. 2004, 41, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Vangelisti, A.; Turrini, A.; Sbrana, C.; Avio, L.; Giordani, T.; Natali, L.; Giovannetti, M.; Cavallini, A. Gene expression in Rhizoglomus irregulare at two different time points of mycorrhiza establishment in Helianthus annuus roots, as revealed by RNA-seq analysis. Mycorrhiza 2020, 30, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.K.; Javot, H.; Deewatthanawong, P.; Torres-Jerez, I.; Tang, Y.; Blancaflor, E.B.; Udvardi, M.K.; Harrison, M.J. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009, 9, 10. [Google Scholar] [CrossRef]
- Seddas, P.M.; Arnould, C.; Tollot, M.; Arias, C.M.; Gianinazzi-Pearson, V. Spatial monitoring of gene activity in extraradical and intraradical developmental stages of arbuscular mycorrhizal fungi by direct fluorescent in situ RT-PCR. Fungal Genet. Biol. 2008, 45, 1155–1165. [Google Scholar] [CrossRef]
- Lanfranco, L.; Novero, M.; Bonfante, P. The mycorrhizal fungus Gigaspora margarita possesses a CuZn superoxide dismutase that is up-regulated during symbiosis with legume hosts. Plant Physiol. 2005, 137, 1319–1330. [Google Scholar] [CrossRef]
- Yao, Q.; Ohtomo, R.; Saito, M. Influence of nitrogen and phosphorus on polyphosphate accumulation in Gigaspora margarita during spore germination. Plant Soil 2010, 330, 303–311. [Google Scholar] [CrossRef]
- Bucking, H.; Shachar-Hill, Y. Phosphate uptake, transport and transfer by the arbuscular mycorrhizal fungus Glomus intraradices is stimulated by increased carbohydrate availability. New Phytol. 2005, 165, 899–911. [Google Scholar] [CrossRef]
- Fellbaum, C.R.; Gachomo, E.W.; Beesetty, Y.; Choudhari, S.; Strahan, G.D.; Pfeffer, P.E.; Kiers, E.T.; Bucking, H. Carbon availability triggers fungal nitrogen uptake and transport in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2012, 109, 2666–2671. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Penmetsa, R.V.; Terzaghi, N.; Cook, D.R.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef] [PubMed]
- Breuillin, F.; Floss, D.S.; Gomez, S.K.; Pumplin, N.; Ding, Y.; Levesque-Tremblay, V.; Noar, R.D.; Daniels, D.A.; Bravo, A.; Eaglesham, J.B.; et al. Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 2015, 27, 1352–1366. [Google Scholar] [CrossRef]
- Bakshi, M.; Vahabi, K.; Bhattacharya, S.; Sherameti, I.; Varma, A.; Yeh, K.W.; Baldwin, I.; Johri, A.K.; Oelmuller, R. WRKY6 restricts Piriformospora indica-stimulated and phosphate-induced root development in Arabidopsis. BMC Plant Biol. 2015, 15, 305. [Google Scholar] [CrossRef]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef]
- Kobae, Y.; Ohmori, Y.; Saito, C.; Yano, K.; Ohtomo, R.; Fujiwara, T. Phosphate treatment strongly inhibits new arbuscule development but not the maintenance of arbuscule in mycorrhizal rice roots. Plant Physiol. 2016, 171, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Bago, B.; Pfeffer, P.E.; Abubaker, J.; Jun, J.; Allen, J.W.; Brouillette, J.; Douds, D.D.; Lammers, P.J.; Shachar-Hill, Y. Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol. 2003, 131, 1496–1507. [Google Scholar] [CrossRef]
- Tang, N.; San Clemente, H.; Roy, S.; Bécard, G.; Zhao, B.; Roux, C. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Front. Microbiol. 2016, 7, 233. [Google Scholar] [CrossRef]
- Bago, B.; Zipfel, W.; Williams, R.M.; Jun, J.; Arreola, R.; Lammers, P.J.; Pfeffer, P.E.; Shachar-Hill, Y. Translocation and utilization of fungal storage lipid in the arbuscular mycorrhizal symbiosis. Plant Physiol. 2002, 128, 108–124. [Google Scholar] [CrossRef]
- Gaspar, L.; Pollero, R.J.; Cabello, M. Triacylglycerol consumption during spore germination of vesiculararcuscular mycorrhizal fungi. J. Am. Oil Chem. Soc. 1994, 71, 449–452. [Google Scholar] [CrossRef]
- Bago, B.; Pfeffer, P.E.; Douds, D.D., Jr.; Brouillette, J.; Bécard, G.; Shachar-Hill, Y. Carbon metabolism in spores of the arbuscular mycorrhizal fungus Glomus intraradices as revealed by nuclear magnetic resonance spectroscopy. Plant Physiol. 1999, 121, 263–272. [Google Scholar] [CrossRef][Green Version]
- Shachar-Hill, Y.; Pfeffer, P.E.; Douds, D.; Osman, S.F.; Doner, L.W.; Ratcliffe, R.G. Partitioning of intermediate carbon metabolism in VAM colonized leek. Plant Physiol. 1995, 108, 7–15. [Google Scholar] [CrossRef]
- Yamauchi, J.; Takayanagi, N.; Komeda, K.; Takano, Y.; Okuno, T. cAMP-PKA signaling regulates multiple steps of fungal infection cooperatively with Cmk1 MAP kinase in Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 2004, 17, 1355–1365. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, H.S.; Park, S.Y.; Lee, S.; Adams, E.L.; Czymmek, K.; Kang, S. Loss of cAMP-dependent Protein Kinase A affects multiple traits important for root pathogenesis by Fusarium oxysporum. Mol. Plant Microbe Interact. 2011, 4, 719–732. [Google Scholar] [CrossRef]
- Tang, K.; Lv, W.; Zhang, Q.; Zhou, C. Coding the alpha-subunit of SNF1 kinase, Snf1 is required for the conidiogenesis and pathogenicity of the Alternaria alternata tangerine pathotype. Fungal Biol. 2020, 124, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Klose, J.; de Sa, M.M.; Kronstad, J.W. Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 2004, 52, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.Q.; Chen, G.Q.; Liu, X.H.; Dong, B.; Shi, H.B.; Lu, J.P.; Lin, F. Crosstalk between SNF1 pathway and the peroxisome-mediated lipid metabolism in Magnaporthe oryzae. PLoS ONE 2014, 9, e103124. [Google Scholar] [CrossRef] [PubMed]
- Schepers, W.; Van Zeebroeck, G.; Pinkse, M.; Verhaert, P.; Thevelein, J.M. In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J. Biol. Chem. 2012, 287, 44130–44142. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Sun, J.; Radhakrishnan, G.V.; Lee, T.; Bozsoki, Z.; Fort, S.; Gavrin, A.; Gysel, K.; Thygesen, M.B.; Andersen, K.R.; et al. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat Commun. 2019, 10, 5047. [Google Scholar] [CrossRef]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, H.; Kuster, H.; Requena, N. Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula. New Phytol. 2010, 185, 716–733. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; Andre, O.; Puech-Pages, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pages, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Becard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J. Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 2005, 59, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Yu, R.; Shen, X.; Liu, M.; Liu, X.; Yin, Z.; Li, X.; Feng, W.; Hu, J.; Zhang, H.; et al. The rice blast fungus MoRgs1 functioning in cAMP signaling and pathogenicity is regulated by casein kinase MoCk2 phosphorylation and modulated by membrane protein MoEmc2. PLoS Pathog. 2021, 17. [Google Scholar] [CrossRef]
- Li, X.; Zhong, K.; Yin, Z.; Hu, J.; Wang, W.; Li, L.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. The seven transmembrane domain protein MoRgs7 functions in surface perception and undergoes coronin MoCrn1-dependent endocytosis in complex with Galpha subunit MoMagA to promote cAMP signaling and appressorium formation in Magnaporthe oryzae. PLoS Pathog. 2019, 15, e1007382. [Google Scholar] [CrossRef]
- Howard, R.J.; Ferrari, M.A.; Roach, D.H.; Money, N.P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 1991, 88, 11281–11284. [Google Scholar] [CrossRef]
- Yi, M.; Park, J.H.; Ahn, J.H.; Lee, Y.H. MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2008, 45, 1172–1181. [Google Scholar] [CrossRef]
- Marroquin-Guzman, M.; Wilson, R.A. GATA-dependent glutaminolysis drives appressorium formation in Magnaporthe oryzae by suppressing TOR inhibition of cAMP/PKA signaling. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006, 312, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Kiers, E.T.; Duhamel, M.; Beesetty, Y.; Mensah, J.A.; Franken, O.; Verbruggen, E.; Fellbaum, C.R.; Kowalchuk, G.A.; Hart, M.M.; Bago, A.; et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 2011, 333, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Doidy, J.; van Tuinen, D.; Lamotte, O.; Corneillat, M.; Alcaraz, G.; Wipf, D. The Medicago truncatula sucrose transporter family: Characterization and implication of key members in carbon partitioning towards arbuscular mycorrhizal fungi. Mol. Plant 2012, 5, 1346–1358. [Google Scholar] [CrossRef]
- An, J.; Zeng, T.; Ji, C.; de Graaf, S.; Zheng, Z.; Xiao, T.T.; Deng, X.; Xiao, S.; Bisseling, T.; Limpens, E.; et al. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol. 2019, 224, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Paszkowski, U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 2017, 39, 50–56. [Google Scholar] [CrossRef]
- Zhang, Q.; Blaylock, L.A.; Harrison, M.J. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 2010, 22, 1483–1497. [Google Scholar] [CrossRef]
- Aono, T.; Maldonado-Mendoza, I.E.; Dewbre, G.R.; Harrison, M.J.; Saito, M. Expression of alkaline phosphatase genes in arbuscular mycorrhizas. New Phytol. 2004, 162, 525–534. [Google Scholar] [CrossRef]
- Ohtomo, R.; Saito, M. Polyphosphate dynamics in mycorrhizal roots during colonization of an arbuscular mycorrhizal fungus. New Phytol. 2005, 167, 571–578. [Google Scholar] [CrossRef]
- Becquer, A.; Garcia, K.; Amenc, L.; Rivard, C.; Dore, J.; Trives-Segura, C.; Szponarski, W.; Russet, S.; Baeza, Y.; Lassalle-Kaiser, B.; et al. The Hebeloma cylindrosporum HcPT2 Pi transporter plays a key role in ectomycorrhizal symbiosis. New Phytol. 2018, 220, 1185–1199. [Google Scholar] [CrossRef]
- Dreyer, I.; Spitz, O.; Kanonenberg, K.; Montag, K.; Handrich, M.R.; Ahmad, S.; Schott-Verdugo, S.; Navarro-Retamal, C.; Rubio-Melendez, M.E.; Gomez-Porras, J.L.; et al. Nutrient exchange in arbuscular mycorrhizal symbiosis from a thermodynamic point of view. New Phytol. 2019, 222, 1043–1053. [Google Scholar] [CrossRef]
- Plassard, C.; Becquer, A.; Garcia, K. Phosphorus transport in mycorrhiza: How far are we? Trends Plant Sci. 2019, 24, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Ezawa, T. Phosphorus Metabolism and Transport in Arbuscular Mycorrhizal Symbiosis; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 197–216. [Google Scholar]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Volpe, V.; Giovannetti, M.; Sun, X.G.; Fiorilli, V.; Bonfante, P. The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non mycorrhizal roots. Plant Cell Environ. 2016, 39, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Pumplin, N.; Harrison, M.J. Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ. 2007, 30, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.; Harrison, M.J. Accumulation of phosphoinositides in distinct regions of the periarbuscular membrane. New Phytol. 2018, 221, 2213–2227. [Google Scholar] [CrossRef]
- Cox, G.C.; Sanders, F.E. Ultrastructure of the host-fungus interface in a vesicular-arbuscular mycorrhiza. New Phytol. 1974, 73, 901–912. [Google Scholar] [CrossRef]
- Kobae, Y.; Gutjahr, C.; Paszkowski, U.; Kojima, T.; Fujiwara, T.; Hata, S. Lipid droplets of arbuscular mycorrhizal fungi emerge in concert with arbuscule collapse. Plant Cell Physiol. 2014, 55, 1945–1953. [Google Scholar] [CrossRef]
- Ivanov, S.; Austin, J., 2nd; Berg, R.H.; Harrison, M.J. Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat. Plants 2019, 5, 194–203. [Google Scholar] [CrossRef]
- Choi, H.S.; Su, W.M.; Han, G.S.; Plote, D.; Xu, Z.; Carman, G.M. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 2012, 287, 11290–11301. [Google Scholar] [CrossRef]
- Su, W.M.; Han, G.S.; Casciano, J.; Carman, G.M. Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 2012, 287, 33364–33376. [Google Scholar] [CrossRef]
- Rajvanshi, P.K.; Arya, M.; Rajasekharan, R. The stress-regulatory transcription factors Msn2 and Msn4 regulate fatty acid oxidation in budding yeast. J. Biol. Chem. 2017, 292, 18628–18643. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Liu, X.; Feng, G.; Zhu, H.; Yao, Q. Linking lipid transfer with reduced arbuscule formation in tomato roots colonized by arbuscular mycorrhizal fungus under low pH stress. Environ. Microbiol. 2020, 22, 1036–1051. [Google Scholar] [CrossRef] [PubMed]
- Dechant, R.; Saad, S.; Ibanez, A.J.; Peter, M. Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol. Cell 2014, 55, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Wilms, T.; Swinnen, E.; Eskes, E.; Dolz-Edo, L.; Uwineza, A.; Van Essche, R.; Rosseels, J.; Zabrocki, P.; Cameroni, E.; Franssens, V.; et al. The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability. PLoS Genet. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Bond, S.; Forgac, M. The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J. Biol. Chem. 2008, 283, 36513–36521. [Google Scholar] [CrossRef] [PubMed]
- Deprez, M.; Eskes, E.; Wilms, T.; Ludovico, P.; Winderickx, J. pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 menage-a-trois to stress tolerance and longevity. Microb. Cell 2018, 5, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Zaman, S.; Broach, J.R.; Klionsky, D.J. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4180–4189. [Google Scholar] [CrossRef] [PubMed]
- Budovskaya, Y.V.; Stephan, J.S.; Reggiori, F.; Klionsky, D.J.; Herman, P.K. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20663–20671. [Google Scholar] [CrossRef]
- Wang, Z.; Wilson, W.A.; Fujino, M.A.; Roach, P.J. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 2001, 21, 5742–5752. [Google Scholar] [CrossRef]
- Sampaio-Marques, B.; Burhans, W.C.; Ludovico, P. Longevity pathways and maintenance of the proteome: The role of autophagy and mitophagy during yeast ageing. Microb. Cell 2014, 1, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, Y.; Akiyama, R.; Tanaka, S.; Yano, K.; Kameoka, H.; Marui, S.; Saito, M.; Kawaguchi, M.; Akiyama, K.; Saito, K. Myristate can be used as a carbon and energy source for the asymbiotic growth of arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 25779–25788. [Google Scholar] [CrossRef] [PubMed]
- Kameoka, H.; Tsutsui, I.; Saito, K.; Kikuchi, Y.; Handa, Y.; Ezawa, T.; Hayashi, H.; Kawaguchi, M.; Akiyama, K. Stimulation of asymbiotic sporulation in arbuscular mycorrhizal fungi by fatty acids. Nat. Microbiol. 2019, 4, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Hashimoto, K.; Kobayashi, Y.; Yano, K.; Maeda, T.; Kameoka, H.; Ezawa, T.; Saito, K.; Akiyama, K.; Kawaguchi, M. Asymbiotic mass production of the arbuscular mycorrhizal fungus Rhizophagus clarus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Wang, H.; Wei, D.; Akhberdi, O.; Liu, Y.; Xiang, B.; Hao, X.; Zhu, X. The AMP-activated protein kinase homolog Snf1 concerts carbon utilization, conidia production and the biosynthesis of secondary metabolites in the taxol-producer Pestalotiopsis microspora. Genes 2018, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Liu, X.; Ye, Z.; Zhou, Q.; Liu, P.; Yin, Z.; Wang, W.; Zheng, X.; Zhang, H.; Zhang, Z. Phosphatase-associated protein MoTip41 interacts with the phosphatase MoPpe1 to mediate crosstalk between TOR and cell wall integrity signaling during infection by the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Liu, X.H.; Gao, H.M.; Xu, F.; Lu, J.P.; Devenish, R.J.; Lin, F.C. Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy 2012, 8, 1415–1425. [Google Scholar] [CrossRef]
- Khan, I.A.; Lu, J.P.; Liu, X.H.; Rehman, A.; Lin, F.C. Multifunction of autophagy-related genes in filamentous fungi. Microbiol. Res. 2012, 167, 339–345. [Google Scholar] [CrossRef]
- Josefsen, L.; Droce, A.; Sondergaard, T.E.; Sørensen, J.L.; Bormann, J.; Schäfer, W.; Giese, H.; Olsson, S. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy 2014, 8, 326–337. [Google Scholar] [CrossRef]
- St-Arnaud, M.; Hamel, C.; Vimard, B.; Caron, M.; Fortin, J.A. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol. Res. 1996, 100, 328–332. [Google Scholar] [CrossRef]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.; Hamby, R.; Heller, J.; Zhao, H.; Louise Glass, N.; Judelson, H.S.; et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef] [PubMed]
| Phylum | Types | Fungal Genomes | References | |
|---|---|---|---|---|
| Mucoromycota | Glomeromycotina | Arbuscular mycorrhiza | Rhizophagus irregularis DAOM197198 | [33,34,40] |
| Rhizophagus clarus HR1 | [35] | |||
| Rhizophagus cerebriforme DAOM227022 | [36] | |||
| Gigaspora margarita BEG34 | [38] | |||
| Gigaspora rosea DAOM194575 | [36] | |||
| Diversispora epigaea IT104 | [37] | |||
| Mucoromycotina | Animal pathogen | Podila verticillata NRRL 6337 | GenBank: KN042421.1 | |
| Mucor lusitanicus MU402 | [41] | |||
| Rhizopus delemar RA 99-880 | [42] | |||
| Basidiomycota | Ectomycorrhizal mycorrhiza | Hebeloma cylindrosporum h7 | [43] | |
| Laccaria bicolor S238N-H82 | [44] | |||
| Pisolithus tinctorius Marx 270 | [43] | |||
| Root endophyte | Serendipita indica DSM 11827 | [45] | ||
| Orchid mycorrhiza | Tulasnella calospora MUT 4182 | [43] | ||
| Plant pathogen | Sporisorium scitamineum SscI8 | [46] | ||
| Ustilago maydis 521 | [47] | |||
| Animal pathogen | Cryptococcus neoformans var. neoformans JEC21 | [48] | ||
| Ascomycota | Ectomycorrhizal mycorrhiza | Tuber melanosporum Mel28 | [49] | |
| Plant pathogen | Fusarium graminearum PH-1 | [50] | ||
| Fusarium oxysporum f. sp. lycopersici 4287 | [51] | |||
| Pyricularia grisea NI907 | [52,53] | |||
| Colletotrichum higginsianum IMI 349063 | [54,55] | |||
| Zymoseptoria tritici IPO323 | [56] | |||
| Verticillium dahliae VdLs.17 | [57] | |||
| Animal pathogen | Aspergillus fumigatus Af293 | [58] | ||
| Saprotroph or pathogen | Neurospora crassa OR74A | [59] | ||
| No | Homolog | Description | Accession Numbers | Conserved Domains | Expression Profile Retrieved from Transcriptome 1 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R. irregularis | G. rosea | |||||||||||||||||||||||
| S. cerevisiae | R. irregularis | R. clarus | G. cerebriforme | G. rosea | G. margarita | D. epigaea | Spores | Mycorrhizas | Spores | Mycorrhizas | ||||||||||||||
| Sucrose non-fermenting 1 (SNF1) protein kinase signaling pathway | ||||||||||||||||||||||||
| 1 | Snf3/Hxts | Sugar sensor or transporter | CAA98771.1 | XP025173015.1 | GBC07484.1 | RIA98081.1 | RIB05796.1 | KAF0510814.1 | RHZ52775.1 | MFS_HXT, Sugar_tr, xylE | 3.6 | 3.5 | 3.6 | 0.0 | 0.0 | 1.9 | ↓ | 4.0 | 4.0 | 4.0 | 8.2 | 8.7 | 8.6 | ↑ |
| 2 | Hxk1 | Hexokinase | KZV11659.1 | XP025172441.1 | GBB98726.1 | RIA97617.1 | RIB22304.1 | KAF0429716.1 | RHZ53334.1 | COG5026 | 6.4 | 6.5 | 6.3 | 6.4 | 6.8 | 6.6 | 6.2 | 6.4 | 6.5 | 5.9 | 5.7 | 5.7 | ||
| 3 | Yck1 | Casein kinase I | KZV10871.1 | XP025174102.1 | GBB87197.1 | RIA91239.1 | RIB06770.1 | KAF0535241.1 | RHZ66640.1 | STKc_CK1_fungal | 6.3 | 6.3 | 6.3 | 6.6 | 6.5 | 6.5 | 7.8 | 7.8 | 7.9 | 7.0 | 7.0 | 7.3 | ||
| 4 | Snf1 | Carbon catabolite derepressing protein kinase | QHB07934.1 | XP025181474.1 | GBB86133.1 | RIA96595.1 | RIB06014.1 | KAF0504798.1 | RHZ87761.1 | STKc_AMPK_alpha, AMPKA_C, UBA_SNF1_fungi | 5.4 | 5.5 | 5.5 | 5.3 | 5.1 | 5.2 | 5.8 | 6.0 | 5.9 | 5.2 | 5.3 | 5.7 | ||
| 5 | Gal83 | 5′-AMP-activated protein kinase SNF1 beta subunit | KZV11779.1 | XP025175162.1 | GBC02790.1 | RIA96007.1 | RIB23376.1 | KAF0492722.1 | AMPKBI, AMPK1_CBM, E_set_AMPKbeta_like_N | 6.1 | 6.2 | 6.1 | 6.6 | 5.5 | 5.6 | 5.7 | 5.6 | 5.6 | 5.1 | 5.4 | 5.5 | |||
| 6 | Sak1 | Calcium/calmodulin-dependent protein kinase kinase | KZV11897.1 | XP025169502.1 | GBB90230.1 | RIA89443.1 | RIB14285.1 | KAF0377796.1 | RHZ56086.1 | STKc_LKB1_CaMKK | 5.5 | 5.6 | 5.8 | 5.5 | 6.0 | 6.0 | 5.7 | 5.8 | 5.7 | 5.3 | 5.2 | 4.8 | ||
| 7 | Snf4 | Nuclear protein SNF4 | CAA96823.1 | XP025168407.1 | GBB93785.1 | RIA84755.1 | RIB14600.1 | KAF0525700.1 | RHZ44230.1 | CBS_euAMPK_gamma-like_repeat2, CBS_euAMPK_gamma-like_repeat1 | 6.0 | 5.9 | 6.0 | 5.8 | 5.9 | 5.5 | 5.2 | 5.1 | 5.1 | 4.1 | 3.7 | 3.3 | ↓ | |
| 8 | Reg1 | REG1-regulatory subunit for protein phosphatase Glc7 | CAA98850.1 | XP025174225.1 | GBB84436.1 | RIA95576.1 | RIB18190.1 | KAF0465234.1 | RHZ64075.1 | DUF1752 | 5.3 | 5.5 | 5.6 | 8.0 | 7.8 | 8.6 | ↑ | 7.7 | 7.9 | 7.8 | 9.1 | 8.5 | 9.2 | ↑ |
| 9 | Rgt1 | Fungal-specific transcription factor domain-containing protein | CAA49301.1 | XP025183341.1 | GBB97255.1 | RIA96158.1 | RIB27603.1 | KAF0395517.1 | RHZ69529.1 | GAL4, Zn_clus, Fungal_trans, fungal_TF_MHR | 3.1 | 2.8 | 2.8 | 4.3 | 2.4 | 3.4 | 4.5 | 4.1 | 4.1 | 3.8 | 4.0 | 4.3 | ||
| 10 | Gal1 | Galactokinase | CAA84962.1 | XP025167943.1 | GBC05682.1 | RIA85840.1 | RIB23881.1 | KAF0523208.1 | RHZ56837.1 | PLN02521 | 3.9 | 3.4 | 3.8 | 5.3 | 5.4 | 6.3 | ↑ | 5.1 | 5.1 | 5.1 | 6.0 | 6.4 | 6.2 | ↑ |
| 11 | Mig1 | DNA-binding protein creA | CAA96736.1 | XP025167185.1 | GBB93790.1 | RIA84743.1 | RIB13066.1 | KAF0519624.1 | RHZ72138.1 | COG5048 (Zn-finger) | 5.0 | 4.8 | 4.0 | 4.6 | 4.6 | 4.4 | 1.2 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| 12 | Ssn6 | General transcriptional repressor | CAA46973.1 | XP025187165.1 | GBB94614.1 | RIA93188.1 | RIB04547.1 | KAF0478375.1 | RHZ44816.1 | TPR | 7.0 | 7.1 | 7.0 | 6.8 | 6.9 | 6.9 | 6.8 | 6.9 | 6.7 | 5.7 | 5.6 | 5.6 | ↓ | |
| 13 | Tup1 | Glucose repression regulatory protein | QHB07212.1 | XP025177165.1 | GBB99361.1 | RIA83988.1 | RIB05835.1 | KAF0554834.1 | RHZ78296.1 | WD40, Tup_N | 7.8 | 7.9 | 7.8 | 7.6 | 7.7 | 8.3 | 6.7 | 6.8 | 6.8 | 6.3 | 6.1 | 6.0 | ||
| cAMP-dependent protein kinase A (cAMP-PKA) signaling pathway | ||||||||||||||||||||||||
| 14 | Cdc25 | Cell division control protein 25 | KZV09553.1 | XP025169177.1 | GBB99424.1 | RIA94624.1 | RIB25226.1 | KAF0428185.1 | RHZ74526.1 | RasGEF, RasGEF_N, SH3_Sdc25 | 6.3 | 6.4 | 6.4 | 6.3 | 5.7 | 6.2 | ↓ | 6.4 | 6.5 | 6.5 | 7.2 | 7.1 | 7.7 | ↑ |
| 15 | Ras1 | Ras-like protein | CAA99298.1 | XP025166744.1 | GBB97044.1 | RIA98161.1 | RIB12382.1 | KAF0440412.1 | RHZ56098.1 | small_GTPase | 9.1 | 9.2 | 9.2 | 8.4 | 8.5 | 8.3 | ↓ | 3.6 | 3.8 | 3.5 | 3.7 | 2.5 | 2.0 | |
| 16 | Ira1 | GTPase-activator protein for Ras-like GTPase | AAA34709.1 | XP025175181.1 | GBC02045.1 | RIA95985.1 | RIB13611.1 | KAF0427762.1 | RHZ89446.1 | RasGAP_Neurofibromin_like, CRAL_TRIO_2, RasGAP, PH-like | 5.1 | 5.1 | 5.0 | 4.9 | 3.7 | 4.4 | ↓ | 5.5 | 5.7 | 5.5 | 4.2 | 4.1 | 4.4 | ↓ |
| 17 | Gpa3 | Guanine nucleotide-binding protein alpha subunit | QHB08090.1 | XP025188707.1 | GBB91367.1 | RIA81730.1 | RIB01335.1 | KAF0408695.1 | RHZ45261.1 | G-alpha | 6.5 | 6.7 | 6.6 | 5.5 | 6.3 | 6.4 | ↓ | 4.6 | 4.6 | 4.4 | 2.9 | 3.7 | 3.2 | ↓ |
| 18 | Cyr1 | Adenylate cyclase | CAA89295.1 | XP025179329.1 | GBB94024.1 | RIA93841.1 | RIB11944.1 | KAF0398164.1 | RHZ86430.1 | PP2Cc, CYCc, RA_CYR1_like | 5.3 | 5.3 | 5.4 | 5.8 | 5.0 | 4.8 | 6.0 | 6.1 | 6.0 | 6.2 | 5.7 | 6.2 | ↑ | |
| 19 | Bcy1 | PKA regulatory subunit | QHB09340.1 | XP025186511.1 | GBC10838.1 | RIA97450.1 | RIB13909.1 | KAF0528101.1 | RHZ83631.1 | CAP_ED | 5.6 | 5.7 | 5.9 | 6.3 | 6.4 | 5.8 | 7.1 | 7.0 | 6.9 | 6.8 | 7.0 | 6.7 | ||
| 20 | Tpk1 | cAMP-dependent protein kinase catalytic subunit | KZV10110.1 | XP025170217.1 | GBB88842.1 | RIA98707.1 | RIB04262.1 | KAF0332898.1 | RHZ57338.1 | STKc_PKA_like | 5.6 | 5.6 | 5.8 | 5.9 | 5.8 | 6.8 | 6.4 | 6.3 | 6.4 | 5.6 | 5.3 | 6.1 | ||
| 21 | Pde2 | cAMP phosphodiesterase | CAA99689.1 | XP025182850.1 | GBC09025.1 | RIA86790.1 | RIB09177.1 | KAF0469187.1 | RHZ67223.1 | PDEase_I | 4.1 | 4.2 | 4.0 | 5.7 | 5.0 | 5.0 | ↑ | 4.5 | 4.6 | 4.9 | 4.0 | 4.8 | 4.9 | |
| 22 | Yak1 | YAK1-ser/thr protein kinase | CAA89437.1 | XP025178972.1 | GBC09926.1 | RIA82219.1 | RIB05244.1 | KAF0349434.1 | RHZ76762.1 | PKc_YAK1 | 5.4 | 5.4 | 5.6 | 6.1 | 5.7 | 5.9 | 6.8 | 6.7 | 6.7 | 5.9 | 5.8 | 5.9 | ↓ | |
| Target of rapamycin (TOR) kinase signaling pathway | ||||||||||||||||||||||||
| 23 | Ssy1 | Amino acid permease | QHB07619.1 | XP025173052.1 | GBB94153.1 | RIA83029.1 | RIB02084.1 | KAF0517317.1 | RHZ74142.1 | LysP, 2A0306 | 3.2 | 3.1 | 2.6 | 8.2 | 7.6 | 8.6 | ↑ | 5.6 | 5.5 | 5.6 | 5.8 | 4.3 | 5.9 | |
| 24 | Mep2 | Ammonium transporter | CAA96025.1 | XP025183237.1 | GBC08321.1 | RIA89994.1 | RIB20969.1 | KAF0484149.1 | RHZ83241.1 | AmtB | 5.2 | 5.2 | 5.5 | 6.1 | 6.1 | 5.8 | ↑ | 8.0 | 7.8 | 7.6 | 3.2 | 3.0 | 3.6 | ↓ |
| 25 | Ure2 | Glutathione S-transferase | CAA96134.1 | XP025188118.1 | GBC09082.1 | RIA99246.1 | RIB13374.1 | KAF0420358.1 | GST_C_, GST_N_Ure2p_like, GstA | 6.7 | 6.8 | 6.6 | 6.9 | 5.3 | 6.3 | ↓ | 6.1 | 5.9 | 6.1 | 5.9 | 5.4 | 5.5 | ||
| 26 | Gat1 | Nitrogen regulatory protein areA | KZV11580.1 | XP025166471.1 | GBC08162.1 | RIA92069.1 | RIB23596.1 | KAF0391989.1 | RHZ89193.1 | ZnF_GATA, DUF1752, GAT1 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.3 | 4.5 | 4.4 | 5.0 | 4.7 | 4.9 | ↑ | |
| 27 | Gtr1 | GTP-binding protein GTR1 | QHB10618.1 | XP025166820.1 | GBC04641.1 | RIA82430.1 | RIB25894.1 | KAF0507139.1 | RHZ45447.1 | RagA_like | 6.1 | 6.0 | 6.0 | 6.1 | 6.1 | 4.4 | 6.2 | 6.2 | 6.2 | 5.8 | 5.9 | 5.5 | ||
| 28 | Gtr2 | Ras-related GTP-binding protein | BAA28781.1 | XP025175556.1 | GBB95986.1 | RIA90228.1 | RIB22850.1 | KAF0500663.1 | RHZ88007.1 | Gtr1_RagA, RagC_like, P-loop_NTPase | 6.3 | 6.4 | 6.4 | 6.0 | 6.1 | 6.0 | 6.4 | 6.3 | 6.4 | 6.1 | 5.8 | 5.7 | ||
| 29 | Tor2 | FKBP12-rapamycin complex-associated protein | CAA50548.1 | XP025188708.1 | GBB93380.1 | RIA95684.1 | RIB03527.1 | P32600.3 | RHZ66605.1 | FAT, TEL1, PIKKc_TOR | 5.8 | 6.0 | 5.9 | 5.6 | 5.5 | 5.7 | ↓ | 6.2 | 6.2 | 6.1 | 5.3 | 5.0 | 5.5 | ↓ |
| 30 | Kog1 | Regulatory-associated protein of TOR | QHB09177.1 | XP025188793.1 | GBC02948.1 | RIA93930.1 | RIB12272.1 | KAF0421509.1 | RHZ51963.1 | Raptor_N, WD40 | 5.6 | 5.8 | 5.6 | 5.7 | 6.1 | 6.2 | 6.6 | 6.4 | 6.4 | 6.4 | 6.4 | 6.6 | ↑ | |
| 31 | Lst8 | Target-rapamycin complex subunit LST8 | QHB11374.1 | XP025168325.1 | GBB93049.1 | RIA94383.1 | RIB13947.1 | KAF0366345.1 | RHZ52815.1 | WD40 | 6.1 | 6.0 | 6.2 | 6.5 | 5.6 | 6.2 | 6.1 | 6.1 | 6.1 | 5.3 | 5.5 | 5.4 | ||
| 32 | Sch9 | Serine/threonine-protein kinase | CAA40853.1 | XP025179023.1 | GBB99293.1 | RIA97083.1 | RIB30460.1 | KAF0544487.1 | RHZ75318.1 | STKc_Sck1_like, C2 surperfamily, PKc_like | 2.1 | 2.5 | 2.0 | 0.0 | 2.5 | 0.0 | 3.4 | 3.7 | 3.4 | 0.9 | 0.0 | 1.7 | ↓ | |
| 33 | Tap42 | TAP42-like protein | KZV08874.1 | XP025174300.1 | GBC04705.1 | RIA84937.1 | RIB23487.1 | KAF0516295.1 | RHZ77344.1 | TAP42 | 5.8 | 5.9 | 5.9 | 5.6 | 5.2 | 5.0 | ↓ | 6.3 | 6.1 | 6.2 | 5.4 | 5.2 | 5.5 | ↓ |
| 34 | Rrd1 | Serine/threonine-protein phosphatase 2A activator | QHB09229.1 | XP025178889.1 | GBC03931.1 | RIA82137.1 | RIB22358.1 | KAF0482349.1 | RHZ86199.1 | PTPA | 5.2 | 5.3 | 5.5 | 5.6 | 4.8 | 5.5 | 5.7 | 5.4 | 5.7 | 5.3 | 5.6 | 5.1 | ||
| 35 | Rrd2 | Serine/threonine-protein phosphatase 2A activator | QHB12111.1 | XP025179635.1 | GBB93114.1 | RIA87981.1 | RIB08558.1 | KAF0386976.1 | RHZ72688.1 | PTPA | 6.7 | 6.7 | 6.8 | 6.4 | 6.1 | 6.9 | 5.7 | 5.7 | 5.9 | 6.0 | 5.2 | 5.8 | ||
| 36 | Tip41 | TIP41-domain-containing protein | KZV07556.1 | XP025181449.1 | GBB97298.1 | RIA84855.1 | RIB12158.1 | KAF0369945.1 | RHZ86053.1 | TIP41 | 4.7 | 4.5 | 4.7 | 5.0 | 5.2 | 4.7 | 5.6 | 5.5 | 5.6 | 4.5 | 4.0 | 3.5 | ↓ | |
| 37 | Pph22 | PP2A catalytic subunit | CAA98765.1 | XP025186163.1 | RIA78842.1 | RIB01551.1 | KAF0479568.1 | RHZ81746.1 | MPP_PP2A_PP4_PP6 | 7.7 | 7.9 | 7.7 | 7.5 | 7.3 | 7.7 | 7.8 | 7.8 | 7.8 | 6.9 | 7.3 | 7.1 | |||
| 38 | Tpd3 | Protein phosphatase PP2A regulatory subunit A | QHB06631.1 | XP025181440.1 | GBB86608.1 | RIA96926.1 | RIB23797.1 | KAF0429327.1 | RHZ79342.1 | HEAT_2, HEAT, PRK13800 | 6.8 | 6.8 | 6.8 | 7.0 | 7.0 | 6.9 | 7.4 | 7.3 | 7.3 | 7.4 | 7.6 | 7.4 | ↑ | |
| 39 | Cdc55 | Protein phosphatase PP2A regulatory subunit B | AAA34482.1 | XP025174227.1 | GBC09955.1 | RIA95590.1 | RIB10219.1 | KAF0437825.1 | RHZ64089.1 | CDC55 | 6.8 | 6.9 | 6.9 | 6.9 | 7.0 | 6.9 | 6.9 | 7.0 | 7.0 | 6.7 | 6.7 | 6.4 | ||
| 40 | Rts1 | Phosphatase 2A regulatory B subunit | CAA99203.1 | XP025171904.1 | GBB94689.1 | RIA88794.1 | RIB06671.1 | KAF0374544.1 | RHZ47872.1 | B56 | 6.3 | 6.4 | 6.3 | 7.3 | 6.9 | 7.1 | ↑ | 6.3 | 6.4 | 6.2 | 6.2 | 6.0 | 6.2 | |
| 41 | Sap190 | SIT4 phosphatase-associated protein | CAA82100.1 | XP025183818.1 | GBC07712.1 | RIA90499.1 | RIB24656.1 | KAF0417990.1 | RHZ82279.1 | SAPS | 5.7 | 5.6 | 5.7 | 6.2 | 6.2 | 6.6 | ↑ | 6.3 | 6.2 | 6.2 | 6.0 | 6.0 | 6.2 | |
| 42 | Fpr1 | FKBP-type peptidyl-prolyl isomerase | CAA96017.1 | XP025188636.1 | GBC03273.1 | RIA79256.1 | RIB11261.1 | KAF0538299.1 | RHZ84133.1 | FkpA, FKBP_C, FkpA | 7.7 | 7.8 | 7.7 | 8.2 | 7.8 | 7.7 | 5.5 | 5.5 | 5.5 | 3.8 | 2.7 | 4.6 | ||
| 43 | Avo1 | Stress activated MAP kinase interacting protein | KZV07803.1 | XP025173883.1 | GBB90387.1 | RIA85683.1 | RIB07950.1 | KAF0552014.1 | RHZ47334.1 | CRIM, SIN1_PH | 5.9 | 5.9 | 6.0 | 5.6 | 4.9 | 5.6 | ↓ | 6.2 | 6.2 | 6.2 | 5.5 | 5.0 | 5.7 | |
| Phosphate (PHO) signalling pathway | ||||||||||||||||||||||||
| 44 | Pho84 | Phosphate transporter | QHB10616.1 | XP025183371.1 | GBC04213.1 | RIA99731.1 | RIB05912.1 | KAF0463572.1 | RHZ61205.1 | 2A0109 | 5.7 | 5.8 | 5.7 | 9.6 | 9.1 | 9.4 | ↑ | 7.3 | 7.4 | 7.5 | 9.1 | 8.9 | 9.3 | ↑ |
| 45 | Pho91 | SPX domain-containing protein similar to Pho87 and Pho90 | KZV08641.1 | XP025171348.1 | GBC10644.1 | RIA85720.1 | RIB09524.1 | KAF0379302.1 | RHZ77664.1 | SLC13_permease, SPX_PHO87_PHO90_like | 5.0 | 5.1 | 5.0 | 5.2 | 5.8 | 5.9 | ↑ | 4.9 | 4.7 | 4.9 | 6.0 | 6.7 | 6.4 | ↑ |
| 46 | Arg82 | ARG82-dual-specificity inositol polyphosphate kinase | QHB07633.1 | XP025175101.1 | GBC08502.1 | RIA86563.1 | RIB03691.1 | KAF0406837.1 | RHZ46527.1 | IPK | 3.9 | 4.0 | 4.5 | 4.9 | 5.0 | 4.1 | 6.9 | 6.9 | 7.0 | 6.6 | 6.6 | 6.5 | ||
| 47 | Kcs1 | Inositol hexaphosphate kinase KCS1 | AAB36234.1 | XP025174322.1 | RIA96250.1 | RIB08669.1 | KAF0502809.1 | IPK | 5.8 | 5.9 | 5.9 | 6.7 | 6.4 | 6.5 | ↑ | 5.5 | 5.5 | 5.3 | 4.9 | 4.8 | 4.6 | |||
| 48 | Adk1 | Adenylate kinase | QHB07687.1 | XP025181716.1 | GBC08115.1 | RIA88751.1 | RIB29553.1 | KAF0391071.1 | RHZ69843.1 | adk | 6.4 | 6.6 | 6.4 | 7.1 | 6.4 | 6.3 | 6.4 | 6.4 | 6.5 | 7.4 | 7.9 | 7.7 | ↑ | |
| 49 | Ado1 | Adenosine kinase | QHB09727.1 | XP025188792.1 | GBC02949.1 | RIA93929.1 | RIB12283.1 | KAF0421504.1 | RHZ51964.1 | ribokinase_pfkB, adenosine_kinase | 7.1 | 7.1 | 7.0 | 7.4 | 8.2 | 8.1 | ↑ | 7.4 | 7.4 | 7.4 | 7.1 | 7.4 | 6.7 | |
| 50 | Pho81 | Cyclin-dependent protein kinase inhibitor | CAA97261.1 | XP025171189.1 | GBB88938.1 | RIA87384.1 | RIB13327.1 | KAF0478836.1 | RHZ82191.1 | SPX_PHO81_NUC-2_like, Ank_2, PI-PLCc_GDPD_SF | 5.3 | 5.3 | 5.3 | 8.4 | 7.8 | 7.6 | ↑ | 5.5 | 5.5 | 5.4 | 5.7 | 5.2 | 5.2 | |
| 51 | Pho80 | Cyclin-dependent protein kinase regulator | CAA99000.1 | XP025168925.1 | GBC00404.1 | RIA96867.1 | RIB28759.1 | KAF0480379.1 | RHZ79637.1 | CYCLIN (Cyclin box fold) | 4.4 | 5.1 | 4.8 | 4.5 | 4.6 | 4.9 | 5.6 | 5.8 | 5.6 | 5.6 | 5.8 | 5.8 | ||
| 52 | Pho85 | Negative regulator of the PHO system | CAA68773.1 | XP025181480.1 | GBB86139.1 | RIA96589.1 | RIB12090.1 | KAF0377917.1 | STKc_PHO85, PKc_like | 7.2 | 7.3 | 7.3 | 6.9 | 7.0 | 7.1 | 5.4 | 5.4 | 5.6 | 5.9 | 6.0 | 6.0 | ↑ | ||
| 53 | Pho2 | Homeodomain transcription factor | CAA64906.1 | XP025175623.1 | GBC04742.1 | RIA91171.1 | RIB14816.1 | KAF0560668.1 | RHZ56225.1 | Homeobox | 5.5 | 5.3 | 5.2 | 5.3 | 7.2 | 5.9 | ↑ | 6.7 | 6.9 | 6.6 | 8.4 | 8.0 | 8.8 | ↑ |
| 54 | Pho4 | HLH-domain-containing protein | QHB08372.1 | XP025170451.1 | GBB94725.1 | RIA91802.1 | RIB24056.1 | KAF0419854.1 | RHZ63527.1 | HLH, bHLHzip_USF_MITF, bHLH_SF | 6.7 | 6.6 | 6.6 | 6.5 | 6.6 | 6.6 | 5.1 | 4.9 | 5.1 | 5.8 | 5.7 | 5.6 | ↑ | |
| 55 | Ino80 | SNF2 family helicase/ATPase Ino80 | QHB08505.1 | XP025176540.1 | GBB97662.1 | RIA93554.1 | RIB09135.1 | KAF0508025.1 | RHZ78787.1 | HepA, SF2_C_SNF | 6.8 | 6.9 | 6.8 | 6.6 | 6.5 | 6.7 | 7.7 | 7.8 | 7.7 | 6.8 | 6.7 | 6.7 | ↓ | |
| Common targets and relevant components | ||||||||||||||||||||||||
| 56 | Rim15 | Serine threonine protein kinase | QHB08304.1 | XP025183815.1 | GBC07710.1 | RIB15849.1 | KAF0546391.1 | RHZ82277.1 | STKc_MAST_like, REC_hyHK_CKI1_RcsC-like | 5.2 | 5.2 | 5.1 | 5.6 | 5.6 | 6.1 | ↑ | 6.4 | 6.6 | 6.5 | 5.7 | 5.7 | 6.0 | ||
| 57 | 14-3-3 | 14-3-3 protein | CAA46959.1 | XP025174150.1 | GBC07263.1 | RIA97653.1 | RIB22291.1 | KAF0420307.1 | RHZ47341.1 | 14-3-3, BMH1 | 8.3 | 8.4 | 8.3 | 8.3 | 8.3 | 8.5 | 9.4 | 9.3 | 9.4 | 8.8 | 9.1 | 9.0 | ||
| 58 | Msn2 | STE12-like C2H2 type Zn-finger transcription factor | KZV08884.1 | XP025172111.1 | GBC01837.1 | RIA98728.1 | RIB08568.1 | KAF0407570.1 | RHZ45449.1 | COG5048 (Zn-finger), STE | 3.3 | 2.9 | 1.8 | 0.0 | 0.0 | 0.0 | 4.1 | 4.1 | 4.6 | 6.4 | 6.2 | 6.5 | ↑ | |
| 59 | Rph1 | Jumonji family transcription factor | KZV11945.1 | XP025184487.1 | GBB85389.1 | RIA98904.1 | RIB17285.1 | KAF0402735.1 | JmjC, JmjN, ePHD | 3.3 | 3.3 | 3.3 | 3.3 | 3.2 | 3.2 | ↓ | 2.9 | 2.9 | 2.9 | 2.7 | 2.8 | 2.8 | ↓ | |
| 60 | Tps2 | Glycosyltransferase | CAA98893.1 | XP025178428.1 | GBB84837.1 | RIA80605.1 | RIB18150.1 | KAF0420413.1 | RHZ45612.1 | PRK14501 | 7.1 | 7.2 | 7.3 | 7.4 | 6.8 | 7.4 | 7.7 | 8.0 | 7.7 | 6.4 | 6.5 | 6.5 | ↑ | |
| 61 | Nth1 | Neutral trehalase | KZV12232.1 | XP025174909.1 | GBC10103.1 | RIA80971.1 | RIB03117.1 | KAF0525729.1 | RHZ89787.1 | Trehalase_Ca-bi, Trehalase | 6.9 | 7.0 | 7.0 | 8.7 | 7.7 | 8.0 | ↑ | 5.8 | 5.7 | 5.8 | 6.8 | 6.3 | 6.5 | ↑ |
| 62 | Fhl1 | Fork head domain-containing protein | QHB12349.1 | XP025186513.1 | GBB95086.1 | RIA97453.1 | RIB29328.1 | KAF0409680.1 | RHZ62376.1 | Forkhead, COG5025 | 5.6 | 5.5 | 5.5 | 6.9 | 6.6 | 6.7 | ↑ | 5.7 | 5.5 | 5.3 | 5.3 | 4.2 | 5.0 | |
| 63 | Maf1 | Repressor of RNA polymerase III transcription MAF1 | KZV12237.1 | XP025179483.1 | GBC06348.1 | RIA93056.1 | RIB05097.1 | KAF0345972.1 | RHZ79416.1 | Maf1 | 5.9 | 5.9 | 5.8 | 6.0 | 5.9 | 5.6 | 5.9 | 6.1 | 6.0 | 5.3 | 5.8 | 4.7 | ||
| 64 | Rps13 | 40S ribosomal protein S13 | CAA98882.1 | XP025181192.1 | GBC02894.1 | RIA87612.1 | RIB04680.1 | KAF0396453.1 | RHZ52793.1 | PTZ00072 | 8.9 | 9.1 | 9.0 | 8.8 | 8.4 | 8.2 | ↓ | 8.7 | 8.7 | 8.7 | 8.2 | 8.1 | 7.7 | |
| 65 | Gcn2 | Serine/threonine-protein kinase GCN2 | AAA34636.1 | XP025188225.1 | GBC08433.1 | RIA89287.1 | RIB06418.1 | KAF0497404.1 | RHZ77885.1 | HGTP_anticodon2, STKc_EIF2AK4_GCN2_rpt2 | 5.5 | 5.4 | 5.5 | 5.0 | 5.4 | 5.5 | 5.7 | 5.7 | 5.7 | 4.7 | 4.5 | 4.6 | ↓ | |
| 66 | Gcn4 | Amino acid starvation-responsive transcription factor GCN4 | QHB08060.1 | XP025166650.1 | GBB92808.1 | RIA86909.1 | RIB09613.1 | KAF0458657.1 | RHZ87741.1 | bZIP_GCN4 | 8.4 | 8.2 | 8.6 | 9.6 | 8.8 | 9.3 | ↑ | 6.9 | 6.7 | 6.9 | 7.5 | 8.3 | 7.0 | ↑ |
| 67 | Atg8 | Autophagy-like protein 8 | QHB06698.1 | XP025175505.1 | GBB98445.1 | RIA88779.1 | RIB13205.1 | KAF0352158.1 | RHZ64615.1 | Ubl_ATG8 | 9.5 | 9.4 | 9.5 | 9.4 | 9.8 | 9.2 | 9.3 | 9.1 | 9.3 | 9.6 | 9.9 | 9.2 | ↑ | |
| 68 | Ssa3 | Heat shock protein 70 | CAA84896.1 | XP025181914.1 | GBB89555.1 | RIA88208.1 | RIB01431.1 | KAF0462691.1 | RHZ75893.1 | PTZ00009, HSP70 | 10.5 | 10.7 | 10.5 | 11.0 | 10.8 | 10.6 | 8.6 | 8.7 | 8.7 | 9.8 | 9.9 | 9.4 | ↑ | |
| 69 | Sod1 | Superoxide dismutase Cu-Zn | CAA89634.1 | XP025178555.1 | GBB95268.1 | RIA94955.1 | RIB09248.1 | KAF0517010.1 | RHZ44790.1 | Sod_Cu | 10.6 | 10.6 | 10.5 | 10.6 | 10.6 | 10.8 | 6.6 | 6.7 | 6.6 | 5.0 | 2.8 | 3.9 | ↓ | |
| 70 | Sod2 | Mitochondrial Mn-superoxide dismutase | QHB08994.1 | XP025168721.1 | GBC00991.1 | RIA91372.1 | RIB04073.1 | KAF0438330.1 | RHZ79502.1 | SodA, PLN02471 | 9.4 | 9.6 | 9.4 | 7.8 | 7.8 | 7.8 | ↓ | 8.6 | 8.8 | 8.6 | 7.6 | 7.3 | 7.2 | ↓ |
| 71 | Rho1 | GTP-binding protein rhoA | QHB12411.1 | XP025181617.1 | GBC08606.1 | RIA85906.1 | RIB06833.1 | KAF0421499.1 | RHZ75862.1 | RhoA_like, RHO, P-loop_NTPase | 9.2 | 9.3 | 9.2 | 9.2 | 9.1 | 9.2 | 6.6 | 6.6 | 6.7 | 8.2 | 8.1 | 8.3 | ↑ | |
| 72 | Hog1 | Mitogen-activated protein kinase HOG1 | CAA97680.1 | XP025185210.1 | GBB87021.1 | RIA92778.1 | RIB04295.1 | KAF0432463.1 | STKc_Sty1_Hog1 | 6.9 | 7.0 | 7.0 | 7.4 | 7.4 | 7.3 | 6.8 | 7.0 | 7.1 | 8.0 | 8.6 | 8.4 | ↑ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Li, J.; Tang, N.; Xie, H.; Fan, X.; Chen, H.; Tang, M.; Xie, X. Genome-Wide Analysis of Nutrient Signaling Pathways Conserved in Arbuscular Mycorrhizal Fungi. Microorganisms 2021, 9, 1557. https://doi.org/10.3390/microorganisms9081557
Zhou X, Li J, Tang N, Xie H, Fan X, Chen H, Tang M, Xie X. Genome-Wide Analysis of Nutrient Signaling Pathways Conserved in Arbuscular Mycorrhizal Fungi. Microorganisms. 2021; 9(8):1557. https://doi.org/10.3390/microorganisms9081557
Chicago/Turabian StyleZhou, Xiaoqin, Jiangyong Li, Nianwu Tang, Hongyun Xie, Xiaoning Fan, Hui Chen, Ming Tang, and Xianan Xie. 2021. "Genome-Wide Analysis of Nutrient Signaling Pathways Conserved in Arbuscular Mycorrhizal Fungi" Microorganisms 9, no. 8: 1557. https://doi.org/10.3390/microorganisms9081557
APA StyleZhou, X., Li, J., Tang, N., Xie, H., Fan, X., Chen, H., Tang, M., & Xie, X. (2021). Genome-Wide Analysis of Nutrient Signaling Pathways Conserved in Arbuscular Mycorrhizal Fungi. Microorganisms, 9(8), 1557. https://doi.org/10.3390/microorganisms9081557







