Re-Emergence of Salmonellosis in Hog Farms: Outbreak and Bacteriological Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Information
2.2. Salmonella Characterization
2.2.1. Serotyping
2.2.2. Phenotypic Antimicrobial Resistance Profile
2.2.3. Pulsed-Field Gel Electrophoresis (PFGE)
2.2.4. Whole Genome Sequencing (WGS)
2.2.5. WGS Analysis
3. Results
3.1. Serotyping
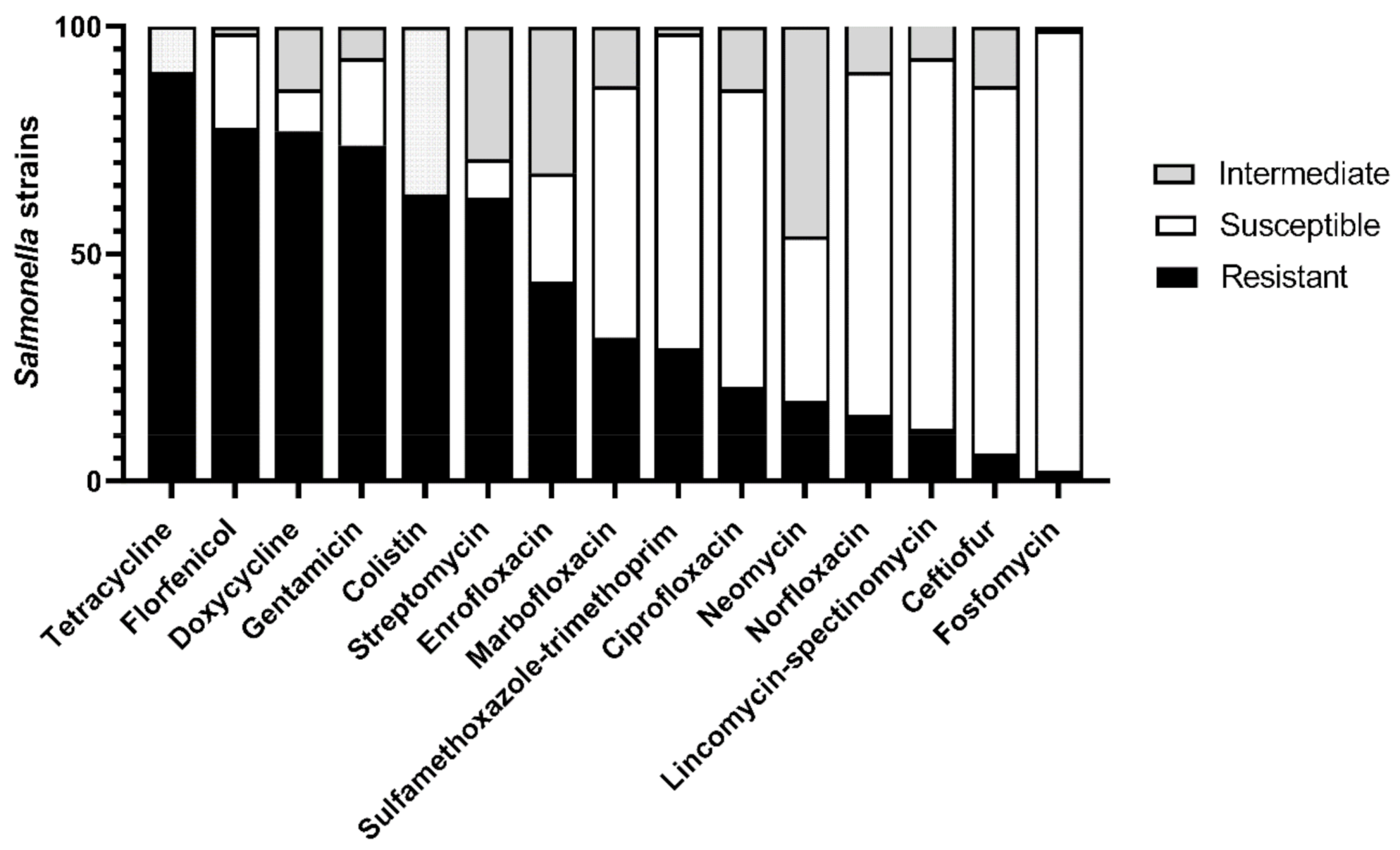
3.2. Phenotypic Antimicrobial Resistance Profile
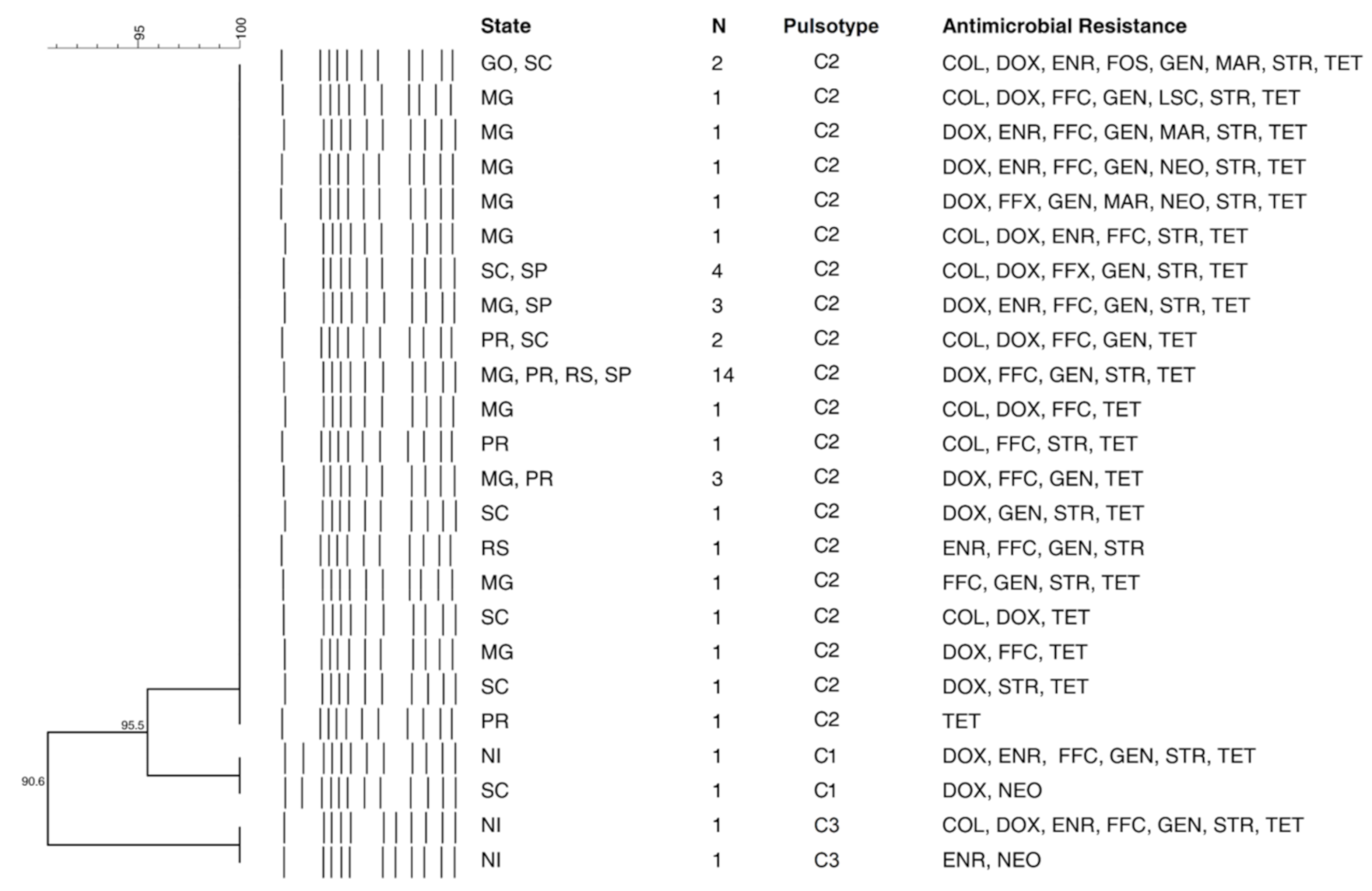
3.3. Pulsed-Field Gel Electrophoresis (PFGE)
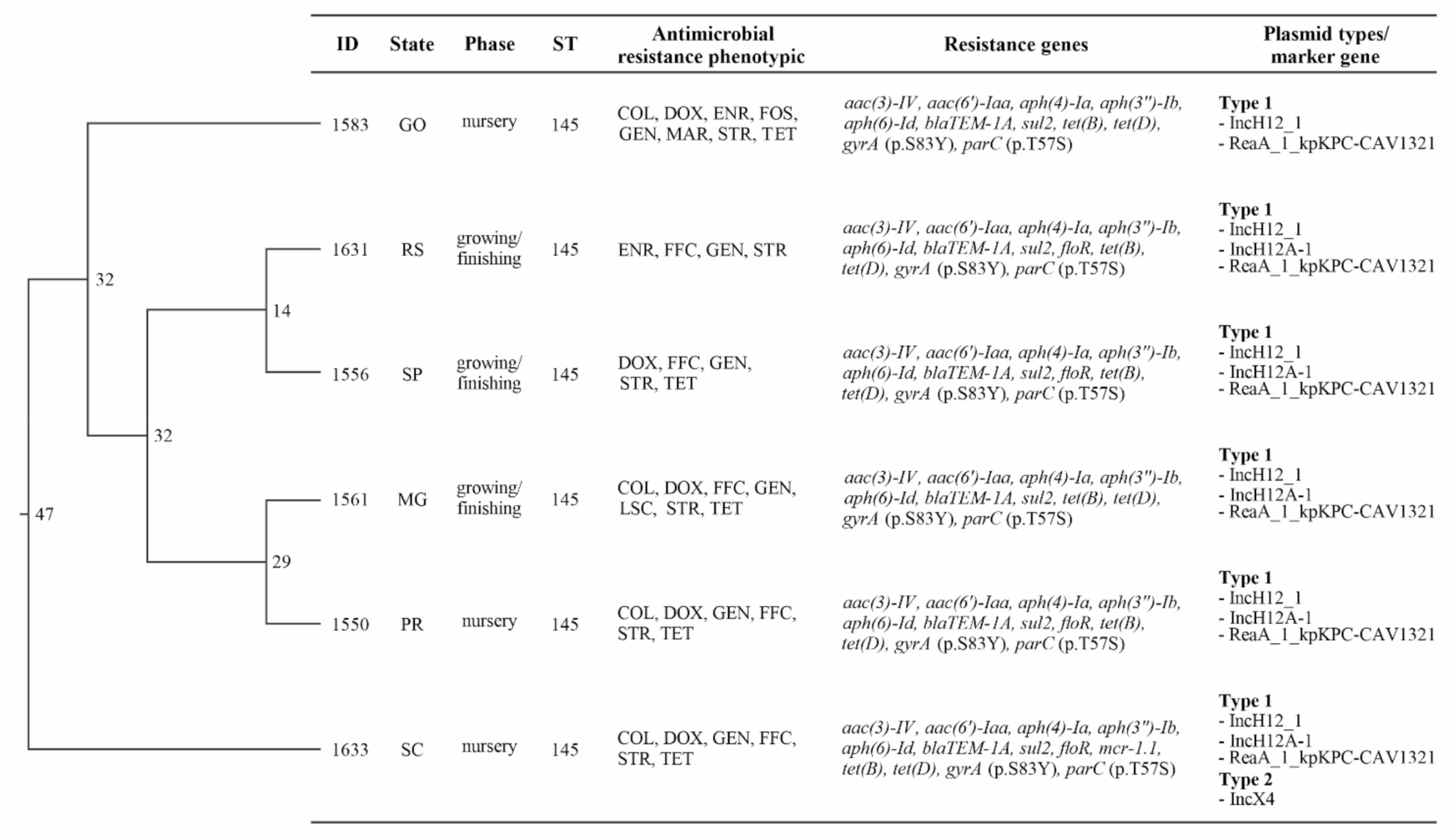
3.4. Whole Genome Sequencing Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gavin, C.; Simons, R.; Berriman, A.; Moorhouse, D.; Snary, E.; Smith, R.; Hill, A. A cost-benefit assessment of Salmonella-control strategies in pigs reared in the United Kingdom. Prev. Veter Med. 2018, 160, 54–62. [Google Scholar] [CrossRef]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.-X. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. Available online: https://apps.who.int/iris/handle/10665/199350 (accessed on 15 April 2021).
- Griffith, R.W.; Carlson, S.A.; Krull, A.C. Salmonellosis. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley Blackwell: Hoboken, NJ, USA, 2019; pp. 912–925. [Google Scholar]
- Hurd, H.S.; Mckean, J.D.; Griffith, R.W.; Wesley, I.V.; Rostagno, M.H. Salmonella enterica Infections in Market Swine with and without Transport and Holding. Appl. Environ. Microbiol. 2002, 68, 2376–2381. [Google Scholar] [CrossRef] [Green Version]
- Argüello, H.; Sørensen, G.; Carvajal, A.; Baggesen, D.L.; Rubio, P.; Pedersen, K. Characterization of the EmergingSalmonella4,[5],12:i:- in Danish Animal Production. Foodborne Pathog. Dis. 2014, 11, 366–372. [Google Scholar] [CrossRef]
- Barco, L.; Barrucci, F.; Cortini, E.; Ramon, E.; Olsen, J.E.; Luzzi, I.; Lettini, A.A.; Ricci, A. Ascertaining the relationship between Salmonella typhimurium and Salmonella 4,[5],12:i:- by MLVA and inferring the sources of human salmonellosis due to the two serovars in Italy. Front. Microbiol. 2015, 6, 301. [Google Scholar] [CrossRef]
- Possebon, F.S.; Casas, M.R.T.; Nero, L.A.; Yamatogi, R.S.; Araújo, J.P., Jr.; Pinto, J.P.D.A.N. Prevalence, antibiotic resistance, PFGE and MLST characterization of Salmonella in swine mesenteric lymph nodes. Prev. Veter Med. 2020, 179, 105024. [Google Scholar] [CrossRef]
- Rau, R.B.; De Lima-Morales, D.; Wink, P.L.; Ribeiro, A.R.; Barth, A.L. Salmonella enterica mcr-1 Positive from Food in Brazil: Detection and Characterization. Foodborne Pathog. Dis. 2020, 17, 202–208. [Google Scholar] [CrossRef]
- Arruda, B.L.; Burrough, E.R.; Schwartz, K.J. Salmonella enterica I 4,[5],12:i:- Associated with Lesions Typical of Swine Enteric Salmonellosis. Emerg. Infect. Dis. 2019, 25, 1377–1379. [Google Scholar] [CrossRef] [Green Version]
- Naberhaus, S.A.; Krull, A.C.; Arruda, B.L.; Arruda, P.; Sahin, O.; Schwartz, K.J.; Burrough, E.R.; Magstadt, D.R.; Ferreyra, F.M.; Gatto, I.R.H.; et al. Pathogenicity and Competitive Fitness of Salmonella enterica Serovar 4,[5],12:i:- Compared to Salmonella typhimurium and Salmonella Derby in Swine. Front. Veter Sci. 2020, 6, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, C.-H.; Su, L.-H.; Chu, C. Salmonella enterica Serotype Choleraesuis: Epidemiology, Pathogenesis, Clinical Disease, and Treatment. Clin. Microbiol. Rev. 2004, 17, 311–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, M.P.; Gray, J.T. Salmonella infections in pigs. In Salmonella in Domestic Animals; Barrow, P.A., Methner, U., Eds.; CABI International: Wallingford, UK, 2013; pp. 263–294. [Google Scholar]
- Asai, T.; Namimatsu, T.; Osumi, T.; Kojima, A.; Harada, K.; Aoki, H.; Sameshima, T.; Takahashi, T. Molecular typing and antimicrobial resistance of Salmonella enterica subspecies enterica serovar Choleraesuis isolates from diseased pigs in Japan. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 109–119. [Google Scholar] [CrossRef]
- Pedersen, K.; Sørensen, G.; Löfström, C.; Leekitcharoenphon, P.; Nielsen, B.; Wingstrand, A.; Aarestrup, F.M.; Hendriksen, R.S.; Baggesen, D.L. Reappearance of Salmonella serovar Choleraesuis var. Kunzendorf in Danish pig herds. Veter Microbiol. 2015, 176, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Methner, U.; Heller, M.; Bocklisch, H. Salmonella enterica subspecies enterica serovar Choleraesuis in a wild boar population in Germany. Eur. J. Wildl. Res. 2009, 56, 493–502. [Google Scholar] [CrossRef]
- Leekitcharoenphon, P.; Sørensen, G.; Löfström, C.; Battisti, A.; Szabo, I.; Wasyl, D.; Slowey, R.; Zhao, S.; Brisabois, A.; Kornschober, C.; et al. Cross-Border Transmission of Salmonella choleraesuis var. Kunzendorf in European Pigs and Wild Boar: Infection, Genetics, and Evolution. Front. Microbiol. 2019, 10, 179. [Google Scholar] [CrossRef] [Green Version]
- Longo, A.; LoSasso, C.; Vitulano, F.; Mastrorilli, E.; Turchetto, S.; Petrin, S.; Mantovani, C.; Pozza, M.C.D.; Ramon, E.; Conedera, G.; et al. Insight into an outbreak of Salmonella Choleraesuis var. Kunzendorf in wild boars. Veter Microbiol. 2019, 238, 108423. [Google Scholar] [CrossRef]
- Gil Molino, M.; Pérez, D.R.; Blanco, P.G.; Llario, P.F.; Molina, A.Q.; Sánchez, A.G.; Gerveno, J.M.C.; Gordo, L.G.; Cano, F.E.M.; Martínez, R.P.; et al. Outbreaks of antimicrobial resistant Salmonella Choleraesuis in wild boars piglets from central-western Spain. Transbound. Emerg. Dis. 2019, 66, 225–233. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Antimicrobial Resistance. Available online: www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 15 April 2021).
- Associação Brasileira de Proteína Animal (ABPA). Annual Reporter; ABPA: São Paulo, Brazil, 2020; 160p. [Google Scholar]
- Vannucci, F.A.; Oliveira, G.; Henriques, M.R.; Reis, K.C.P.; Bouillet, L.E.M.; Guimaraes, W.V.; Santos, D.L.; Santos, L.F.; Santos, J.S. Retrospective Study and Antimicrobial Susceptibilities of S enterica Serovar Choleraesuis Isolated from Swine Salmonellosis Outbreaks during 2013 in Brazil. In Proceedings of the 23rd International Pig Veterinary Society (IPVS) Congress, Cancun, Mexico, 8–11 June 2014; Volume II, p. 269. [Google Scholar]
- Santos, L.D.; Teixeira, R.; Santos, D.; Guimaraes, W.; Santos, J.L. Comparative Study of the Occurrence of S. enterica Serovar Choleraesuis Isolated from Swine Salmonellosis Outbreaks during 2013 to 2015 in Brazil. In Proceedings of the 24th International Pig Veterinary Society (IPVS) Congress, Dublin, Ireland, 7–10 June 2016; p. 276. [Google Scholar]
- Olsen, J.; Brown, D.; Skov, M.; Christensen, J. Bacterial typing methods suitable for epidemiological analysis. applications in investigations of salmonellosis among livestock. Veter Q. 1993, 15, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; McDermott, P.; White, D.; Qaiyumi, S.; Friedman, S.; Abbott, J.; Glenn, A.; Ayers, S.; Post, K.; Fales, W. Characterization of multidrug resistant Salmonella recovered from diseased animals. Veter Microbiol. 2007, 123, 122–132. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weil, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007. [Google Scholar]
- Tennant, S.M.; Diallo, S.; Levy, H.; Livio, S.; Sow, S.O.; Tapia, M.; Fields, P.I.; Mikoleit, M.; Tamboura, B.; Kotloff, K.L.; et al. Identification by PCR of Non-typhoidal Salmonella enterica Serovars Associated with Invasive Infections among Febrile Patients in Mali. PLoS Negl. Trop. Dis. 2010, 4, e621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanegas, R.A.; Joys, T.M. Molecular analyses of the phase-2 antigen complex 1,2,. of Salmonella spp. J. Bacteriol. 1995, 177, 3863–3864. [Google Scholar] [CrossRef] [Green Version]
- Echeita, M.A.; Herrera, S.; Usera, M.A. Atypical, fljB-Negative Salmonella enterica subsp. enterica Strain of Serovar 4,5,12:i: Appears To Be a Monophasic Variant of Serovar Typhimurium. J. Clin. Microbiol. 2001, 39, 2981–2983. [Google Scholar] [CrossRef] [Green Version]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing, 2019a Antimicrobial Susceptibility Testing EUCAST Disk Diffusion Method, 1–21. Available online: www.eucast.org (accessed on 10 December 2020).
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing, 2019b Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 9.0. Available online: www.eucast.org (accessed on 10 December 2020).
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing, Recommendations for MIC Determination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Available online: www.eucast.org (accessed on 10 December 2020).
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Carrico, J.A.; Pinto, F.R.; Simas, C.; Nunes, S.; Sousa, N.G.; Frazão, N.; De Lencastre, H.; Almeida, J.S. Assessment of Band-Based Similarity Coefficients for Automatic Type and Subtype Classification of Microbial Isolates Analyzed by Pulsed-Field Gel Electrophoresis. J. Clin. Microbiol. 2005, 43, 5483–5490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Kaiser, B.L.D.; Dinsmore, B.A.; Fitzgerald, C.; Fields, P.I.; Deng, X. Salmonella Serotype Determination Utilizing High-Throughput Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In SilicoDetection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, L.; Mo, K.; Mark, S. A Curated, Comprehensive Database of Plamid Sequences. Microbiol. Resour. Announc. 2019, 8, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- van Bloois, L.G.; Wagenaar, J.A.; Zomer, A.L. RFPlasmid: Predicting plasmid sequences from short read assembly data using machine learning. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; Van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemother. 2010, 65, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Rovira, A.; Davies, P.; Ahlstrom, C.; Muellner, P.; Rendahl, A.; Olsen, K.; Bender, J.B.; Wells, S.; Perez, A.; et al. Serotypes and Antimicrobial Resistance in Salmonella enterica Recovered from Clinical Samples from Cattle and Swine in Minnesota. PLoS ONE 2016, 11, e0168016. [Google Scholar] [CrossRef] [PubMed]
- Shippy, D.C.; Bearson, B.L.; Holman, D.B.; Brunelle, B.W.; Allen, H.K.; Bearson, S.M. Porcine Response to a Multidrug-Resistant Salmonella enterica serovar I 4,[5],12:i:- Outbreak Isolate. Foodborne Pathog. Dis. 2018, 15, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.; Mourão, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastrorilli, E.; Pietrucci, D.; Barco, L.; Ammendola, S.; Petrin, S.; Longo, A.; Mantovani, C.; Battistoni, A.; Ricci, A.; Desideri, A.; et al. A Comparative Genomic Analysis Provides Novel Insights Into the Ecological Success of the Monophasic Salmonella Serovar 4,[5],12:i:-. Front. Microbiol. 2018, 9, 715. [Google Scholar] [CrossRef] [PubMed]
- Brunelle, B.W.; Bearson, S.M.; Bearson, B.L. Tetracycline accelerates the temporally-regulated invasion response in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013, 13, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edrington, T.S.; Harvey, R.B.; Farrington, L.A.; Nisbet, D.J. Evaluation of Subtherapeutic Use of the Antibiotics Apramycin and Carbadox on the Prevalence of Antimicrobial-Resistant Salmonella Infection in Swine. J. Food Prot. 2001, 64, 2067–2070. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.S.; Robinson, T.P.; Teillant, A.A.; Laxminarayan, R.R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Wang, J.; Yassin, A.K.; Butaye, P.; Kelly, P.; Gong, J.; Guo, W.; Li, J.; Li, M.; et al. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Brasil, Instrução Normativa N° 45, de 22 de Novembro de 2016, Ministério da Agricultura, Pecuária e Abastecimento (MAPA). Available online: https://www.in.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/22078290/do1-2016-11-30-instrucao-normativa-n-45-de-22-de-novembro-de-2016-22078259 (accessed on 21 April 2021).
- Li, P.; Zhu, T.; Zhou, D.; Lu, W.; Liu, H.; Sun, Z.; Ying, J.; Lu, J.; Lin, X.; Li, K.; et al. Analysis of Resistance to Florfenicol and the Related Mechanism of Dissemination in Different Animal-Derived Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 369. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, R.; Bhattacharyya, T.; Bhando, T.; Pathania, R. Fosfomycin resistance inAcinetobacter baumanniiis mediated by efflux through a major facilitator superfamily (MFS) transporter—AbaF. J. Antimicrob. Chemother. 2017, 72, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Rehman, M.A.; Yin, X.; Persaud-Lachhman, M.G.; Diarra, M.S. First Detection of a Fosfomycin Resistance Gene, fosA7, in Salmonella enterica Serovar Heidelberg Isolated from Broiler Chickens. Antimicrob. Agents Chemother. 2017, 61, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Fang, T.; Zhou, X.; Zhang, D.-F.; Shi, X.; Shi, C. IncHI2 Plasmids Are Predominant in Antibiotic-Resistant Salmonella Isolates. Front. Microbiol. 2016, 7, 1566. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.A.; Cunha, M.P.V.; Bertani, A.M.D.J.; De Almeida, E.A.; Gonçalves, C.R.; Sacchi, C.T.; De Paiva, J.B.; Camargo, C.H.; Tiba-Casas, M.R. Detection of multidrug- and colistin-resistant Salmonella Choleraesuis causing bloodstream infection. J. Antimicrob. Chemother. 2020, 75, 2009–2010. [Google Scholar] [CrossRef]
- Mathys, D.A.; Mollenkopf, D.F.; Feicht, S.M.; Adams, R.J.; Albers, A.L.; Stuever, D.M.; Grooters, S.V.; Ballash, G.A.; Daniels, J.B.; Wittum, T.E. Carbapenemase-producing Enterobacteriaceae and Aeromonas spp. present in wastewater treatment plant effluent and nearby surface waters in the US. PLoS ONE 2019, 14, e0218650. [Google Scholar] [CrossRef] [PubMed]
- Prussing, C.; Snavely, E.A.; Singh, N.; Lapierre, P.; Lasek-Nesselquist, E.; Mitchell, K.; Haas, W.; Owsiak, R.; Nazarian, E.; Musser, K.A. Nanopore MinION Sequencing Reveals Possible Transfer of blaKPC–2 Plasmid Across Bacterial Species in Two Healthcare Facilities. Front. Microbiol. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Keefer, A.B.; Xiaoli, L.; M’Ikanatha, N.M.; Yao, K.; Hoffmann, M.; Dudley, E.G. Retrospective whole-genome sequencing analysis distinguished PFGE and drug-resistance-matched retail meat and clinical Salmonella isolates. Microbiology 2019, 165, 270–286. [Google Scholar] [CrossRef]
- Schürch, A.; Arredondo-Alonso, S.; Willems, R.; Goering, R. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin. Microbiol. Infect 2018, 24, 350–354. [Google Scholar] [CrossRef] [Green Version]
- Kich, J.D.; Coldebella, A.; Morés, N.; Nogueira, M.G.; Cardoso, M.; Fratamico, P.M.; Call, J.E.; Fedorka-Cray, P.; Luchansky, J.B. Prevalence, distribution, and molecular characterization of Salmonella recovered from swine finishing herds and a slaughter facility in Santa Catarina, Brazil. Int. J. Food Microbiol. 2011, 151, 307–313. [Google Scholar] [CrossRef] [Green Version]
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic Resistance in Bacteria Associated with Food Animals: A United States Perspective of Livestock Production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef] [Green Version]

| Farming Phase | N | Clinical-Pathological Classification | N | Serovars | N |
|---|---|---|---|---|---|
| Suckling | 8 | Enteric | 4 | Newport | 1 |
| Rissen | 1 | ||||
| * Monophasic Typhimurium | 2 | ||||
| Hepato-biliary invasive | 1 | Choleraesuis | 1 | ||
| Nodal | 1 | * Monophasic Typhimurium | 1 | ||
| Septicemic | 2 | Choleraesuis | 2 | ||
| Nursery | 53 | Enteric | 20 | Typhimurium | 4 |
| * Monophasic Typhimurium | 16 | ||||
| Hepato-biliary invasive | 4 | Choleraesuis | 2 | ||
| * Monophasic Typhimurium | 2 | ||||
| Nodal | 3 | Choleraesuis | 2 | ||
| Panama | 1 | ||||
| Septicemic | 26 | Anatum | 1 | ||
| Choleraesuis | 19 | ||||
| Typhimurium | 1 | ||||
| * Monophasic Typhimurium | 5 | ||||
| Growing/Finishing | 53 | Enteric | 22 | Infantis | 1 |
| Rissen | 1 | ||||
| Typhimurium | 5 | ||||
| * Monophasic Typhimurium | 15 | ||||
| Hepato-biliary invasive | 11 | Choleraesuis | 2 | ||
| London | 1 | ||||
| Oslo | 1 | ||||
| Typhimurium | 1 | ||||
| * Monophasic Typhimurium | 6 | ||||
| Nodal | 8 | Choleraesuis | 2 | ||
| Derby | 1 | ||||
| London | 1 | ||||
| Typhimurium | 2 | ||||
| * Monophasic Typhimurium | 2 | ||||
| Septicemic | 12 | Choleraesuis | 7 | ||
| Grupo D | 1 | ||||
| * Monophasic Typhimurium | 4 | ||||
| No information | 16 | Enteric | 4 | Grupo E4 (O:19:-) | 1 |
| Panama | 1 | ||||
| Typhimurium | 1 | ||||
| * Monophasic Typhimurium | 1 | ||||
| Hepato-biliary invasive | 1 | Rissen | 1 | ||
| Nodal | 1 | Choleraesuis | 1 | ||
| Septicemic | 8 | Choleraesuis | 8 | ||
| NI | 2 | Bovismorbificans | 1 | ||
| * Monophasic Typhimurium | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meneguzzi, M.; Pissetti, C.; Rebelatto, R.; Trachsel, J.; Kuchiishi, S.S.; Reis, A.T.; Guedes, R.M.C.; Leão, J.A.; Reichen, C.; Kich, J.D. Re-Emergence of Salmonellosis in Hog Farms: Outbreak and Bacteriological Characterization. Microorganisms 2021, 9, 947. https://doi.org/10.3390/microorganisms9050947
Meneguzzi M, Pissetti C, Rebelatto R, Trachsel J, Kuchiishi SS, Reis AT, Guedes RMC, Leão JA, Reichen C, Kich JD. Re-Emergence of Salmonellosis in Hog Farms: Outbreak and Bacteriological Characterization. Microorganisms. 2021; 9(5):947. https://doi.org/10.3390/microorganisms9050947
Chicago/Turabian StyleMeneguzzi, Mariana, Caroline Pissetti, Raquel Rebelatto, Julian Trachsel, Suzana Satomi Kuchiishi, Adrienny Trindade Reis, Roberto Maurício Carvalho Guedes, Joice Aparecida Leão, Caroline Reichen, and Jalusa Deon Kich. 2021. "Re-Emergence of Salmonellosis in Hog Farms: Outbreak and Bacteriological Characterization" Microorganisms 9, no. 5: 947. https://doi.org/10.3390/microorganisms9050947
APA StyleMeneguzzi, M., Pissetti, C., Rebelatto, R., Trachsel, J., Kuchiishi, S. S., Reis, A. T., Guedes, R. M. C., Leão, J. A., Reichen, C., & Kich, J. D. (2021). Re-Emergence of Salmonellosis in Hog Farms: Outbreak and Bacteriological Characterization. Microorganisms, 9(5), 947. https://doi.org/10.3390/microorganisms9050947






