Comparative Characteristics and Zoonotic Potential of Avian Pathogenic Escherichia coli (APEC) Isolates from Chicken and Duck in South Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. O-Serogrouping
2.3. DNA Extraction and Phylogenetic Group Determination
2.4. Detection of Virulence-Associated Genes (VAGs)
2.5. Antibiotic Susceptibility Test
2.6. Detection of Antimicrobial Resistance Genes
2.7. Multi-Locus Sequence Typing (MLST) Analysis
2.8. Statistical Analysis
3. Results
3.1. O-Serogroup Distribution of APEC Isolates
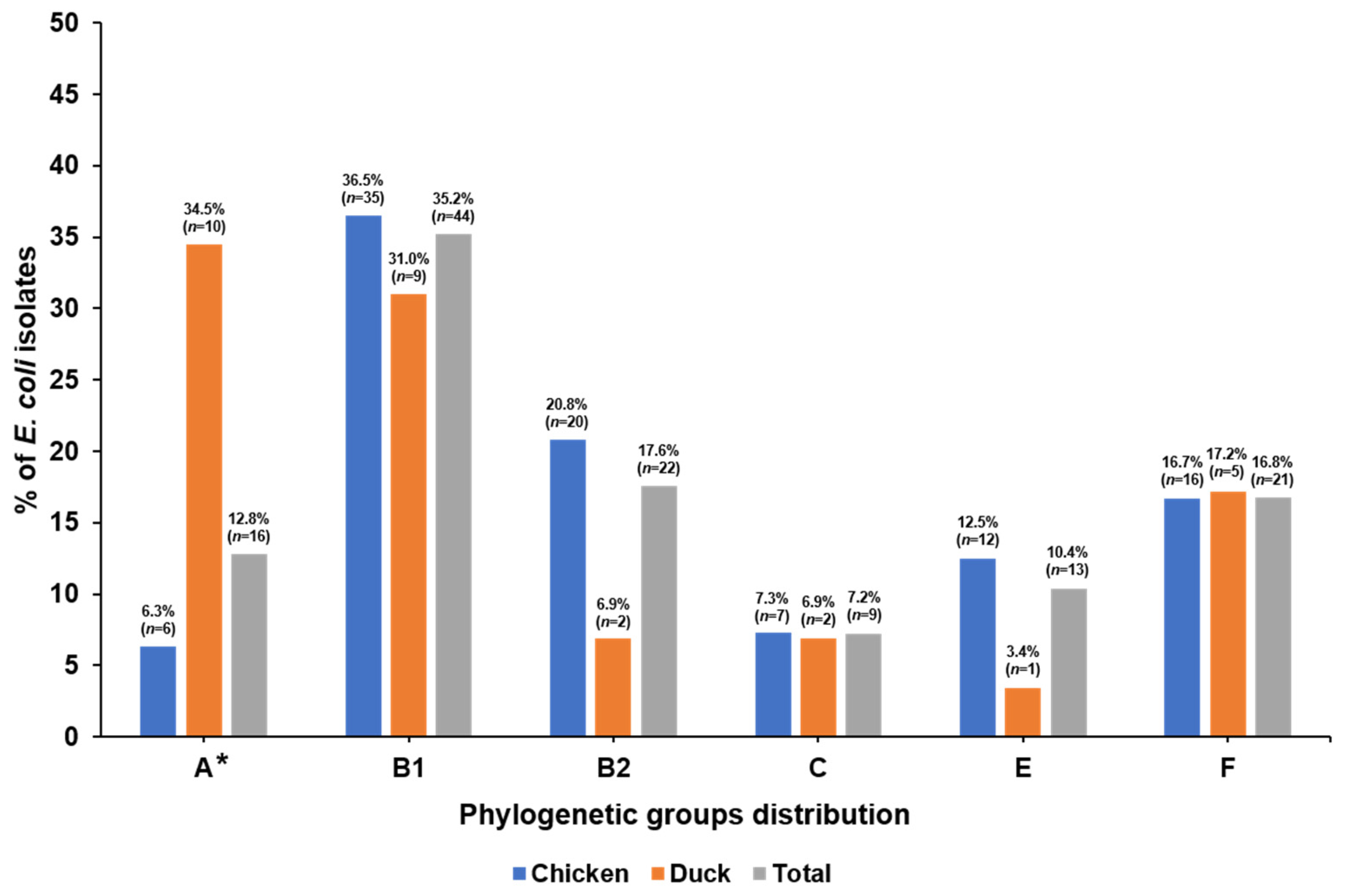
3.2. Phylogenetic Groups and VAGs
3.3. Antimicrobial Resistance Profiles of APEC Isolates
3.4. Prevalence of Antimicrobial Resistance Genes
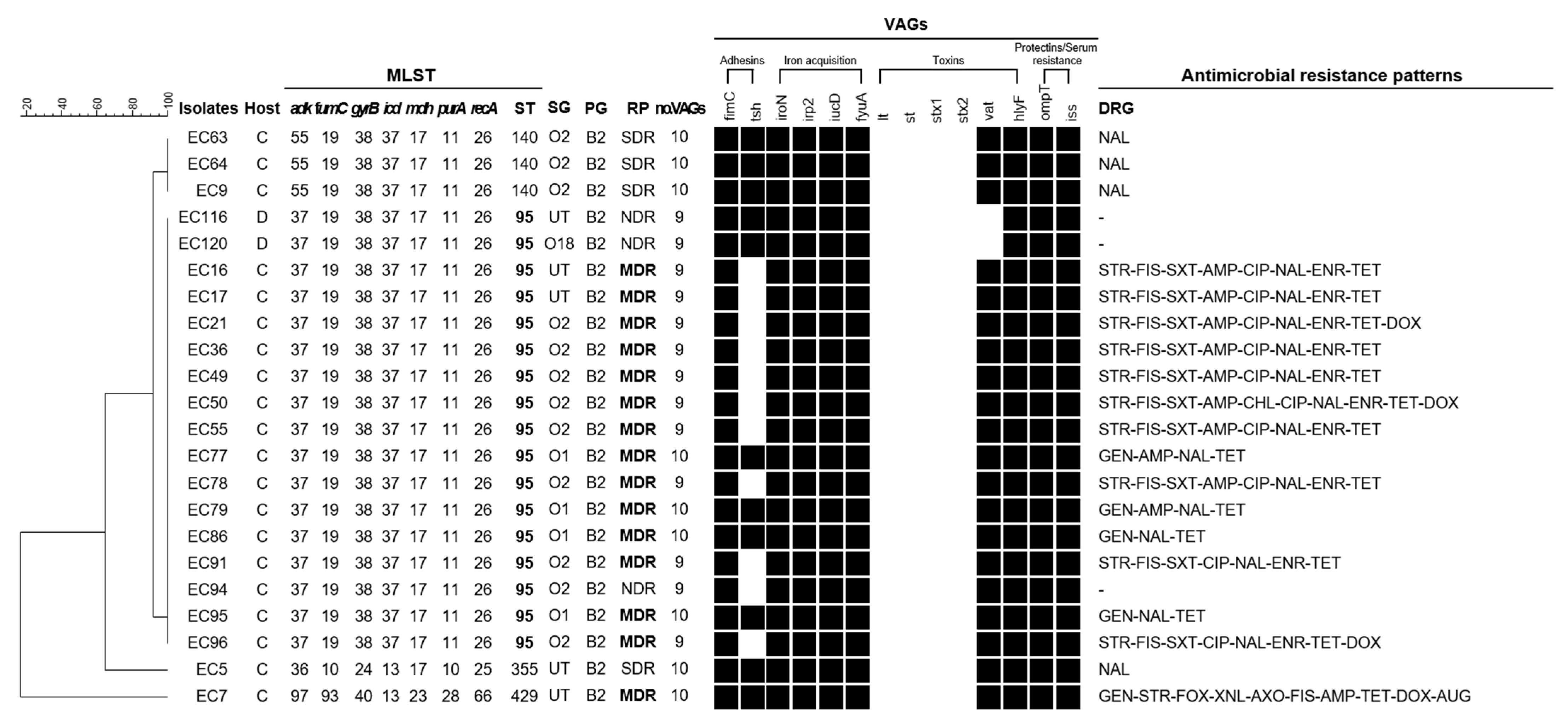
3.5. Comprehensive Characteristics as Measure of A Zoonotic Potential in Isolates Belonging to Phylogenetic Group B2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venkitanarayanan, K.; Thakur, S.; Ricke, S.C. Food Safety in Poultry Meat Production; Springer: Cham, Switzerland, 2019; pp. 261–279. [Google Scholar]
- Russo, T.A.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.-D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Nolan, L.K.; Vaillancourt, J.P.; Barbieri, N.L.; Logue, C.M. Colibacillosis. Dis. Poult. 2020, 770–830. [Google Scholar] [CrossRef]
- Dziva, F.; Stevens, M.P. Colibacillosis in poultry: Unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 2008, 37, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Li, G.; Wilking, H.; Kieβling, S.; Alt, K.; Antáo, E.-M.; Laturnus, C.; Diehl, I.; Glodde, S.; Homeier, T. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int. J. Med. Microbiol. 2007, 297, 163–176. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Logue, C.M.; Wannemuehler, Y.; Nicholson, B.A.; Doetkott, C.; Barbieri, N.L.; Nolan, L.K. Comparative Analysis of Phylogenetic Assignment of Human and Avian ExPEC and Fecal Commensal Escherichia coli Using the (Previous and Revised) Clermont Phylogenetic Typing Methods and its Impact on Avian Pathogenic Escherichia coli (APEC) Classification. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Coura, F.M.; Diniz, S.A.; Silva, M.X.; Arcebismo, T.L.; Minharro, S.; Feitosa, A.C.; Lage, A.P.; Knöbl, T.; Mussi, J.M.S.; Heinemann, M.B. Phylogenetic group of Escherichia coli isolates from broilers in Brazilian poultry slaughterhouse. Sci. World J. 2017, 2017, 5898701. [Google Scholar] [CrossRef]
- Papouskova, A.; Masarikova, M.; Valcek, A.; Senk, D.; Cejkova, D.; Jahodarova, E.; Cizek, A. Genomic analysis of Escherichia coli strains isolated from diseased chicken in the Czech Republic. BMC Vet. Res. 2020, 16, 1–10. [Google Scholar] [CrossRef]
- Yun, K.W.; Do Soo Kim, W.K.; Lim, I.S. Molecular typing of uropathogenic Escherichia coli isolated from Korean children with urinary tract infection. Korean J. Pediatrics 2015, 58, 20. [Google Scholar] [CrossRef]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Nolan, L.K. Characterizing the APEC pathotype. Vet. Res. 2005, 36, 241–256. [Google Scholar] [CrossRef]
- Ewers, C.; Janßen, T.; Kießling, S.; Philipp, H.-C.; Wieler, L.H. Rapid detection of virulence-associated genes in avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian Dis. 2005, 49, 269–273. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Fakhr, M.K.; Nolan, L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 2005, 151, 2097–2110. [Google Scholar] [CrossRef]
- Jeong, Y.-W.; Kim, T.-E.; Kim, J.-H.; Kwon, H.-J. Pathotyping avian pathogenic Escherichia coli strains in Korea. J. Vet. Sci. 2012, 13, 145. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 1–16. [Google Scholar] [CrossRef]
- Azam, M.; Mohsin, M.; Saleemi, M.K. Virulence-associated genes and antimicrobial resistance among avian pathogenic Escherichia coli from colibacillosis affected broilers in Pakistan. Trop. Anim. Health Prod. 2019, 51, 1259–1265. [Google Scholar] [CrossRef]
- Mellata, M. Human and avian extraintestinal pathogenic Escherichia coli: Infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 2011, 17, 1216. [Google Scholar] [CrossRef]
- Giufrè, M.; Graziani, C.; Accogli, M.; Luzzi, I.; Busani, L.; Cerquetti, M. Escherichia coli of human and avian origin: Detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J. Antimicrob. Chemother. 2012, 67, 860–867. [Google Scholar] [CrossRef]
- Fricke, W.F.; McDermott, P.F.; Mammel, M.K.; Zhao, S.; Johnson, T.J.; Rasko, D.A.; Fedorka-Cray, P.J.; Pedroso, A.; Whichard, J.M.; LeClerc, J.E. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 2009, 75, 5963–5971. [Google Scholar] [CrossRef]
- Thomrongsuwannakij, T.; Blackall, P.J.; Djordjevic, S.P.; Cummins, M.L.; Chansiripornchai, N. A comparison of virulence genes, antimicrobial resistance profiles and genetic diversity of avian pathogenic Escherichia coli (APEC) isolates from broilers and broiler breeders in Thailand and Australia. Avian Pathol. 2020, 49, 457–466. [Google Scholar] [CrossRef]
- Xu, X.; Sun, Q.; Zhao, L. Virulence factors and antibiotic resistance of avian pathogenic Escherichia coli in eastern China. J. Vet. Res. 2019, 63, 317–320. [Google Scholar] [CrossRef]
- Solà-Ginés, M.; Cameron-Veas, K.; Badiola, I.; Dolz, R.; Majó, N.; Dahbi, G.; Viso, S.; Mora, A.; Blanco, J.; Piedra-Carrasco, N. Diversity of multi-drug resistant avian pathogenic Escherichia coli (APEC) causing outbreaks of colibacillosis in broilers during 2012 in Spain. PLoS ONE 2015, 10, e0143191. [Google Scholar]
- Saha, O.; Hoque, M.N.; Islam, O.K.; Rahaman, M.; Sultana, M.; Hossain, M.A. Multidrug-resistant avian pathogenic Escherichia coli strains and association of their virulence genes in Bangladesh. Microorganisms 2020, 8, 1135. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, C.; Yu, X.; Xia, M.; Yue, H. Distribution of serotypes and virulence-associated genes in pathogenic Escherichia coli isolated from ducks. Avian Pathol. 2010, 39, 297–302. [Google Scholar] [CrossRef]
- Watts, J.L.; Salmon, S.A.; Yancey Jr, R.J.; Nersessian, B.; Kounev, Z.V. Minimum inhibitory concentrations of bacteria isolated from septicemia and airsacculitis in ducks. J. Vet. Diagn. Investig. 1993, 5, 625–628. [Google Scholar] [CrossRef]
- Sung, M.S.; Kim, J.H.; Ha, J.S.; Cho, J.K.; Seol, S.Y.; Kim, K.S. Biochemical properties and serotypes of pathogenic Escherichia coli isolated from poultry in Korea. Korean J. Vet. Res. 2008, 48, 145–151. [Google Scholar]
- Sung, M.S.; Kim, J.H.; Cho, J.K.; Seol, S.Y.; Kim, K.S. Antimicrobial resistance transfer of R plasmid of pathogenic Escherichia coli isolated from poultry in Korea. Korean J. Vet. Res. 2008, 48, 275–285. [Google Scholar]
- Kim, Y.B.; Yoon, M.Y.; Ha, J.S.; Seo, K.W.; Noh, E.B.; Son, S.H.; Lee, Y.J. Molecular characterization of avian pathogenic Escherichia coli from broiler chickens with colibacillosis. Poult. Sci. 2020, 99, 1088–1095. [Google Scholar] [CrossRef]
- El-Shaer, S.; Abdel-Rhman, S.H.; Barwa, R.; Hassan, R. Virulence Characteristics, Serotyping and Phylogenetic Typing of Clinical and Environmental Escherichia coli Isolates. Jundishapur J. Microbiol. 2018, 11, e82835. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI Supplement VET01; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- CDC National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report); US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2018.
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters. European Committee on Antimicrobial Susceptibility Testing Växjö, Sweden. 2018. Available online: https://eucast.org/eucast_news/news_singleview/?tx_ttnews%5Btt_news%5D=248&cHash=91e3ef09a79b333746462d8854ee016d (accessed on 14 February 2021).
- Liu, X.; Thungrat, K.; Boothe, D.M. Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PLoS ONE 2015, 10, e0143335. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.t.; Carmeli, Y.; Falagas, M.t.; Giske, C.t.; Harbarth, S.; Hindler, J.T.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Nguyen, M.C.P.; Woerther, P.-L.; Bouvet, M.; Andremont, A.; Leclercq, R.; Canu, A. Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 2009, 15, 1648. [Google Scholar] [CrossRef] [PubMed]
- Ciesielczuk, H.; Hornsey, M.; Choi, V.; Woodford, N.; Wareham, D. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med Microbiol. 2013, 62, 1823–1827. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Hasman, H.; Xia, S.; Aarestrup, F.M. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 2009, 53, 603–608. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Wareham, D.; Umoren, I.; Khanna, P.; Gordon, N. Allele-specific polymerase chain reaction (PCR) for rapid detection of the aac (6′)-Ib-cr quinolone resistance gene. Int. J. Antimicrob. Agents 2010, 36, 476–477. [Google Scholar] [CrossRef]
- Yamane, K.; Wachino, J.-I.; Suzuki, S.; Arakawa, Y. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 2008, 52, 1564–1566. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17–00672. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi, B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- Monstein, H.J.; Östholm-Balkhed, Å.; Nilsson, M.; Nilsson, M.; Dornbusch, K.; Nilsson, L. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; Hossain, A.; Hanson, N.D. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 2004, 42, 5715–5721. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, Y.; Briñas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef]
- Guerra, B.; Junker, E.; Miko, A.; Helmuth, R.; Mendoza, M. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 2004, 10, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Dijkshoorn, L.; Collard, J.-M.; Olsen, J.; Dalsgaard, A. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 2000, 49, 929–936. [Google Scholar] [CrossRef]
- Guillaume, G.; Verbrugge, D.; Chasseur-Libotte, M.-L.; Moens, W.; Collard, J.-M. PCR typing of tetracycline resistance determinants (Tet A–E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 2000, 32, 77–85. [Google Scholar]
- Zhao, J.; Aoki, T. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol. Immunol. 1992, 36, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Sunde, M.; Norström, M. The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J. Antimicrob. Chemother. 2005, 56, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Lariviere, S.; Harel, J. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149: K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Maluta, R.P.; Logue, C.M.; Casas, M.R.T.; Meng, T.; Guastalli, E.A.L.; Rojas, T.C.G.; Montelli, A.C.; Sadatsune, T.; de Carvalho Ramos, M.; Nolan, L.K. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE 2014, 9, e105016. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Johnson, S.J.; Stell, A.L.; Doetkott, C.; Johnson, J.R.; Kim, K.S.; Spanjaard, L.; Nolan, L.K. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 2008, 74, 7043–7050. [Google Scholar] [CrossRef]
- Jørgensen, S.L.; Stegger, M.; Kudirkiene, E.; Lilje, B.; Poulsen, L.L.; Ronco, T.; Dos Santos, T.P.; Kiil, K.; Bisgaard, M.; Pedersen, K. Diversity and population overlap between avian and human Escherichia coli belonging to sequence type 95. MSphere 2019, 4. [Google Scholar] [CrossRef]
- Jakobsen, L.; Spangholm, D.J.; Pedersen, K.; Jensen, L.B.; Emborg, H.-D.; Agersø, Y.; Aarestrup, F.M.; Hammerum, A.M.; Frimodt-Møller, N. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int. J. Food Microbiol. 2010, 142, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, L.; Garneau, P.; Bruant, G.; Harel, J.; Olsen, S.; Porsbo, L.J.; Hammerum, A.; Frimodt-Møller, N. Is Escherichia coli urinary tract infection a zoonosis? Proof of direct link with production animals and meat. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Yassin, A.K.; Gong, J.; Kelly, P.; Lu, G.; Guardabassi, L.; Wei, L.; Han, X.; Qiu, H.; Price, S.; Cheng, D. Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock, China. PLoS ONE 2017, 12, e0185326. [Google Scholar] [CrossRef] [PubMed]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; de Brito, K.C.T.; de Brito, B.G.; Nakazato, G.; Kobayashi, R.K.T. Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 2019, 9, 3254. [Google Scholar] [CrossRef]
- Lambrecht, E.; Van Meervenne, E.; Boon, N.; Van de Wiele, T.; Wattiau, P.; Herman, L.; Heyndrickx, M.; Van Coillie, E. Characterization of cefotaxime-and ciprofloxacin-resistant commensal Escherichia coli originating from Belgian farm animals indicates high antibiotic resistance transfer rates. Microb. Drug Resist. 2018, 24, 707–717. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Shimabukuro, H.; Shimamoto, T. Isolation and molecular characterization of multidrug-resistant strains of Escherichia coli and Salmonella from retail chicken meat in Japan. J. Food Sci. 2009, 74, M405–M410. [Google Scholar] [CrossRef]
- Cummins, M.L.; Reid, C.J.; Chowdhury, P.R.; Bushell, R.N.; Esbert, N.; Tivendale, K.A.; Noormohammadi, A.H.; Islam, S.; Marenda, M.S.; Browning, G.F. Whole genome sequence analysis of Australian avian pathogenic Escherichia coli that carry the class 1 integrase gene. Microb. Genom. 2019, 5, e000250. [Google Scholar] [CrossRef] [PubMed]
- Ammar, A.; El-Hamid, M.; Eid, S.E.; El Oksh, A.S. Insight into antimicrobial resistance and virulence genes of emergent multidrug resistant avian pathogenic Escherichia coli in Egypt: How closely related are they. Rev. De Med. Vet. 2015, 166, 304–314. [Google Scholar]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Wang, J.; Klumpp, J.; Nüesch-Inderbinen, M.; Fanning, S.; Stephan, R. Vertical transmission of highly similar blaCTX-M-1-harbouring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front. Microbiol. 2014, 5, 519. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 289–316. [Google Scholar]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-K.; Kang, H.Y.; Lee, K.; Moon, D.-C.; Lee, H.-S.; Jung, S.-C. First detection of the mcr-1 gene in Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrob. Agents Chemother. 2016, 60, 6991–6993. [Google Scholar] [CrossRef][Green Version]
- Manges, A.R.; Johnson, J.R. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 2012, 55, 74. [Google Scholar] [CrossRef]
| Chicken (n = 96) | Duck (n = 29) | Total (n = 125) | |||
|---|---|---|---|---|---|
| No. of Isolates (%) | O-Serogroups | No. of Isolates (%) | O-Serogroups | No. of Isolates (%) | O-Serogroups |
| 12 (12.5) | O2 | 3 (10.3) | O78 | 13 (10.4) | O2 |
| 6 (6.3) | O1, O45 | 2 (6.9) | O15, O25, O150 | 7 (5.6) | O1, O78 |
| 5 (5.2) | O88 | 1 (3.4) | O1, O2, O9, O18, O22, O24, O34, O39, O81, O84, O89, O129, O149, O156, O160, O81, O84, O89 | 6 (4.8) | O45 |
| 4 (4.2) | O78 | 5 (17.2) | Non-typable | 5 (4.0) | O88 |
| 3 (3.1) | O102 | - | - | 4 (3.2) | O25 |
| 2 (2.1) | O8, O20, O24, O25, O177, O182 | - | - | 3 (2.4) | O24, O102 |
| 1 (1.0) | O5, O7, O21, O22, O29, O51, O55, O68, O115, O140, O143, O154, O166, O184 | - | - | 2 (1.6) | O8, O15, O20, O22, O150, O177, O182 |
| 34 (35.4) | Non-typable | - | - | 1 (0.8) | O5, O7, O9, O18, O21, O29, O34, O39, O51, O55, O68, O81, O84, O89, O115, O129, O140, O143, O149, O154, O156, O160, O166, O184 |
| - | - | - | - | 39 (31.2) | Non-typable |
| Virulence-Associated Gene | No. (%) of Positive Isolates | ||
|---|---|---|---|
| Chicken (n = 96) | Duck (n = 29) | Total (n = 125) | |
| Adhesin | |||
| fimC | 93 (96.9) | 25 (86.2) | 118 (94.4) |
| tsh | 46 (47.9) | 15 (51.7) | 61 (48.8) |
| Iron acquisition | |||
| iucD * | 81 (84.4) | 15 (51.7) | 96 (76.8) |
| iroN * | 80 (83.3) | 13 (44.8) | 93 (74.4) |
| irp2 | 51 (53.1) | 17 (58.6) | 68 (54.4) |
| fyuA | 46 (47.9) | 13 (44.8) | 59 (47.2) |
| Toxins | |||
| hlyF * | 84 (87.5) | 16 (55.2) | 100 (80.0) |
| vat * | 31 (32.3) | 1 (3.4) | 32 (25.6) |
| lt | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| st | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| stx1A | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| stx2A | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Protectins/serum resistance | |||
| iss * | 78 (81.3) | 14 (48.3) | 92 (73.6) |
| ompT | 67 (69.8) | 18 (62.1) | 85 (68.0) |
| Phylogenetic Group | MVS a (No. of Isolates) | ||
|---|---|---|---|
| Chicken Isolates (n = 96) | Duck Isolates (n = 29) | All Isolates (n = 125) | |
| Total | 6.9 (96) | 5.1 (29) | 6.4 (125) |
| A | 2.7 (6) | 3.5 (10) | 3.2 (16) |
| B1 | 5.8 (35) | 3.7 (9) | 5.3 (44) |
| B2 | 9.5 (20) | 9.0 (2) | 9.4 (22) |
| C | 8.3 (7) | 9.0 (2) | 8.4 (9) |
| E | 4.6 (12) | 3.0 (1) | 4.5 (13) |
| F | 8.6 (16) | 8.0 (5) | 8.4 (21) |
| Antibiotics | Break Point (μg/mL) | Chicken (n = 96) | Duck (n = 29) | Total (n = 125) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC Range (μg/mL) | MIC50 1 (μg/mL) | MIC90 2 (μg/mL) | R 3 (%) | MIC Range (μg/mL) | MIC50 (μg/mL) | MIC90 (μg/mL) | R (%) | MIC Range (μg/mL) | MIC50 (μg/mL) | MIC90 (μg/mL) | R (%) | ||
| Gentamicin | ≥16 a | 0.5–16 | 1 | >16 | 26.0 * | 0.5–2 | 1 | 1 | 0.0 | 0.5–16 | 1 | >16 | 20.0 |
| Streptomycin | ≥32 b | 4–64 | >64 | >64 | 62.5 | 8–64 | 16 | >64 | 44.8 | 4–64 | >64 | >64 | 58.4 |
| Ampicillin | >32 a | ≤1–32 | >32 | >32 | 75.0 * | 2–32 | 4 | >32 | 44.8 | ≤1–32 | >32 | >32 | 68.0 |
| Amoxicillin/ Clavulanic Acid | ≥32/16 a | 2–32 | 8 | 16 | 5.2 | 2–16 | 8 | 16 | 0.0 | 2–32 | 8 | 16 | 4.0 |
| Cefoxitin | ≥32 a | 2–32 | 4 | 8 | 5.2 | 2–16 | 4 | 8 | 0.0 | 2–32 | 4 | 8 | 4.0 |
| Ceftiofur | ≥8 b | 0.25–8 | 0.5 | >8 | 18.8 * | 0.25–1 | 0.5 | 1 | 0.0 | 0.25–8 | 0.5 | >8 | 14.4 |
| Ceftriaxone | ≥4 a | ≤0.25–64 | ≤0.25 | 64 | 19.8 * | ≤0.25 | ≤0.25 | ≤0.25 | 0.0 | ≤0.25–64 | ≤0.25 | 16 | 15.2 |
| Sulfisoxazole | >512 a | ≤16–256 | >256 | >256 | 71.9 | ≤16–256 | >256 | >256 | 55.2 | ≤16–256 | >256 | >256 | 68.0 |
| Trimethoprim/ Sulfamethoxazole | ≥4/76 a | ≤0.12–4 | 0.5 | >4 | 46.9 | ≤0.12–4 | >4 | >4 | 51.7 | ≤0.12–4 | 0.5 | >4 | 48.0 |
| Azithromycin | ≥32 a | 2–16 | 4 | 16 | 3.1 | 2–16 | 4 | >16 | 6.9 | 2–16 | 4 | 16 | 4.0 |
| Chloramphenicol | ≥32 a | ≤2–32 | 8 | >32 | 38.5 | 4–32 | 32 | >32 | 51.7 | ≤2–32 | 8 | >32 | 41.6 |
| Colistin | ≥16 c | 0.5–16 | 0.5 | 0.5 | 1.0 | 0.5–1 | 0.5 | 0.5 | 0.0 | 0.5–16 | 0.5 | 0.5 | 0.8 |
| Ciprofloxacin | ≥1 a | ≤0.015–4 | 4 | >4 | 64.6 | ≤0.015–4 | 2 | >4 | 55.2 | ≤0.015–4 | 4 | >4 | 62.4 |
| Enrofloxacin | ≥2 d | ≤0.12–32 | 8 | 32 | 63.5 | ≤0.12–32 | 4 | >32 | 58.6 | ≤0.12–32 | 8 | 32 | 62.4 |
| Nalidixic Acid | ≥32 a | 1–32 | >32 | >32 | 85.6 | 2–32 | >32 | >32 | 82.8 | 1–32 | >32 | >32 | 84.8 |
| Doxycycline | ≥16 a | 0.5–64 | 8 | 64 | 45.8 | 1–64 | 16 | 64 | 51.7 | 0.5–64 | 8 | 64 | 47.2 |
| Tetracycline | ≥16 a | ≤4–32 | >32 | >32 | 70.8 | ≤4–32 | >32 | >32 | 62.1 | ≤4–32 | >32 | >32 | 68.8 |
| Antibiotics | Antimicrobial Class | Chicken | Duck | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Resistance Isolates | Associated Genes | No. of Positive Isolates | (%) | No. of Resistance Isolates | Associated Genes | No. of Positive Isolates | (%) | No. of Resistance Isolates | Associated Genes | No. of Positive Isolates | (%) | ||
| Ampicillin | β-lactam | 72 (72.5%) | blaTEM | 64 | 88.9 | 13 (44.8%) | blaTEM | 11 | 84.6 | 85 (68.0%) | blaTEM | 75 | 88.2 |
| blaCTX-M group Ⅰ | 7 | 9.7 | blaCTX-M group Ⅰ | 0 | 0.0 | blaCTX-M group Ⅰ | 7 | 8.2 | |||||
| blaCTX-M group Ⅳ | 8 | 11.1 | blaCTX-M group Ⅳ | 0 | 0.0 | blaCTX-M group Ⅳ | 8 | 9.4 | |||||
| Azithromycin | Macrolides | 3(3.1%) | mphA | 3 | 100.0 | 2(6.9%) | mphA | 1 | 50.0 | 5 (4.0%) | mphA | 4 | 80.0 |
| Chloramphenicol | Phenicols | 38 (39.2%) | cmlA | 2 | 5.3 | 15 (51.7%) | clmA | 3 | 20.0 | 53 (42.4%) | clmA | 5 | 9.4 |
| Cat | 36 | 94.7 | Cat | 11 | 73.3 | Cat | 47 | 88.7 | |||||
| floR | 35 | 92.1 | floR | 13 | 86.7 | floR | 48 | 90.6 | |||||
| Colistin | Polymyxins | 1(1.0%) | mcr1 | 1 | 100.0 | 0(0.0%) | mcr1 | 0 | 0.0 | 1 (0.8%) | mcr1 | 1 | 100.0 |
| Nalidixic acid | Quinolones | 82 (85.4%) | qnrB | 0 | 0.0 | 24 (82.8%) | qnrB | 1 | 4.2 | 106(84.8%) | qnrB | 1 | 0.9 |
| qnrS | 7 | 8.5 | qnrS | 4 | 16.7 | qnrS | 11 | 10.4 | |||||
| aac6’-1b-cr | 2 | 2.4 | aac6’-1b-cr | 0 | 0.0 | aac6’-1b-cr | 2 | 1.9 | |||||
| Streptomycin | Aminoglycosides | 60 (61.9%) | strA-B | 54 | 90.0 | 13 (44.8%) | strA-B | 10 | 76.9 | 73 (58.4%) | strA-B | 64 | 87.7 |
| aadA | 25 | 41.7 | aadA | 6 | 46.2 | aadA | 31 | 42.5 | |||||
| Sulfisoxazole | Folate pathway inhibitors | 69 (71.9%) | sul1 | 19 | 27.5 | 16 (55.2%) | sul1 | 5 | 31.3 | 85 (68.0%) | sul1 | 24 | 28.2 |
| sul2 | 62 | 89.9 | sul2 | 12 | 75.0 | sul2 | 74 | 87.1 | |||||
| Tetracycline | Tetracyclines | 68 (70.8%) | tetA | 61 | 89.7 | 18 (62.1%) | tetA | 13 | 72.2 | 86 (68.8%) | tetA | 74 | 86.0 |
| tetB | 17 | 25.0 | tetB | 4 | 22.2 | tetB | 21 | 24.4 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, J.; Lee, J.-Y.; Kang, M.-S.; Lee, H.-J.; Kang, S.-I.; Lee, O.-M.; Kwon, Y.-K.; Kim, J.-H. Comparative Characteristics and Zoonotic Potential of Avian Pathogenic Escherichia coli (APEC) Isolates from Chicken and Duck in South Korea. Microorganisms 2021, 9, 946. https://doi.org/10.3390/microorganisms9050946
Jeong J, Lee J-Y, Kang M-S, Lee H-J, Kang S-I, Lee O-M, Kwon Y-K, Kim J-H. Comparative Characteristics and Zoonotic Potential of Avian Pathogenic Escherichia coli (APEC) Isolates from Chicken and Duck in South Korea. Microorganisms. 2021; 9(5):946. https://doi.org/10.3390/microorganisms9050946
Chicago/Turabian StyleJeong, Jiyeon, Ji-Youn Lee, Min-Su Kang, Hye-Jin Lee, Seong-Il Kang, O-Mi Lee, Yong-Kuk Kwon, and Jin-Hyun Kim. 2021. "Comparative Characteristics and Zoonotic Potential of Avian Pathogenic Escherichia coli (APEC) Isolates from Chicken and Duck in South Korea" Microorganisms 9, no. 5: 946. https://doi.org/10.3390/microorganisms9050946
APA StyleJeong, J., Lee, J.-Y., Kang, M.-S., Lee, H.-J., Kang, S.-I., Lee, O.-M., Kwon, Y.-K., & Kim, J.-H. (2021). Comparative Characteristics and Zoonotic Potential of Avian Pathogenic Escherichia coli (APEC) Isolates from Chicken and Duck in South Korea. Microorganisms, 9(5), 946. https://doi.org/10.3390/microorganisms9050946






