Impact of Chronic Exposure to Sublethal Doses of Glyphosate on Honey Bee Immunity, Gut Microbiota and Infection by Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Solutions
2.2. Spore Suspension of Nosema ceranae
2.3. Experimental Design
2.4. Bee Microbiota Analysis
- –
- DNA extraction: After seven days of chronic exposure to glyphosate (or sucrose syrup), ten honey bees per treatment (three/four bees per cage from the three cages) were sampled to analyze their gut bacterial community. Honey bees were externally sterilized using a chlorine solution 1% [36]. Guts were extracted and individually homogenized in 500 μL of PBS using ceramic beads and a FastPrep-24™ during 40 s at 6.0 m/s. Then, samples were centrifuged for 1 min at 5000× g and the supernatants were transferred to sterile tubes. DNA was extracted from the supernatants using the SDS-CTAB method as previously reported [37]. Then, it was quantified using a NanoDrop1000 spectrophotometer (Thermo Scientific™, Waltham, MA, USA) and concentrations were normalized to 10 ng/μL.
- –
- 16S rRNA amplicon sequencing: DNA was sent to Macrogen (Seoul, Korea) for library construction and 16S rRNA amplicon sequencing, as described by Illumina preparation guide (https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf, accessed on 31 July 2020). Briefly, sequencing libraries were built using a two-step PCR strategy. The V3-V4 region of the 16S rRNA gene was amplified using primers Bakt_341F (5′-CCTACGGGNGGCWGCAG-3) and Bakt_805R (5′-GACTACHVGGGTATCTAATCC-3′) with overhang adapters attached and 25 amplification cycles. Then, a second PCR was performed to attach dual indices and sequencing adapters using the Nextera XT Index Kit. In both cases, PCR conditions and programs recommended by Illumina preparation guide were used. Sequencing was carried out using Illumina MiSeq 2 × 300 bp.
2.5. Immune Gene Expression Analyses and DWV Quantification
- –
- RNA extraction and cDNA synthesis: At 0, 7 and 14 days of chronic exposure to glyphosate (or sucrose syrup), we sampled twelve bees per treatment (four bees per cage, per three cages) to analyze the expression of genes associated with the immune response, as well as for the quantification of DWV, and stored them at −80 °C. Individual bees were homogenized in lysis buffer (Invitrogen) and subjected to RNA extraction using the Mini Kit PureLink RNA (Ambion, Carlsbad, CA, USA), according to the manufacturer’s instructions. One microliter of total RNA was treated with DNAse I (Invitrogen, Carlsbad, CA, USA) and used to generate first-strand cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystem, Foster City, CA, USA), also according to the manufacturer’s instructions.
- –
- qPCR: Relative expression of different genes was assessed by qPCR using previously reported primers. Genes included lysozyme [38], glucose dehydrogenase [38], hymenoptaecin [38], vitellogenin [39] and prophenoloxidase [MC-PPO-F: CGCAACTTAGATGAAAATAGACC and MC-PPO-R: TTGAGGCATCCTTACAACCA, Corona M., personal comm.]. We also quantified infection titers of DWV by qPCR [40]. Ribosomal protein S5 (RPS5) was used as a housekeeping gene to normalize the variation in the amounts of cDNA [41]. The reaction mix consisted of 1X Power SYBR® Green Master Mix (Invitrogen), 0.3 μM of each primer, RNAse-free water and 2 μL of 1:10 diluted cDNA in a final volume of 20 μL. PCR reactions were carried out using a BIO-RAD CFX96™ Real-Time system and the cycling program consisted of an initial 95 °C for 15 min, and 40 cycles of three-step PCR at 94 °C for 15 s, 52 °C for 30 s and 72 °C for 30 s. Specificity of the reaction was checked by analysis of the melting curve of the final amplified product (from 65 to 95 °C, with increments of 0.5 °C every 0.05 s). Fluorescence was measured during the elongation step. In each reaction run, we included negative controls (without DNA) and a standard curve which consisted of four dilution points of a mixture of all cDNA samples, to calculate the reaction efficiency.
2.6. Quantification of Nosema Ceranae Spores
2.7. Statistical Analyses
- –
- Bioinformatic analysis: Illumina sequence reads were processed using R Studio Software version 4.0.2 [43] and Divisive Amplicon Denoising Algorithm 2 (DADA2) package (version 1.12.1, [44]). Low quality raw reads were discarded from obtained data and primer sequences were removed using cutadapt [45]. Then, reads were truncated to 280 bp, filtered based on length, representative sequences were obtained and denoised, and chimeric reads were removed. Then, paired reads were merged. Taxonomy was assigned to amplicon sequence variants (ASVs) using the SILVA_132 database by assignTaxonomy. Reads belonging to mitochondria, chloroplast, and eukaryotes were excluded from further analyses (“phyloseq” package version 1.28.0 [46], “subset_taxa” function). To facilitate the visualization of the barplots (relative ASVs abundance), we retained only ASVs that have at least 1% relative abundance in minimum 2 samples (“genefilter” package version 1.66.0 [47], “filterfun_sample” function). Alpha and beta diversity were calculated using the “Vegan” package [48] with the complete ASVs table. To evaluate alpha diversity, we calculated the number of observed ASVs and the Shannon index [49]. Then, we evaluated beta diversity by using Bray–Curtis, UniFrac weighted (by the relative abundance of ASVs), UniFrac unweighted and Jaccard (presence/absence of ASVs) indexes (“vegdist” function) [49]. To test the effect of treatments on community structure, we used permutational multivariate analysis of variance (ADONIS, “adonis” function) on beta diversity data. We then used the function “betadisper” to test for homogeneity of multivariate dispersions [49,50] and compared the distances of individual samples to group centroids in multidimensional space using “permutest”. The “metaMDS” function was used to plot ordinations. Then, differences between the relative abundance of different ASVs were examined using the DESeq2 software [51], as described by Jones et al. [52]. Generalized linear mixed models (GLMMs) were used to evaluate the effect of treatments (as independent variable) on alpha diversity (number of ASVs and Shannon diversity index as dependent variables) considering the cages as random effects.
- –
- Gene expression analyses and quantification of DWV: The Ct values (threshold cycle number) of the RPS5 reference gene were used for normalization. The expression ratio between genes of interest or DWV levels and the RPS5 gene was analyzed as described by Pfaffl [53]. We used GLMMs to evaluate the effect of treatments and time on the expression of different genes or DWV levels (as dependent variables), considering the cage of origin as a random effect.
- –
- Honey bee survival: We analyzed the effect of treatments on survival by building survival curves using the Kaplan–Meier method. Survival curves were statistically compared using the Log-rank test [54]. We then used the Cox model [55] to assess the mortality risk of the bees subjected to the different treatments.
- –
- Food consumption: We measured daily food consumption per bee in the three cages of each treatment during the first 15 days of trials. GLMMs were used to evaluate the effect of treatments (as independent variable) on food consumption considering the time as a random effect.
3. Results
3.1. Impact of Glyphosate on the Gut Microbiota of Honey Bees
3.2. Impact of Glyphosate on Honey Bee Immunity
3.3. Impact of Glyphosate on the Dynamics of Pathogens DWV and N. ceranae
3.4. Impact of Glyphosate on the Survival of Honey Bees
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, A.M.; Vaissière, E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.; Adjlane, N.; Arab, A.; Ballis, A.; Brusbardis, V.; Charrière, J.-D.; Chlebo, R.; Coffey, M.F.; Cornelissen, B.; Amaro da Costa, C.; et al. Honey bee colony winter loss rates for 35 countries participating in the COLOSS survey for winter 2018–2019, and the effects of a new queen on the risk of colony winter loss. J. Apic. Res. 2020, 59, 744–751. [Google Scholar] [CrossRef]
- Kulhanek, K.; Steinhauer, N.; Rennich, K.; Caron, D.M.; Sagili, R.R.; Pettis, J.S.; Ellis, J.D.; Wilson, M.E.; Wilkes, J.T.; Tarpy, D.R.; et al. A national survey of managed honey bee 2015-2016 annual colony losses in the USA. J. Apic. Res. 2017, 56, 328–340. [Google Scholar] [CrossRef]
- Antúnez, K.; Invernizzi, C.; Mendoza, Y.; van Engelsdorp, D.; Zunino, P. Honeybee colony losses in Uruguay during 2013–2014. Apidologie 2017, 48, 364–370. [Google Scholar] [CrossRef]
- Requier, F.; Antúnez, K.; Morales, C.L.; Sánchez, P.; Castilhos, D.; Garrido, M.; Giacobino, A.; Reynaldi, F.J.; Londoño, J.M.; Santos, E.; et al. Trends in beekeeping and honey bee colony losses in Latin America. J. Apic. Res. 2018, 57, 657–662. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botias, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Steinhauer, N.; Kulhanek, K.; Antúnez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.P.; van Engelsdorp, D. Drivers of Colony Losses. Curr. Opin. Insect. Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef]
- Berg, C.J.; King, P.K.; Delenstarr, G.; Kumar, R.; Rubio, F.; Glaze, T. Glyphosate residue concentrations in honey attributed through geospatial analysis to proximity of large-scale agriculture and transfer off-site by bees. PLoS ONE 2018, 13, e0198876. [Google Scholar] [CrossRef]
- Battaglin, W.A.; Meyer, M.T.; Kuivila, K.M.; Dietze, J.E. Glyphosate and Its Degradation Product AMPA Occur Frequently and Widely in US Soils, Surface Water, Groundwater, and Precipitation. J. Am. Water Resour. Assoc. 2014, 50, 275–290. [Google Scholar] [CrossRef]
- Rubio, F.; Guo, E.; Kamp, L. Survey of glyphosate residues in honey, corn and soy products. J. Environ. Anal. Toxicol. 2014, 4, 249. [Google Scholar]
- Mitchell, E.A.D.; Mulhauser, B.; Mulot, M.; Mutabazi, A.; Glauser, G.; Aebi, A. A worldwide survey of neonicotinoids in honey. Science 2017, 358, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Benbrook, C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016, 28, 3. [Google Scholar] [CrossRef] [PubMed]
- Farina, W.M.; Balbuena, M.S.; Herbert, L.T.; Mengoni Goñalon, C.; Vázquez, D.E. Effects of the Herbicide Glyphosate on Honey Bee Sensory and Cognitive Abilities: Individual Impairments with Implications for the Hive. Insects 2019, 10, 354. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Mak, M.; Tyler, K.; De Jong, J.; Powell, E.; O´Donnell, A.; Suhr, K.J.; Riddington, I.A.; Moran, N.A. Oral or Topical Exposure to Glyphosate in Herbicide Formulation Impacts the Gut Microbiota and Survival Rates of Honey Bees. Appl. Environ. Microbiol. 2020, 86, e01150-20. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Moran, N.A. Impact of Glyphosate on the Honey Bee Gut Microbiota: Effects of Intensity, Duration, and Timing of Exposure. Msystems 2020. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef]
- Blot, N.; Vaillat, L.; Rouzé, R.; Delatte, H. Glyphosate, but not its metabolite AMPA, alters the honeybee gut microbiota. PLoS ONE 2019, 14, e0215466. [Google Scholar] [CrossRef]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Scott Cornman, R.; Dainat, J.; de Miranda, J.R.; Doublet, V.; et al. The bee microbiome: Impact on bee health and model for evolution and ecology of host-microbe interactions. MBio 2016, 7, e02164-15. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect. Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Meana, A.; Bartolomé, C.; Botías, C.; Martín-Hernández, R. Nosema ceranae, a controversial 21st century honey bee pathogen. Environ. Microbiol. Rep. 2013, 5, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Zunino, P.; Higes, M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae. Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef]
- Chaimanee, V.; Chantawannakul, P.; Chen, Y.; Evans, J.D.; Pettis, J.S. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J. Insect. Physiol. 2012, 58, 1090–1095. [Google Scholar] [CrossRef]
- Beaurepaire, A.; Piot, N.; Doublet, V.; Antúnez, K.; Campbell, E.; Chantawannakul, P.; Chejanovsky, N.; Gajda, A.; Heerman, M.; Panziera, D.; et al. Diversity and Global Distribution of Viruses of the Western Honey Bee, Apis mellifera. Insects 2020, 11, 239. [Google Scholar] [CrossRef]
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010, 103, 96–119. [Google Scholar] [CrossRef] [PubMed]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noel, L.; Hall, M.-L.J.; Martin, S.J.; Schoroeder, D.C. Deformed Wing Virus Implicated in Overwintering Honeybee Colony Losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Spivak, M. Socialized Medicine: Individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010, 103, S62–S72. [Google Scholar] [CrossRef]
- Doublet, V.; Labarussias, M.; de Miranda, J.R.; Moritz, R.F.A.; Paxton, R.J. Bees under stress: Sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ. Microbiol. 2015, 17, 969–983. [Google Scholar] [CrossRef]
- Naggar, Y.A.; Baer, B. Consequences of a short time exposure to a sublethal dose of Flupyradifurone (Sivanto) pesticide early in life on survival and immunity in the honeybee (Apis mellifera). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Tesovnik, T.; Zorc, M.; Ristanić, M.; Glavinić, U.; Stevanović, J.; Narat, M.; Stanimirović, Z. Exposure of honey bee larvae to thiamethoxam and its interaction with Nosema ceranae infection in adult honey bees. Environ. Pollut. 2020, 256, 113443. [Google Scholar] [CrossRef]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.A.; Tzilivakis, J.; Warner, D.J.; Green, A. An international database for pesticide risk assessments and management. Human and Ecological Risk Assessment. Hum. Ecol. Risk Assess. 2016, 22, 1050–1064. [Google Scholar] [CrossRef]
- Van Bruggen, A.H.C.; He, M.M.; Shin, K.; Mai, V.; Jeong, K.C.; Finckh, M.R.; Morris, J.G. Environmental and health effects of the herbicide glyphosate. Sci. Total Environ. 2018, 616, 255–268. [Google Scholar] [CrossRef]
- Fries, I.; Chauzat, M.-P.; Chen, Y.-P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard methods for Nosema research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Martínez Salvador, A.; Garrido-Bailón, E.; Higes, M. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; James, R.R.; Koga, R.; Kwong, W.K.; McFrederick, Q.S.; Moran, N.A. Standard methods for research on Apis mellifera gut symbionts. J. Apic. Res. 2013, 52, 1–24. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cox-Foster, D.L. Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 2005, 102, 7470–7475. [Google Scholar] [CrossRef]
- Corona, M.; Velarde, R.; Remolina, S.; Moran-Lauter, A.; Wang, Y.; Hughes, K.A.; Robinson, G.E. Vitellogenin, juvenile hormone, insulin signalling, and queen honey bee longevity. Proc. Natl. Acad. Sci. USA 2007, 104, 7128–7133. [Google Scholar] [CrossRef]
- Kukielka, D.; Esperón, F.; Higes, M.; Sánchez-Vizcaíno, J.M. A sensitive one-step real-time RT-PCR method for detection of deformed wing virus and black queen cell virus in honeybee Apis mellifera. J. Virol. Methods 2008, 147, 275–281. [Google Scholar] [CrossRef]
- Evans, J.D. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invertebr. Pathol. 2006, 93, 135–139. [Google Scholar] [CrossRef]
- Human, H.; Brodschneider, R.; Dietemann, V.; Dively, G.; Ellis, J.D.; Forsgren, E.; Fries, I.; Hatjina, F.; Hu, F.-L.; Jaffé, R.; et al. Miscellaneous standard methods for Apis mellifera research. J. Apic. Res. 2013, 52, 1–53. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 1 April 2020).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Gentleman, R.; Carey, V.; Huber, W.; Hahne, F. Genefilter: Methods for Filtering Genes from High-Throughput Experiments. R Package Version 1.66.0. 2019. Available online: https://bioconductor.org/packages/release/bioc/html/genefilter.html (accessed on 1 April 2020).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.L.P.; Minchin, P.; O’Hara, R.B.; Simpson, G.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Vegan, Vers. 2.2-1. 2015. Available online: https://www.worldagroforestry.org/publication/vegan-community-ecology-package-r-package-vegan-vers-22-1 (accessed on 16 March 2020).
- Anderson, M.J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef]
- Anderson, M.J.; Ellingsen, K.E.; McArdle, B.H. Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 2006, 9, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Toufalilia, H.A.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.W.; Hughes, W.O. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 2018, 8, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M.; Altman, D.G. The logrank test. BMJ 2004, 328, 1073. [Google Scholar] [CrossRef]
- Cox, D.R. Regression models and life tables. J. R. Stat. Soc. B. 1972, 34, 187–220. [Google Scholar] [CrossRef]
- Powell, J.E.; Leonard, S.P.; Kwong, W.K.; Engel, P.; Moran, N.A. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. USA 2016, 113, 13887–13892. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Mancedino, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef]
- Anjuma, S.I.; Shahb, A.H.; Aurongzebc, M.; Koric, J.; Azim, M.K.; Ansari, M.J. Characterization of gut bacterial flora of Apis mellifera from north-west Pakistan, Saudi. J. Biol. Sci. 2018, 25, 388–392. [Google Scholar]
- Priestman, M.A.; Funke, T.; Singh, I.M.; Crupper, S.S.; Schonbrunn, E. 5-Enolpyruvylshikimate-3-phosphate synthase from Staphylococcus aureus is insensitive to glyphosate. FEBS Lett. 2005, 579, 728–732. [Google Scholar] [CrossRef]
- Hove-Jensen, B.; Zechel, D.L.; Jochimsen, B. Utilization of glyphosate as phosphate source: Biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol. Mol. Biol. Rev. 2014, 78, 176–197. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.M. Purification and properties of three lysozymes from hemolymph of the cricket, Gryllus bimaculatus (De Geer). Insect Biochem. 1985, 15, 463–470. [Google Scholar] [CrossRef]
- Bucekova, M.; Valachova, I.; Kohutova, L.; Prochazka, E.; Klaudiny, J.; Majtan, J. Honeybee glucose oxidase—Its expression in honeybee workers and comparative analyses of its content and H2O2-mediated antibacterial activity in natural honeys. Naturwissenschaften 2014, 101, 661–670. [Google Scholar] [CrossRef]
- Alaux, C.; Ducloz, F.; Crauser, D.; Le Conte, Y. Diet effects on honeybee immunocompetence. Biol. Lett. 2010, 6, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, G.; Guo, D.; Wang, Y.; Liu, Q.; Gao, Z.; Wang, H.; Liu, Z.; Guo, X.; Xu, B. Transcriptomic and metabolomic landscape of the molecular effects of glyphosate commercial formulation on Apis mellifera ligustica and Apis cerana cerana. Sci. Total Environ. 2020, 744, 140819. [Google Scholar] [CrossRef] [PubMed]
- Amdam, G.V.; Omholt, W. The hive bee to forager transition in honeybee colonies: The double repressor hypothesis. J. Theor. Biol. 2003, 223, 451–464. [Google Scholar] [CrossRef]
- Nelson, C.M.; Ihle, K.E.; Fondrk, M.K.; Page, R.E.; Amdam, G.V. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007, 5, 673–677. [Google Scholar] [CrossRef]
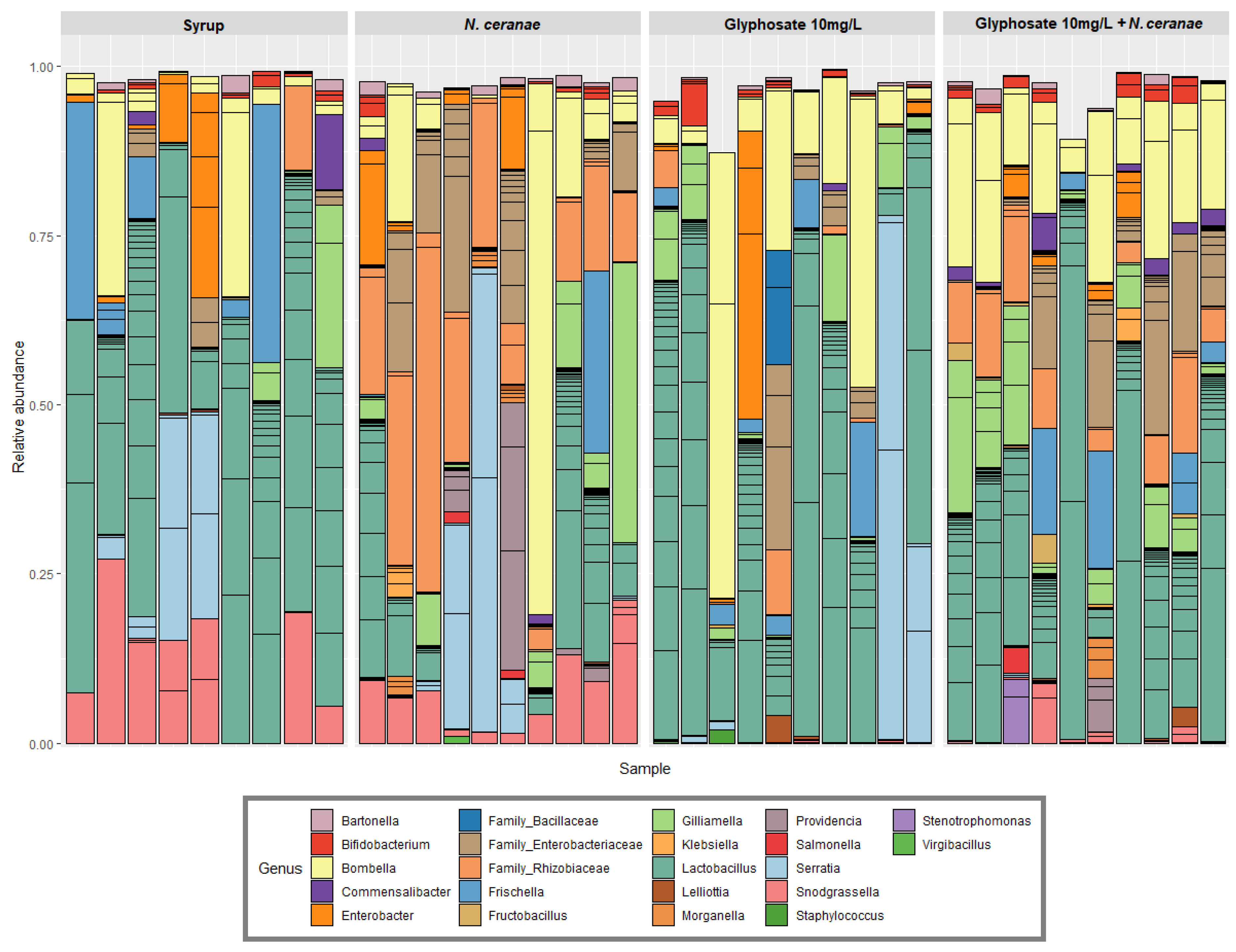


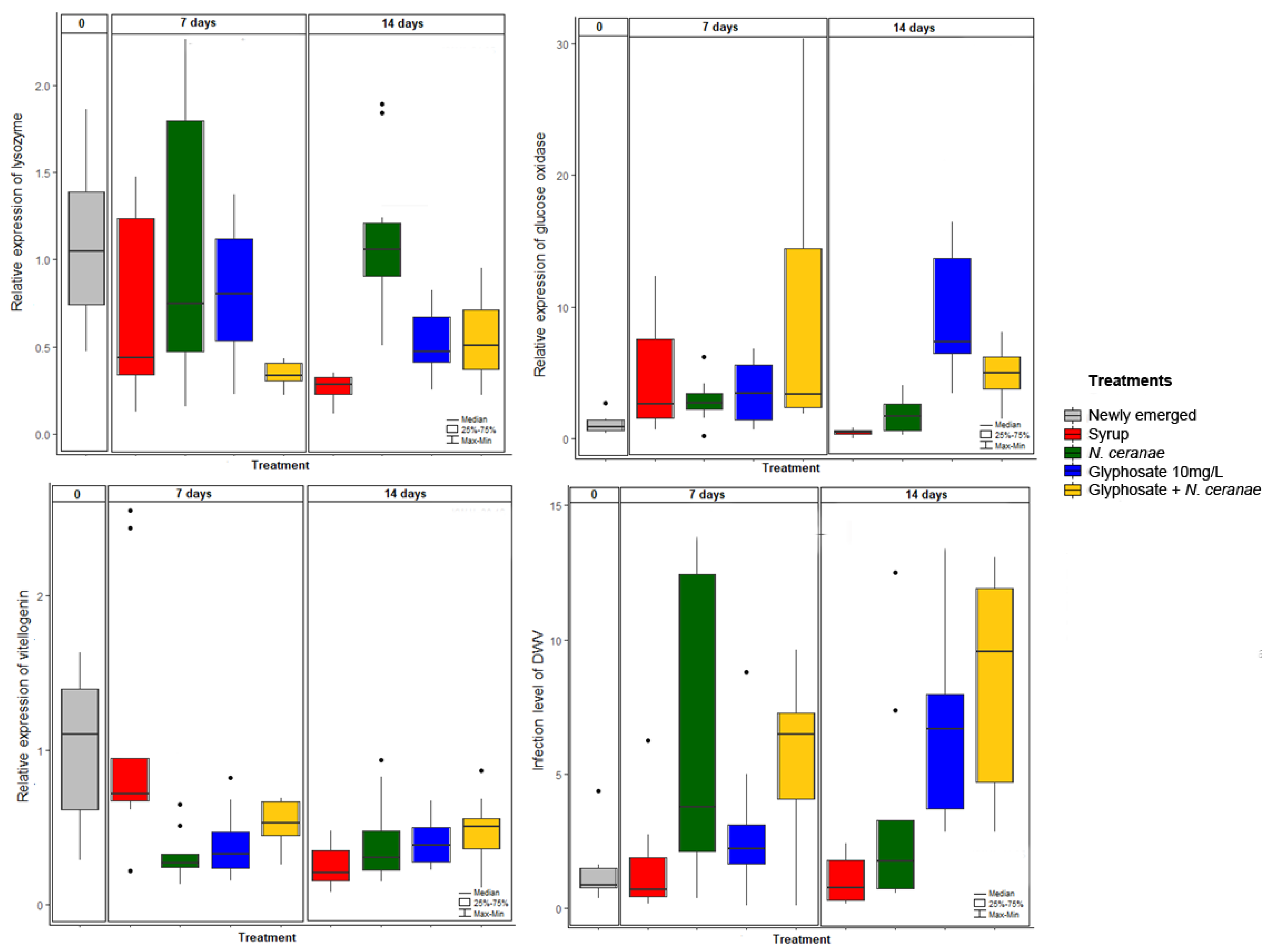
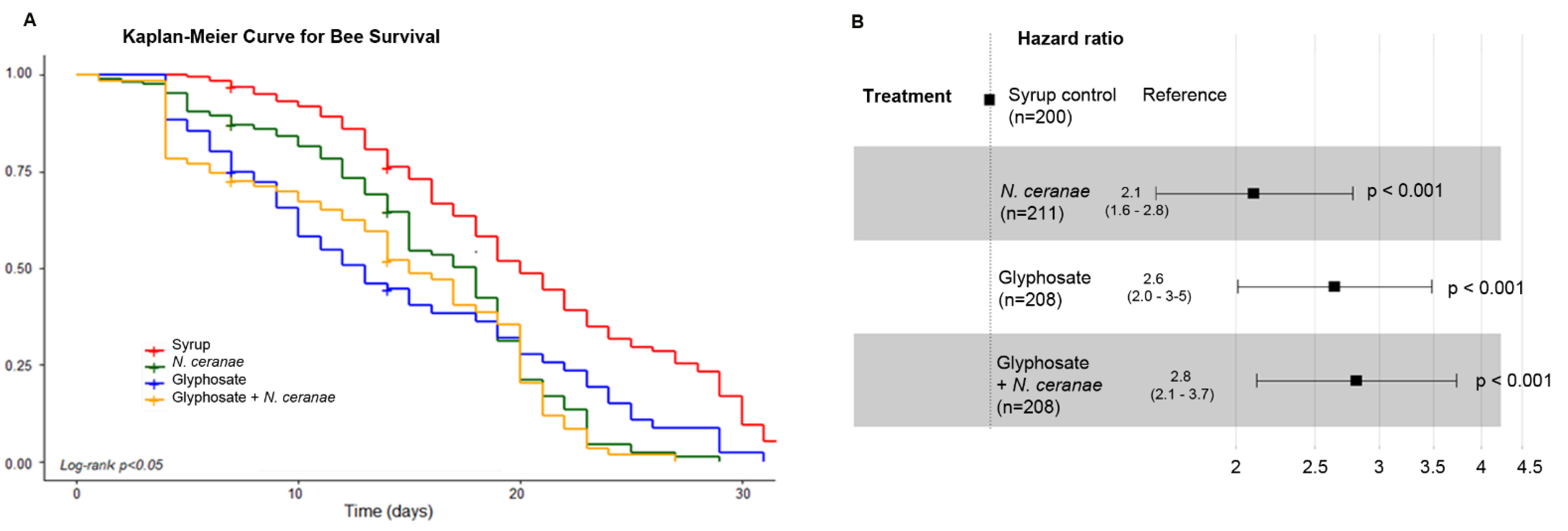
| Dependent Variable | Independent Variable | Coefficient Value | Intercept Value | p Value |
|---|---|---|---|---|
| Observed ASVs | N. ceranae | 0.2 | 4.01 | 0.004 |
| Glyphosate | 0.46 | ≤0.001 | ||
| N. ceranae + Glyphosate | 0.62 | ≤0.001 | ||
| Shannon diversity index | N. ceranae | −0.005 | 0.83 | 0.95 |
| Glyphosate | 0.01 | 0.86 | ||
| N. ceranae + Glyphosate | 0.19 | 0.03 | ||
| Lysozyme | N. ceranae | 1.38 | −1.29 | ≤0.001 |
| Glyphosate | 0.69 | ≤0.001 | ||
| N. ceranae + Glyphosate | 0.67 | ≤0.001 | ||
| Time | - | ns | ||
| Glucose oxidase | N. ceranae | 0.07 | −0.24 | 0.16 |
| Glyphosate | 0.32 | ≤0.001 | ||
| N. ceranae + Glyphosate | 0.25 | ≤0.001 | ||
| Time | - | ns | ||
| Prophenol oxidase | N. ceranae | 0.04 | −0.37 | 0.24 |
| Glyphosate | −0.007 | 0.83 | ||
| N. ceranae + Glyphosate | 0.04 | 0.17 | ||
| Time | 0.15 | ≤0.001 | ||
| Hymenoptaecin | N. ceranae | 0.77 | 0.99 | 0.05 |
| Glyphosate | 0.24 | 0.57 | ||
| N. ceranae + Glyphosate | 0.33 | 0.43 | ||
| Time | 1.67 | ≤0.001 | ||
| Vitellogenin | N. ceranae | −0.16 | −0.16 | ≤0.001 |
| Glyphosate | −0.14 | ≤0.001 | ||
| N. ceranae + Glyphosate | −0.12 | ≤0.001 | ||
| Time | −0.08 | ≤0.001 | ||
| N. ceranae * Time | 0.09 | ≤0.001 | ||
| Glyphosate * Time | 0.08 | ≤0.001 | ||
| N. ceranae + Glyphosate * Time | 0.07 | ≤0.001 | ||
| Deformed Wing Virus | N. ceranae | 0.44 | −0.24 | 0.07 |
| Glyphosate | −0.11 | 0.01 | ||
| N. ceranae + Glyphosate | 0.03 | ≤0.001 | ||
| Time | - | ns | ||
| Syrup consumption | N. ceranae | −0.02 | 2.65 | 0.73 |
| Glyphosate | −0.07 | 0.28 | ||
| N. ceranae + Glyphosate | 0.08 | 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castelli, L.; Balbuena, S.; Branchiccela, B.; Zunino, P.; Liberti, J.; Engel, P.; Antúnez, K. Impact of Chronic Exposure to Sublethal Doses of Glyphosate on Honey Bee Immunity, Gut Microbiota and Infection by Pathogens. Microorganisms 2021, 9, 845. https://doi.org/10.3390/microorganisms9040845
Castelli L, Balbuena S, Branchiccela B, Zunino P, Liberti J, Engel P, Antúnez K. Impact of Chronic Exposure to Sublethal Doses of Glyphosate on Honey Bee Immunity, Gut Microbiota and Infection by Pathogens. Microorganisms. 2021; 9(4):845. https://doi.org/10.3390/microorganisms9040845
Chicago/Turabian StyleCastelli, Loreley, Sofía Balbuena, Belén Branchiccela, Pablo Zunino, Joanito Liberti, Philipp Engel, and Karina Antúnez. 2021. "Impact of Chronic Exposure to Sublethal Doses of Glyphosate on Honey Bee Immunity, Gut Microbiota and Infection by Pathogens" Microorganisms 9, no. 4: 845. https://doi.org/10.3390/microorganisms9040845
APA StyleCastelli, L., Balbuena, S., Branchiccela, B., Zunino, P., Liberti, J., Engel, P., & Antúnez, K. (2021). Impact of Chronic Exposure to Sublethal Doses of Glyphosate on Honey Bee Immunity, Gut Microbiota and Infection by Pathogens. Microorganisms, 9(4), 845. https://doi.org/10.3390/microorganisms9040845






