RNA Interference-Mediated Knockdown of Genes Encoding Spore Wall Proteins Confers Protection against Nosema ceranae Infection in the European Honey Bee, Apis mellifera
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Nosema ceranae Spore Purification
2.3. dsRNA Synthesis
2.4. RNAi Feeding Bioassays
2.5. Data Analysis
3. Results
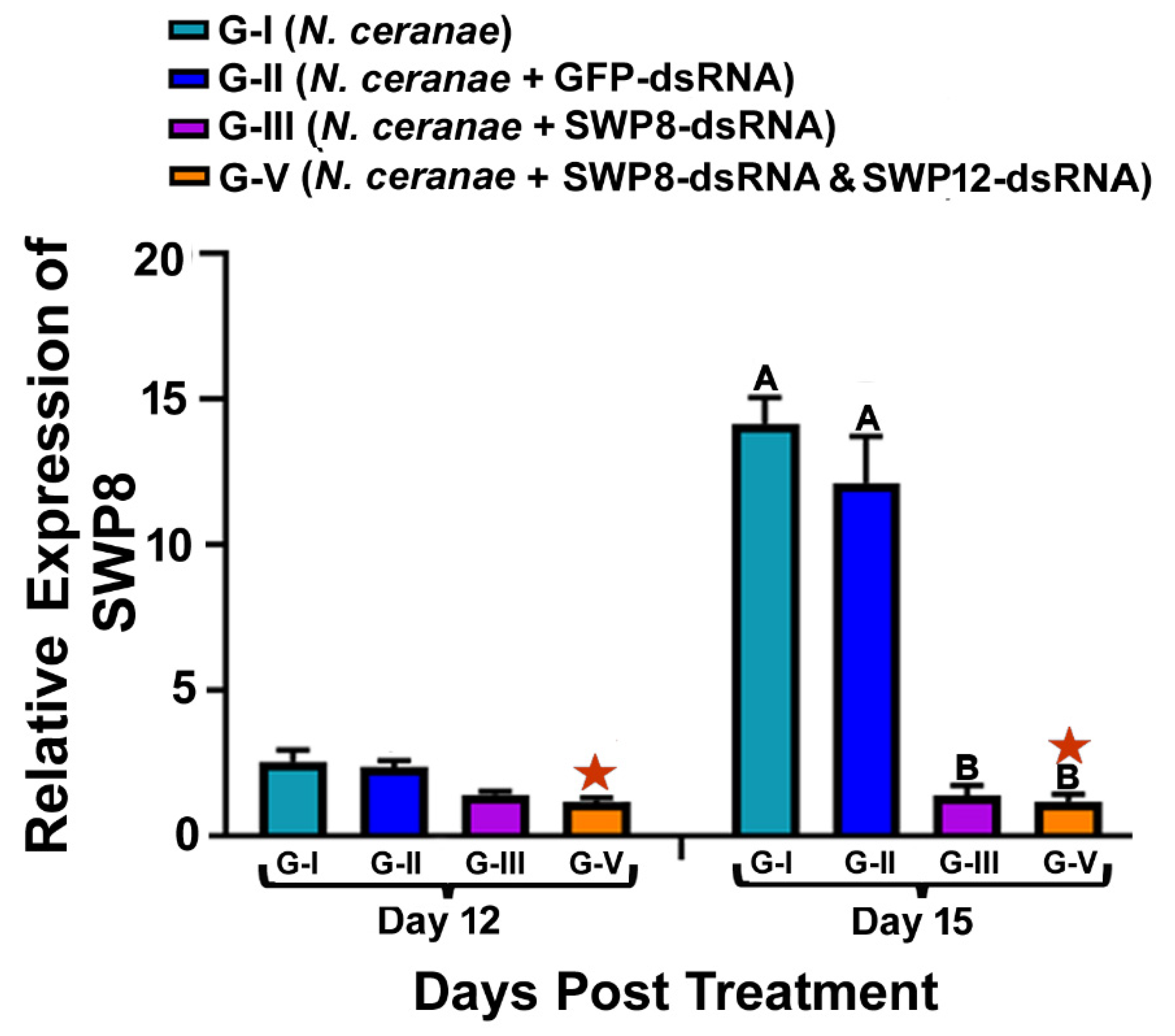
3.1. Ingestion of dsRNAs Corresponding to the Sequences of N. ceranae SWP8 Separately or in Combination with SWP12 Suppresses the Gene Expression SWP8 in Infected Bees
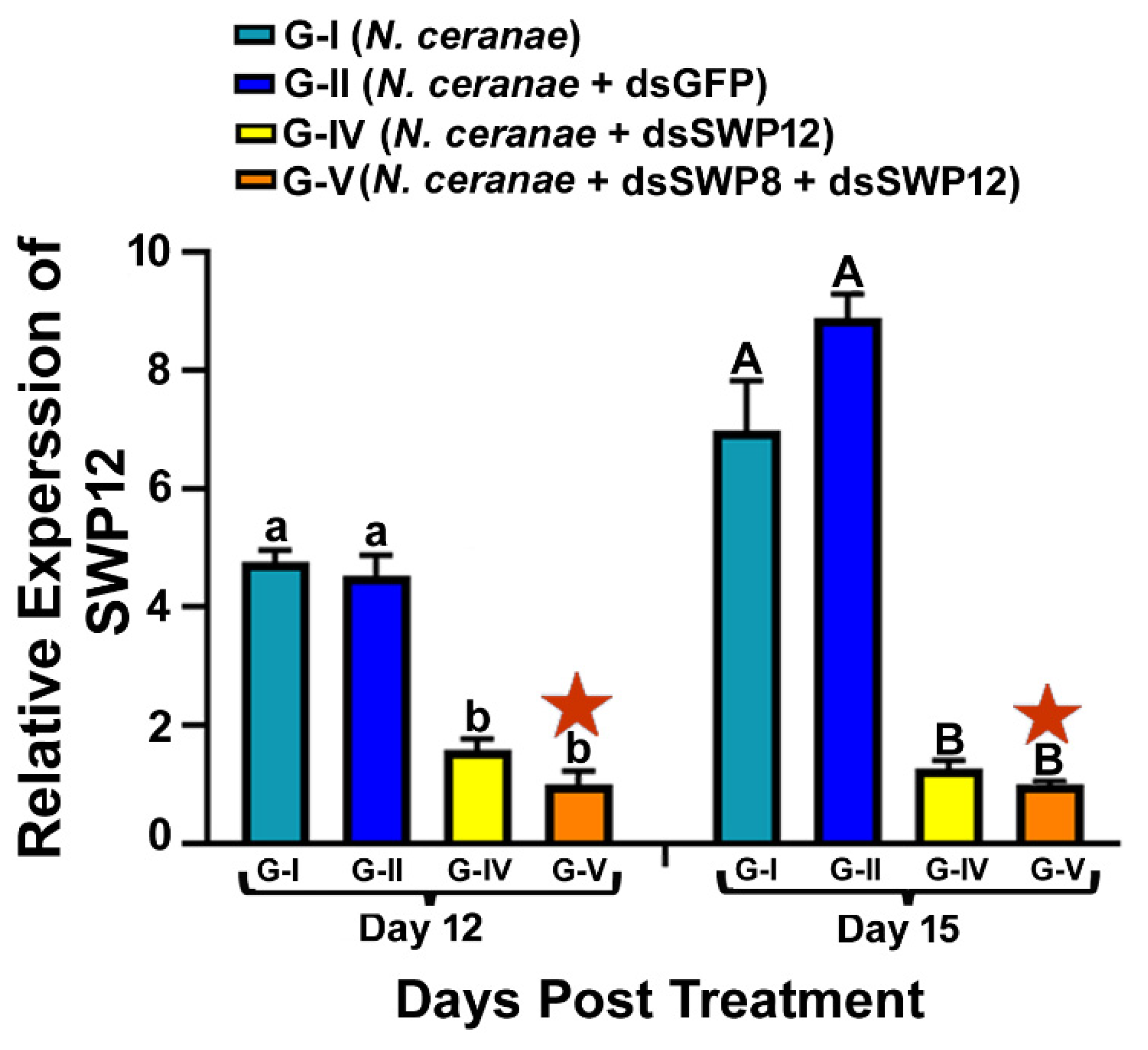
3.2. Ingestion of dsRNAs Corresponding to the Sequences of N. ceranae SWP12 Alone or in Combination with SWP8 Suppresses the Gene Expression of SWP12 in Infected Bees
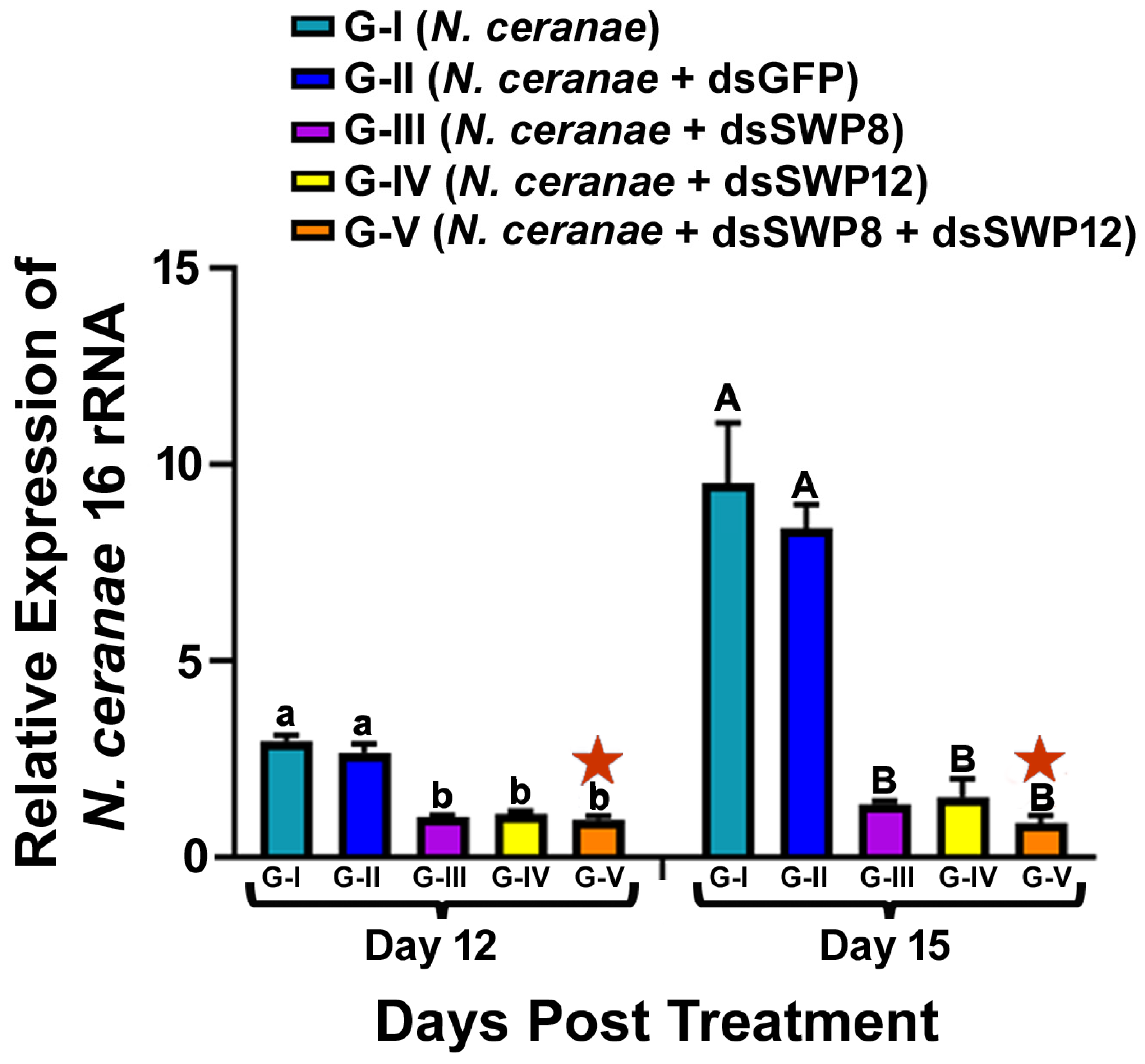
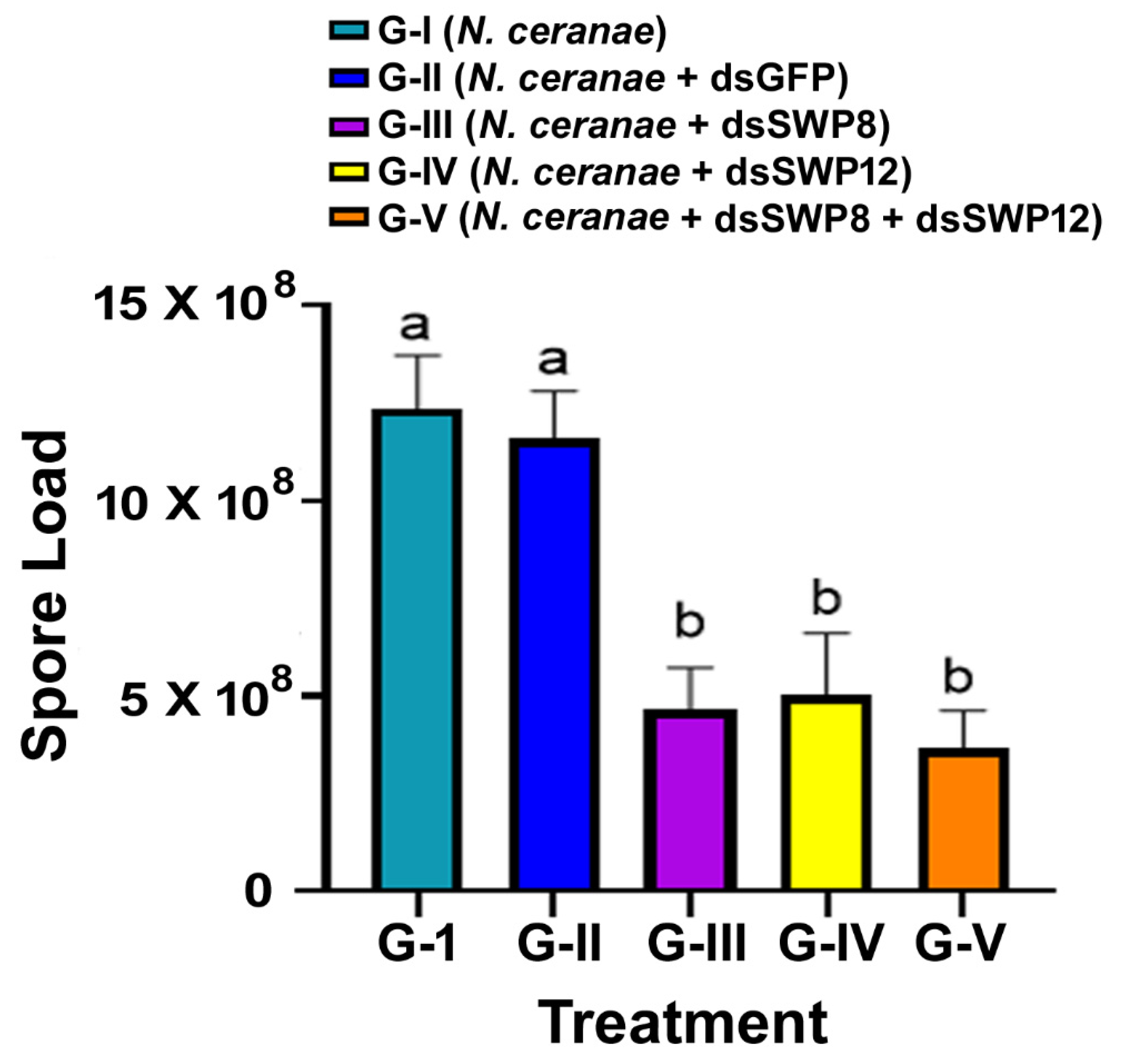
3.3. Silencing of the SWP8 and/or SWP12 Transcript Levels Led to the Significant Reduction in N. ceranae 16S rRNA Level and the Spore Load
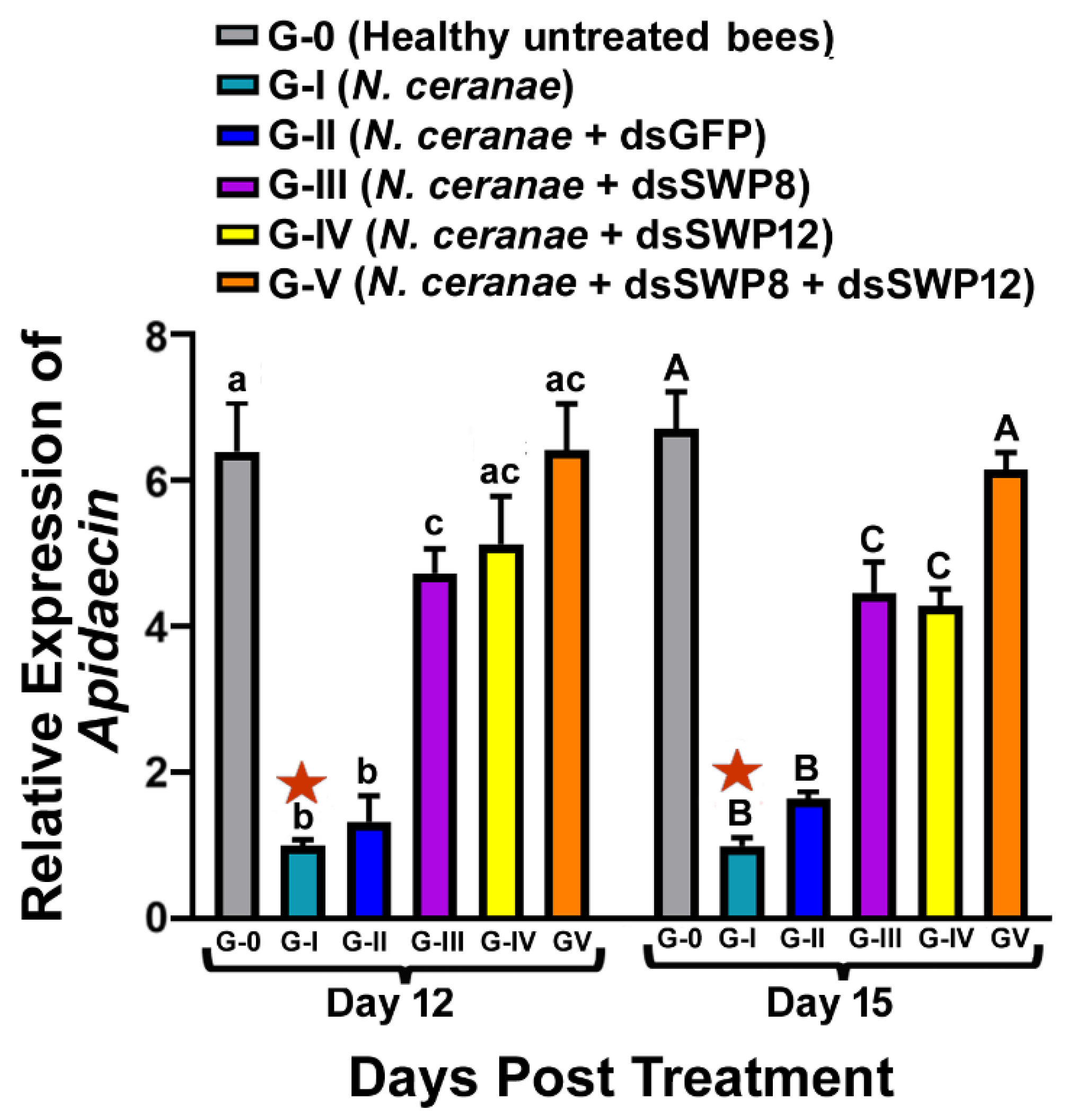
3.4. Effects of SWP8 and/or SWP12 Silencing on Apidaecin Expression
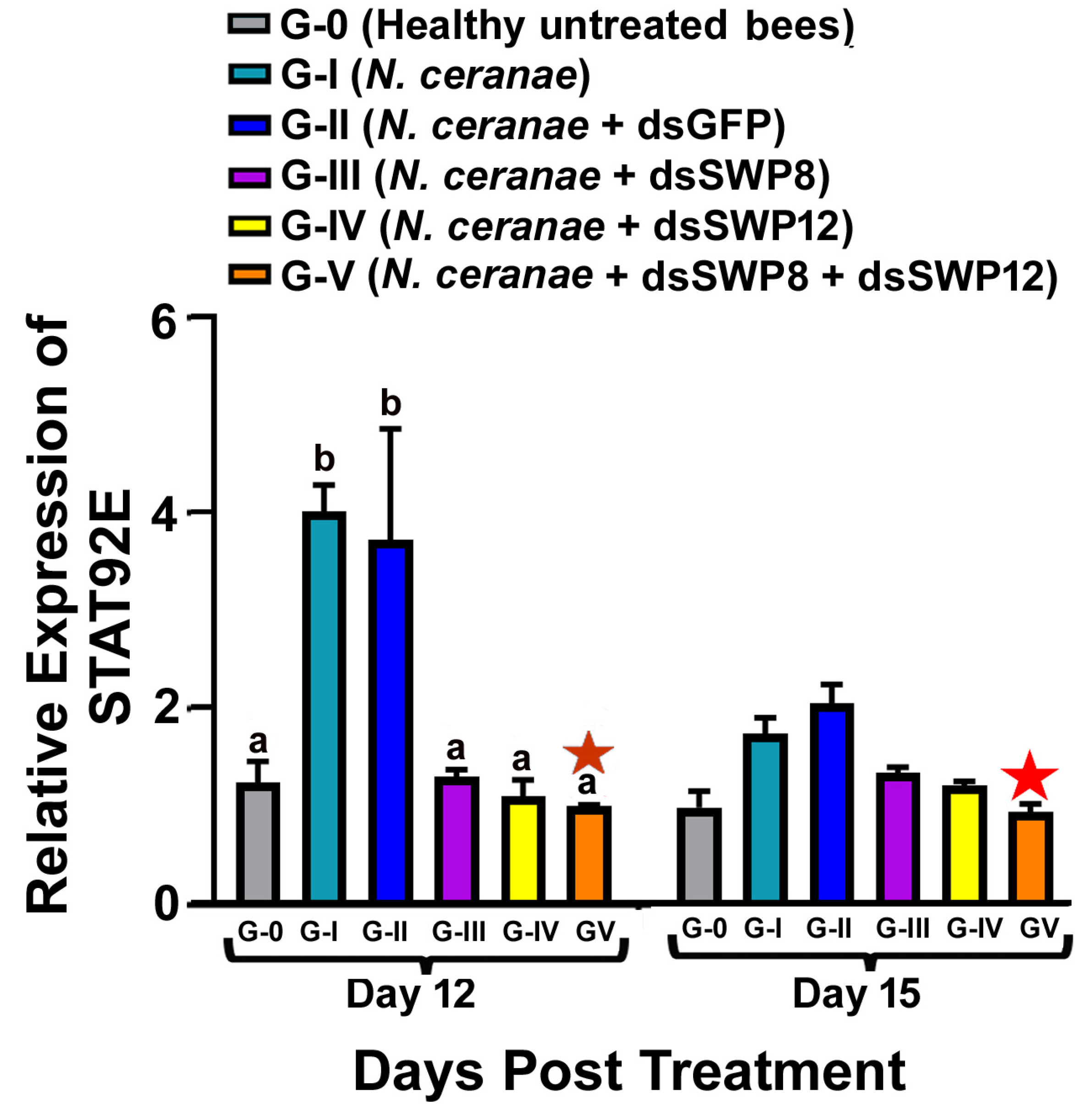
3.5. Effects of SWP8 and SWP12 Silencing on STAT92E Expression
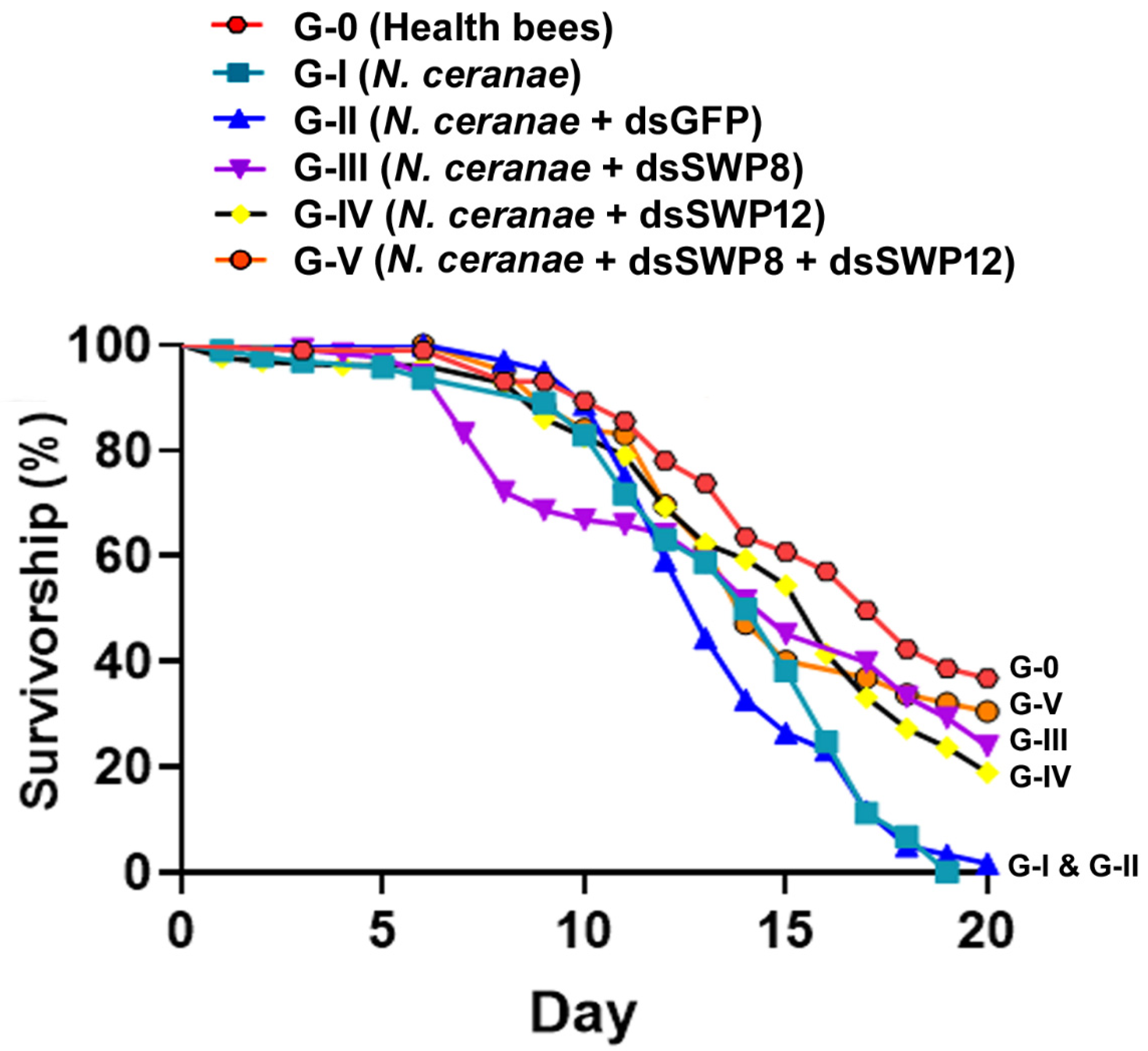
3.6. Silencing of SWP8 and SWP12 Gene Expression Contributed to Improved Survivorship
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, A.M.; Vaissière, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 2007, 274, 303–313. [Google Scholar] [CrossRef]
- Jones Ritten, C.; Peck, D.; Ehmke, M.; Patalee, M.A.B. Firm Efficiency and Returns-to-Scale in the Honey Bee Pollination Services Industry. J. Econ. Entomol. 2018, 111, 1014–1022. [Google Scholar] [CrossRef]
- Gallai, N.; Salles, J.-M.; Settele, J.; Vaissière, B.E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 2009, 68, 810–821. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Kulhanek, K.; Steinhauer, N.; Rennich, K.; Caron, D.M.; Sagili, R.R.; Pettis, J.S.; Ellis, J.D.; Wilson, M.E.; Wilkes, J.T.; Tarpy, D.R.; et al. A national survey of managed honey bee 2015–2016 annual colony losses in the USA. J. Apic. Res. 2017, 56, 328–340. [Google Scholar] [CrossRef]
- Potts, S.G.; Roberts, S.P.M.; Dean, R.; Marris, G.; Brown, M.A.; Jones, R.; Neumann, P.; Settele, J. Declines of managed honey bees and beekeepers in Europe. J. Apic. Res. 2010, 49, 15–22. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Nicholls, E.; Botias, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef]
- Bollan, K.A.; Hothersall, J.D.; Moffat, C.; Durkacz, J.; Saranzewa, N.; Wright, G.A.; Raine, N.E.; Highet, F.; Connolly, C.N. The microsporidian parasitesNosema ceranae and Nosema apisare widespread in honeybee (Apis mellifera) colonies across Scotland. Parasitol. Res. 2013, 112, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Becher, M.A.; Osborne, J.L.; Thorbek, P.; Kennedy, P.J.; Grimm, V. Towards a systems approach for understanding honeybee decline: A stocktaking and synthesis of existing models. J. Appl. Ecol. 2013, 50, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Fries, I.; Feng, F.; da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp.(Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Fries, I. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 103, S73–S79. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Smith, I.B.; Pettis, J.S. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008, 97, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Paxton, R.J.; Klee, J.; Korpela, S.; Fries, I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 2007, 38, 558–565. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; Botías, C.; Meana, A. The presence of Nosema ceranae (Microsporidia) in North African honey bees (Apis mellifera intermissa). J. Apic. Res. 2009, 48, 217–219. [Google Scholar] [CrossRef]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef]
- Katinka, M.D.; Duprat, S.; Cornillot, E.; Méténier, G.; Thomarat, F.; Prensier, G.; Barbe, V.; Peyretaillade, E.; Brottier, P.; Wincker, P.; et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414, 450–453. [Google Scholar] [CrossRef]
- Chen, Y.P.; Evans, J.D.; Murphy, C.; Gutell, R.; Zuker, M.; Gundensen-Rindal, D.; Pettis, J.S. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J. Eukaryot. Microbiol. 2009, 56, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L. Honey Bee Pathology, 2nd ed.; Academic Press: London, UK, 1991. [Google Scholar]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Higes, M. Immune-suppression in the honeybee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef]
- Higes, M.; Juarranz, A.; Dias-Almeida, J.; Lucena, S.; Botias, C.; Meana, A.; Garcia-Palencia, P.; Martin-Hernandez, R. Apoptosis in the pathogenesis of Nosema ceranae (Microsporidia: Nosematidae) in honey bees (Apis mellifera). Environ. Microbiol. Rep. 2013, 5, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Mayack, C.; Natsopoulou, M.; McMahon, D.P. Nosema ceranae alters a highly conserved hormonal stress pathway in honeybees. Insect Mol. Biol. 2015, 24, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; McMahon, D.P.; Lim, K.S.; Pull, C.D.; Clark, S.J.; Paxton, R.J.; Osborne, J.L. So near and yet so far: Harmonic radar reveals reduced homing ability of Nosema infected honeybees. PLoS ONE 2014, 9, e103989. [Google Scholar] [CrossRef]
- Zheng, H.-Q.; Lin, Z.-G.; Huang, S.-K.; Sohr, A.; Wu, L.; Chen, Y.P. Spore loads may not be used alone as a direct indicator of the severity of Nosema ceranae infection in honey bees Apis mellifera (Hymenoptera: Apidae). J. Econ. Entomol. 2014, 107, 2037–2044. [Google Scholar] [CrossRef]
- Dussaubat, C.; Maisonnasse, A.; Crauser, D.; Beslay, D.; Costagliola, G.; Soubeyrand, S.; Kretzchmar, A.; Le Conte, Y. Flight behavior and pheromone changes associated to Nosema ceranae infection of honey bee workers (Apis mellifera) in field conditions. J. Invertebr. Pathol. 2013, 113, 42–51. [Google Scholar] [CrossRef]
- Bromenshenk, J.J.; Henderson, C.B.; Wick, C.H.; Stanford, M.F.; Zulich, A.W.; Jabbour, R.E.; Deshpande, S.V.; McCubbin, P.E.; Seccomb, R.A.; Welch, P.M.; et al. Iridovirus and Microsporidian Linked to Honey Bee Colony Decline. PLoS ONE 2013, 5, e13181. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef]
- Retschnig, G.; Williams, G.R.; Schneeberger, A.; Neumann, P. Cold ambient temperature promotes Nosema spp. intensity in honey bees (Apis mellifera). Insects 2017, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Chung, W.-P.; Wang, C.-H.; Solter, L.F.; Huang, W.-F. Nosema ceranae infection intensity highly correlates with temperature. J. Invertebr. Pathol. 2012, 111, 264–267. [Google Scholar] [CrossRef]
- Jack, C.J.; Uppala, S.S.; Lucas, H.M.; Sagili, R.R. Effects of pollen dilution on infection of Nosema ceranae in honey bees. J. Insect Physiol. 2016, 87, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Pettis, J.S.; Lichtenberg, E.M.; Andree, M.; Stitzinger, J.; Rose, R. Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS ONE 2013, 8, e70182. [Google Scholar]
- Aufauvre, J.; Biron, D.G.; Vidau, C.; Fontbonne, R.; Roudel, M.; Diogon, M.; Viguès, B.; Belzunces, L.P.; Delbac, F.; Blot, N. Parasite-insecticide interactions: A case study of Nosema ceranae and fipronil synergy on honeybee. Sci. Rep. 2012, 2, 326. [Google Scholar] [CrossRef]
- Zheng, H.-Q.; Gong, H.-R.; Huang, S.-K.; Sohr, A.; Hu, F.-L.; Chen, Y.P. Evidence of the synergistic interaction of honey bee pathogens Nosema ceranae and deformed wing virus. Vet. Microbiol. 2015, 177, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Smart, M.D.; Anelli, C.M.; Sheppard, W.S. Honey bees (Apis mellifera) reared in brood combs containing high levels of pesticide residues exhibit increased susceptibility to Nosema (Microsporidia) infection. J. Invertebr. Pathol. 2012, 109, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Paris, L.; Roussel, M.; Pereira, B.; Delbac, F.; Diogon, M. Disruption of oxidative balance in the gut of the western honeybee Apis mellifera exposed to the intracellular parasite Nosema ceranae and to the insecticide fipronil. Microb. Biotechnol. 2017, 10, 1702–1717. [Google Scholar] [CrossRef]
- Alaux, C.; Folschweiller, M.; McDonnell, C.; Beslay, D.; Cousin, M.; Dussaubat, C.; Brunet, J.L.; Le Conte, Y. Pathological effects of the microsporidium Nosema ceranae on honey bee queen physiology (Apis mellifera). J. Invertebr. Pathol. 2011, 106, 380–385. [Google Scholar] [CrossRef]
- Loskotova, J.; Peroutka, M.; Vesely, V. Nosema disease of honey bee queens (Apis mellifera). Apidologie 1980, 12, 53–61. [Google Scholar]
- Martin-Hernandez, R.; Bartolome, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; Garcia-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef]
- Yang, D.; Pan, L.; Chen, Z.; Du, H.; Luo, B.; Luo, J.; Pan, G. The roles of microsporidia spore wall proteins in the spore wall formation and polar tube anchorage to spore wall during development and infection processes. Exp. Parasitol. 2018, 187, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Pan, G.; Dang, X.; Shi, Y.; Li, C.; Peng, P.; Luo, B.; Bian, M.; Song, Y.; Ma, C.; et al. Interaction and assembly of two novel proteins in the spore wall of the microsporidian species Nosema bombycis and their roles in adherence to and infection of host cells. Infect. Immun. 2015, 83, 1715–1731. [Google Scholar] [CrossRef]
- Hayman, J.R.; Hayes, S.F.; Amon, J.; Nash, T.E. Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infect. Immun. 2001, 69, 7057–7066. [Google Scholar] [CrossRef]
- Southern, T.R.; Jolly, C.E.; Lester, M.E.; Hayman, J.R. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot. Cell 2007, 6, 1354–1362. [Google Scholar] [CrossRef]
- Cornman, R.S.; Chen, Y.P.; Schatz, M.C.; Street, C.; Zhao, Y.; Desany, B.; Egholm, M.; Hutchison, S.; Pettis, J.S.; Lipkin, W.I.; et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog 2009, 5, e1000466. [Google Scholar] [CrossRef]
- Pelin, A.; Selman, M.; Aris-Brosou, S.; Farinelli, L.; Corradi, N. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 2015, 17, 4443–4458. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.F.; Solter, L.F.; Yau, P.M.; Imai, B.S. Nosema ceranae escapes fumagillin control in honey bees. PLoS Pathog 2013, 9, e1003185. [Google Scholar] [CrossRef]
- Maggi, M.; Negri, P.; Plischuk, S.; Szawarski, N.; De Piano, F.; De Feudis, L.; Eguaras, M.; Audisio, C. Effects of the organic acids produced by a lactic acid bacterium in Apis mellifera colony development, Nosema ceranae control and fumagillin efficiency. Vet. Microbiol. 2013, 167, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, Y.; Diaz-Cetti, S.; Ramallo, G.; Santos, E.; Porrini, M.; Invernizzi, C. Nosema ceranae Winter Control: Study of the Effectiveness of Different Fumagillin Treatments and Consequences on the Strength of Honey Bee (Hymenoptera: Apidae) Colonies. J. Econ. Entomol. 2017, 110, 1–5. [Google Scholar] [CrossRef]
- Williams, G.R.; Sampson, M.A.; Shutler, D.; Rogers, R.E. Does fumagillin control the recently detected invasive parasite Nosema ceranae in western honey bees (Apis mellifera)? J. Invertebr. Pathol. 2008, 99, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Nozal, M.J.; Alvaro, A.; Barrios, L.; Meana, A.; Martín-Hernández, R.; Bernal, J.L.; Bernal, J. The stability and effectiveness of fumagillin in controlling Nosema ceranae (Microsporidia) infection in honey bees (Apis mellifera) under laboratory and field conditions. Apidologie 2011, 42, 364–377. [Google Scholar] [CrossRef]
- Burnham, A.J. Scientific Advances in Controlling Nosema ceranae (Microsporidia) Infections in Honey Bees (Apis mellifera). Front. Vet. Sci. 2019, 6, 79. [Google Scholar] [CrossRef]
- Mura, A.; Pusceddu, M.; Theodorou, P.; Angioni, A.; Floris, I.; Paxton, R.J.; Satta, A. Propolis Consumption Reduces Nosema ceranae Infection of European Honey Bees (Apis mellifera). Insects 2020, 11, 124. [Google Scholar] [CrossRef] [PubMed]
- Suwannapong, G.; Maksong, S.; Phainchajoen, M.; Benbow, M.E.; Mayack, C. Survival and health improvement of Nosema infected Apis florea (Hymenoptera: Apidae) bees after treatment with propolis extract. J. Asia-Pac. Entomol. 2018, 21, 437–444. [Google Scholar] [CrossRef]
- Yemor, T.; Phiancharoen, M.; Benbow, E.M.; Suwannapong, G. Effects of stingless bee propolis on Nosema ceranae infected Asian honey bees, Apis cerana. J. Apic. Res. 2015, 54, 468–473. [Google Scholar] [CrossRef]
- Borges, D.; Guzman-Novoa, E.; Goodwin, P.H. Control of the microsporidian parasite Nosema ceranae in honey bees (Apis mellifera) using nutraceutical and immuno-stimulatory compounds. PLoS ONE 2020, 15, e0227484. [Google Scholar] [CrossRef]
- Ptaszyńska, A.A.; Trytek, M.; Borsuk, G.; Buczek, K.; Rybicka-Jasińska, K.; Gryko, D. Porphyrins inactivate Nosema spp. microsporidia. Sci. Rep. 2018, 8, 5523. [Google Scholar] [CrossRef]
- Buczek, K.; Trytek, M.; Deryło, K.; Borsuk, G.; Rybicka-Jasińska, K.; Gryko, D.; Cytryńska, M.; Tchórzewski, M. Bioactivity studies of porphyrinoids against microsporidia isolated from honeybees. Sci. Rep. 2020, 10, 11553. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Chen, Y.; Pettis, J.S.; Zhao, Y.; Liu, X.; Tallon, L.J.; Sadzewicz, L.D.; Li, R.; Zheng, H.; Huang, S.; Zhang, X.; et al. Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genomics 2013, 14, 451. [Google Scholar] [CrossRef] [PubMed]
- Paldi, N.; Glick, E.; Oliva, M.; Zilberberg, Y.; Aubin, L.; Pettis, J.; Chen, Y.; Evans, J.D. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 2010, 76, 5960–5964. [Google Scholar] [CrossRef]
- Rodriguez-Garcia, C.; Evans, J.D.; Li, W.; Branchiccela, B.; Li, J.H.; Heerman, M.C.; Banmeke, O.; Zhao, Y.; Hamilton, M.; Higes, M.; et al. Nosemosis control in European honey bees, Apis mellifera, by silencing the gene encoding Nosema ceranae polar tube protein 3. J. Exp. Biol. 2018, 221. [Google Scholar] [CrossRef]
- Huang, Q.; Li, W.; Chen, Y.; Retschnig-Tanner, G.; Yanez, O.; Neumann, P.; Evans, J.D. Dicer regulates Nosema ceranae proliferation in honeybees. Insect Mol. Biol. 2019, 28, 74–85. [Google Scholar] [CrossRef]
- Li, W.; Evans, J.D.; Huang, Q.; Rodríguez-García, C.; Liu, J.; Hamilton, M.; Grozinger, C.M.; Webster, T.C.; Su, S.; Chen, Y.P. Silencing the Honey Bee (Apis mellifera) Naked Cuticle Gene (nkd) Improves Host Immune Function and Reduces Nosema ceranae Infections. Appl. Environ. Microbiol. 2016, 82, 6779–6787. [Google Scholar] [CrossRef]
- Cantwell, G.E. Standard methods for counting Nosema spores. Am. Bee J. 1970, 110, 222–223. [Google Scholar]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; DeGrandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef]
- Horn, T.; Boutros, M. E-RNAi: A web application for the multi-species design of RNAi reagents—2010 update. Nucleic Acids Res. 2010, 38, W332–W339. [Google Scholar] [CrossRef]
- Amdam, G.V.; Norberg, K.; Hagen, A.; Omholt, S.W. Social exploitation of vitellogenin. Proc. Natl. Acad. Sci. USA 2003, 100, 1799–1802. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Higgins, J.A.; Feldlaufer, M.F. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 2005, 71, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Chen, Y.P.; Di Prisco, G.; Pettis, J.; Williams, V. Bee cups: Single-use cages for honey bee experiments. J. Apic. Res. 2009, 48, 300–302. [Google Scholar] [CrossRef]
- Huang, S.K.; Csaki, T.; Doublet, V.; Dussaubat, C.; Evans, J.D.; Gajda, A.M.; Gregorc, A.; Hamilton, M.C.; Kamler, M.; Lecocq, A.; et al. Evaluation of cage designs and feeding regimes for honey bee (Hymenoptera: Apidae) laboratory experiments. J. Econ. Entomol. 2014, 107, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2018, 9, 1912. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, L.M.; Flenniken, M.L. RNAi and Antiviral Defense in the Honey Bee. J. Immunol. Res. 2015, 2015, 941897. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B.; et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics 2014, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [CrossRef]
- Maori, E.; Paldi, N.; Shafir, S.; Kalev, H.; Tsur, E.; Glick, E.; Sela, I. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol. Biol. 2009, 18, 55–60. [Google Scholar] [CrossRef]
- Hunter, W.; Ellis, J.; Vanengelsdorp, D.; Hayes, J.; Westervelt, D.; Glick, E.; Williams, M.; Sela, I.; Maori, E.; Pettis, J.; et al. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog 2010, 6, e1001160. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Evans, J.D. RNAi in treating honey bee diseases. Bee Cult. 2012, 140, 27–29. [Google Scholar]
- Desai, S.D.; Eu, Y.J.; Whyard, S.; Currie, R.W. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol. Biol. 2012, 21, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Flenniken, M.L.; Andino, R. Non-specific dsRNA-mediated antiviral response in the honey bee. PLoS ONE 2013, 8, e77263. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Sci. Rep. 2017, 7, 6448. [Google Scholar] [CrossRef] [PubMed]
- Garbian, Y.; Maori, E.; Kalev, H.; Shafir, S.; Sela, I. Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog 2012, 8, e1003035. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Chaimanee, V.; Pettis, J.S.; Chen, Y.; Evans, J.D.; Khongphinitbunjong, K.; Chantawannakul, P. Susceptibility of four different honey bee species to Nosema ceranae. Vet. Parasitol. 2013, 193, 260–265. [Google Scholar] [CrossRef]
- Chaimanee, V.; Chantawannakul, P.; Chen, Y.; Evans, J.D.; Pettis, J.S. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J. Insect. Physiol. 2012, 58, 1090–1095. [Google Scholar] [CrossRef]
- Li, W.; Chen, Y.; Cook, S.C. Chronic Nosema ceranae infection inflicts comprehensive and persistent immunosuppression and accelerated lipid loss in host Apis mellifera honey bees. Int. J. Parasitol. 2018, 48, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Danihlík, J.; Aronstein, K.; Petřivalský, M. Antimicrobial peptides: A key component of honey bee innate immunity. J. Apic. Res. 2015, 54, 123–136. [Google Scholar] [CrossRef]
- Agaisse, H.; Perrimon, N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 2004, 198, 72–82. [Google Scholar] [CrossRef]
- Leonard, W.J. Role of Jak kinases and STATs in cytokine signal transduction. Int. J. Hematol. 2001, 73, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Chang, Y.S.; Wang, A.H.; Kou, G.H.; Lo, C.F. White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 2007, 81, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
| Purpose | Gene Name | Primer | Sequence (5′---3′) | Reference | Length (bp) |
|---|---|---|---|---|---|
| dsRNA | SWP8 | SWP8-F | taatacgactcactatagggagaCCATCAGTCATAACTTTGCC | This study | 347 |
| (N. ceranae) | SWP8-R | taatacgactcactatagggagaTCTGCAAACTCTCCAACAAC | |||
| dsRNA | SWP12 | SWP12-F | taatacgactcactatagggagaAGGTAGGTTCAAGTATAGCC | This study | 467 |
| (N. ceranae) | SWP12-R | taatacgactcactatagggagaCTAGTGCTTCTAGGCCAACT | |||
| dsRNA | GFP | SWP12-F | taatacgactcactatagggagaATTCCATGGCCAACACTTGTCC | [67] | 502 |
| SWP12-R | taatacgactcactatagggagaTCAAGAAGGACCATGTGGTC | ||||
| qPCR | SWP8 | SWP8-qF | ACAAGTACTGCAGCAAATATAGGTT | This study | 114 |
| (N. ceranae) | SWP8-qR | AATTGGCAAAGTTATGACTGATGGA | |||
| qPCR | SWP12 | SWP12-qF | AGTCAGAAGAATTGAATACAAGCAT | This study | 148 |
| (N. ceranae) | SWP12-qR | CTTTGCATTACCCCCATGTTCA | |||
| qPCR | 16S rRNA | 16S rRNA-qF | CGGATAAAAGAGTCCGTTACC | [14] | 250 |
| (N. ceranae) | 16S rRNA-qR | TGAGCAGGGTTCTAGGGAT | |||
| qPCR | STAT92E | STAT92E-qF | TGAACCTGGAAGAGTGCCAT | This study | 218 |
| (A.mellifera) | STAT92E-qR | TCTTGTCCGCTTGCATTTCC | |||
| qPCR | Apidaecin | Apidaecin-qF | TTTTGCCTTAGCAATTCTTGTTG | [63] | 80 |
| (A.mellifera) | Apidaecin-qR | GCAGGTCGAGTAGGCGGATCT | |||
| qPCR | ß-actin | ß-actin-qF | AGGAATGGAAGCTTGCGGTA | [68] | 181 |
| (A.mellifera) | ß-actin-qR | AATTTTCATGGTGGATGGTGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, N.; Zhang, Y.; Duan, X.L.; Li, J.H.; Huang, W.-F.; Evans, J.D.; DeGrandi-Hoffman, G.; Chen, Y.P.; Huang, S.K. RNA Interference-Mediated Knockdown of Genes Encoding Spore Wall Proteins Confers Protection against Nosema ceranae Infection in the European Honey Bee, Apis mellifera. Microorganisms 2021, 9, 505. https://doi.org/10.3390/microorganisms9030505
He N, Zhang Y, Duan XL, Li JH, Huang W-F, Evans JD, DeGrandi-Hoffman G, Chen YP, Huang SK. RNA Interference-Mediated Knockdown of Genes Encoding Spore Wall Proteins Confers Protection against Nosema ceranae Infection in the European Honey Bee, Apis mellifera. Microorganisms. 2021; 9(3):505. https://doi.org/10.3390/microorganisms9030505
Chicago/Turabian StyleHe, Nan, Yi Zhang, Xin Le Duan, Jiang Hong Li, Wei-Fone Huang, Jay D. Evans, Gloria DeGrandi-Hoffman, Yan Ping Chen, and Shao Kang Huang. 2021. "RNA Interference-Mediated Knockdown of Genes Encoding Spore Wall Proteins Confers Protection against Nosema ceranae Infection in the European Honey Bee, Apis mellifera" Microorganisms 9, no. 3: 505. https://doi.org/10.3390/microorganisms9030505
APA StyleHe, N., Zhang, Y., Duan, X. L., Li, J. H., Huang, W.-F., Evans, J. D., DeGrandi-Hoffman, G., Chen, Y. P., & Huang, S. K. (2021). RNA Interference-Mediated Knockdown of Genes Encoding Spore Wall Proteins Confers Protection against Nosema ceranae Infection in the European Honey Bee, Apis mellifera. Microorganisms, 9(3), 505. https://doi.org/10.3390/microorganisms9030505








