Symphyonema bifilamentata sp. nov., the Right Fischerella ambigua 108b: Half a Decade of Research on Taxonomy and Bioactive Compounds in New Light
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Strain
2.2. Morphological Characterization
2.3. DNA Extraction, Amplification and Sequencing
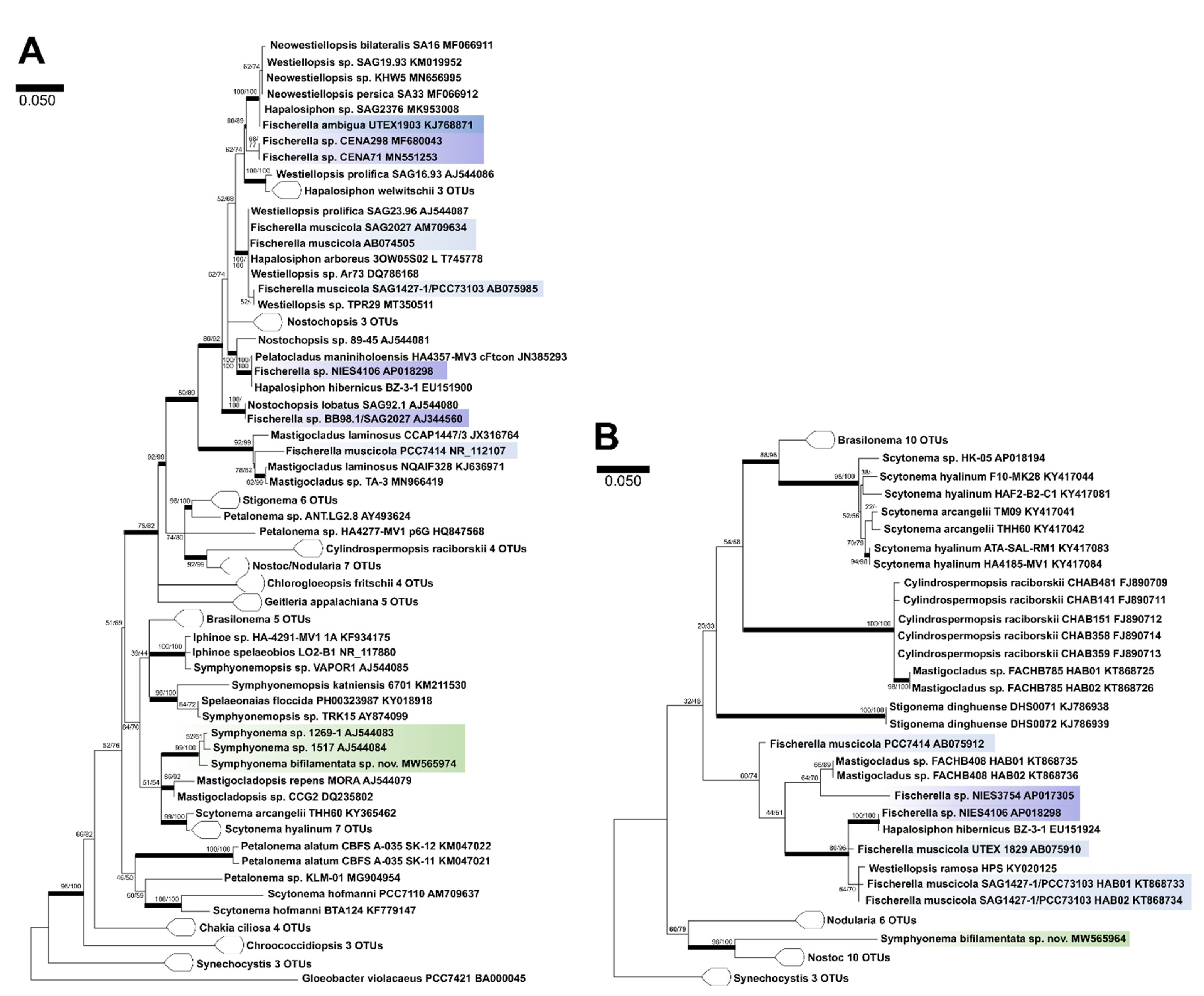
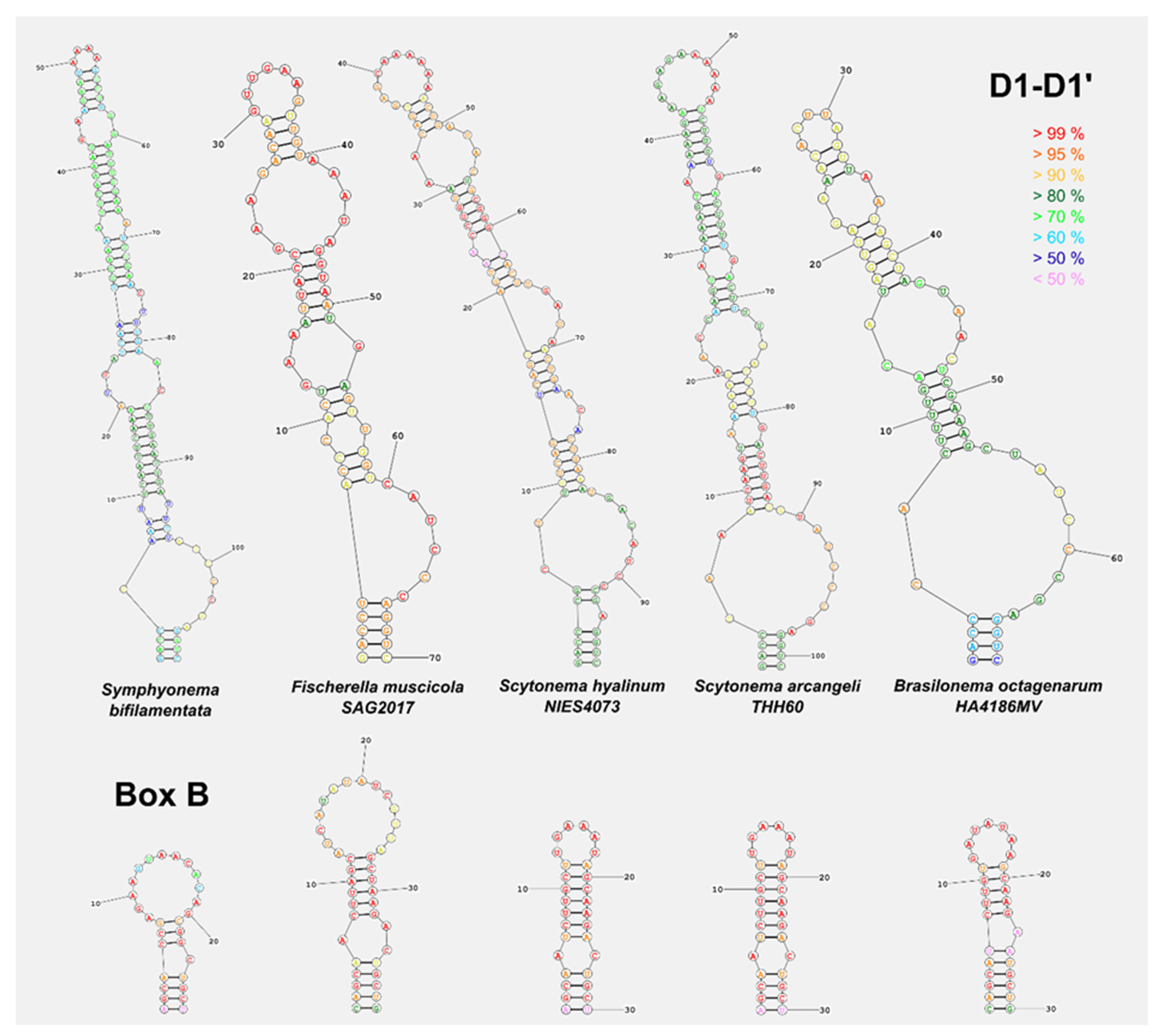
2.4. Molecular Characterization
2.5. Holotype Preparation
3. Results
3.1. Taxonomic treatment
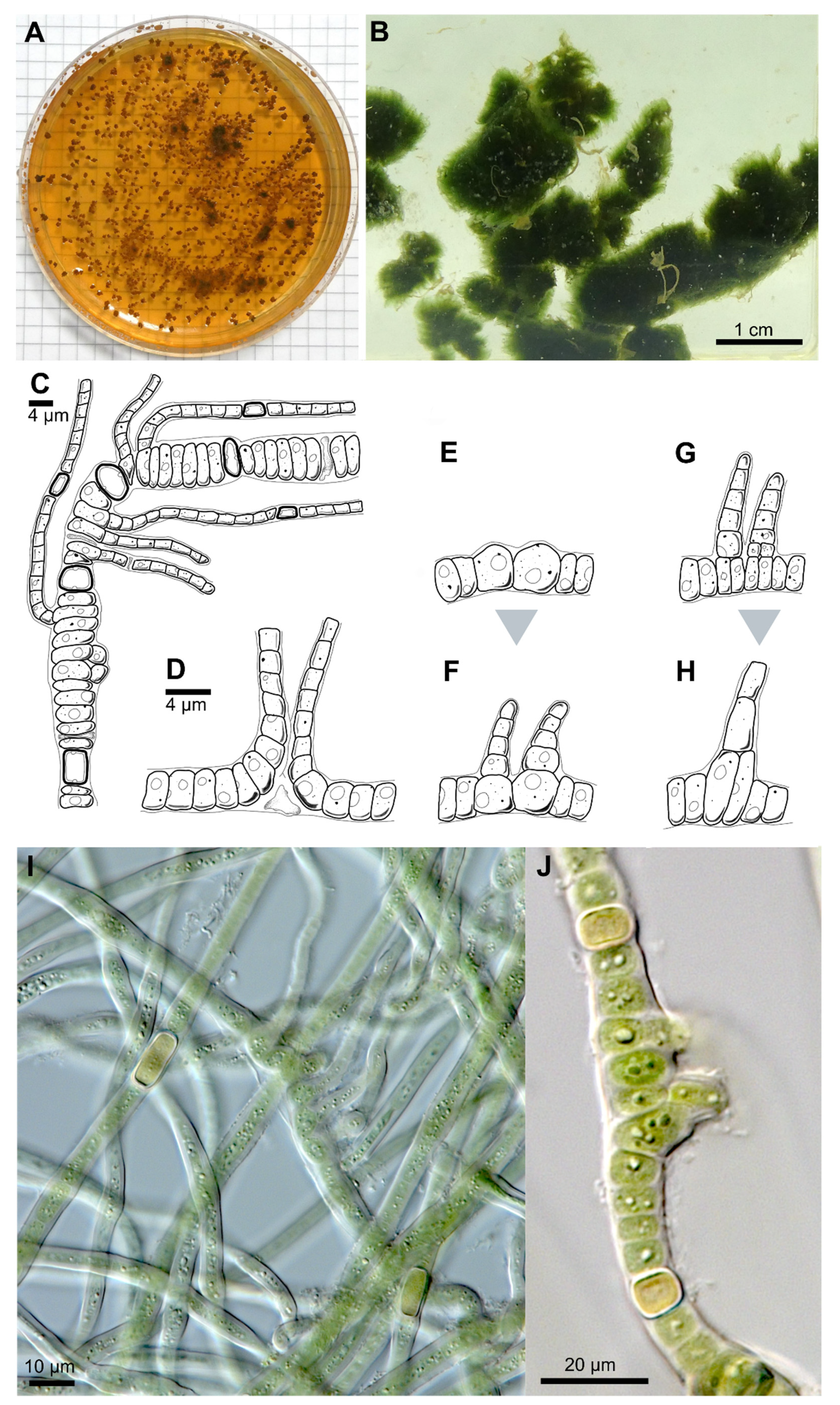
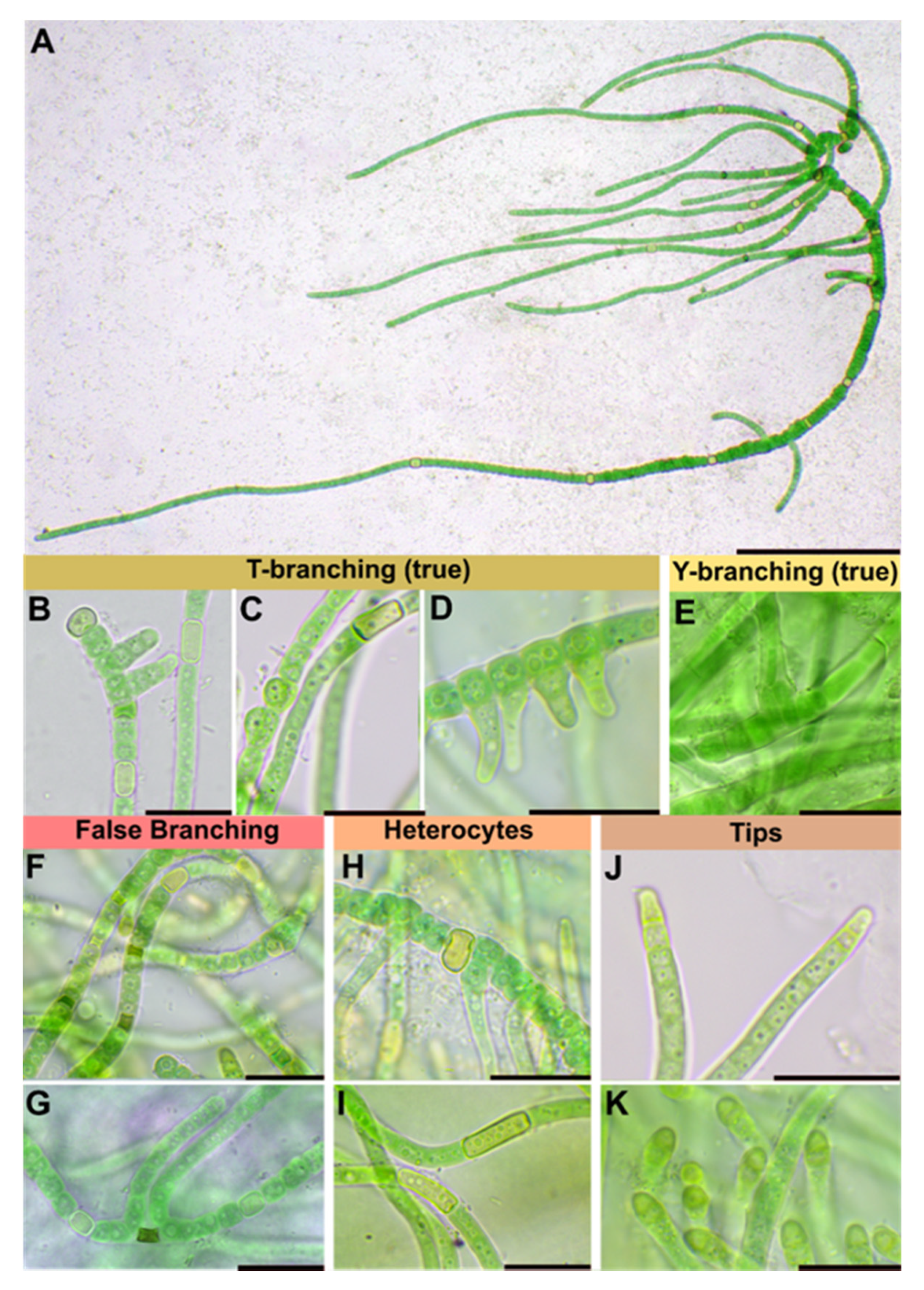
3.1.1. Symphyonema C-C. Jao 1944; Emend. P. Jung, B. Buedel et M. Lakatos
3.1.2. Symphyonema bifilamentata sp. nov. P. Jung, B. Buedel et M. Lakatos
4. Discussion
4.1. Taxonomic Significance
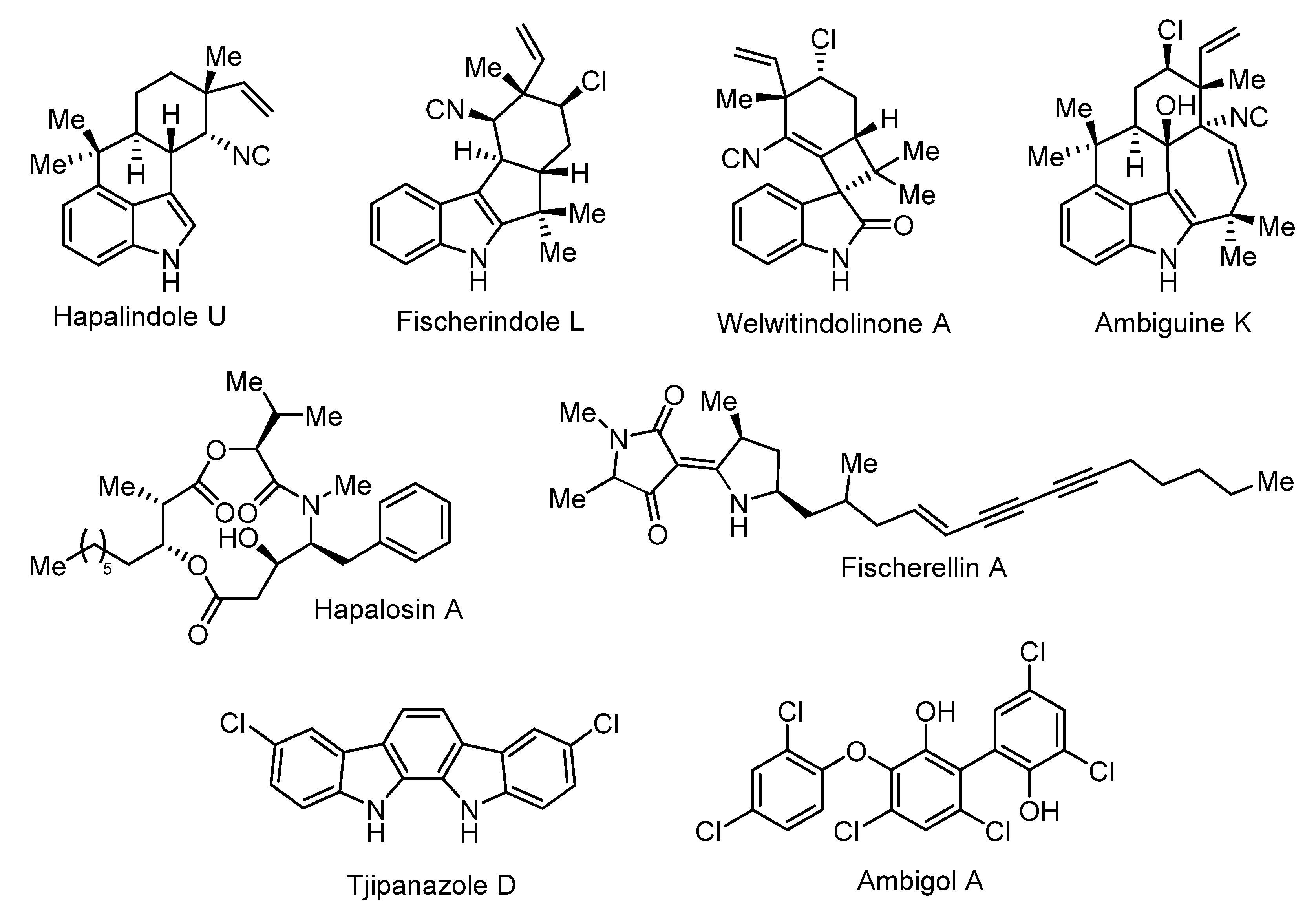
4.2. Bioactive Compound Biosynthesis: Symphyonema vs. Fischerella
| Compound Family | Producing Organism | Natural Product Class | Proposed Bioactivity | Reference |
|---|---|---|---|---|
| Ambigols | Symphyonema bifilamentata97.28 | Polyhalogenated aromatics | Antibiotic, antifungal, molluscicidal | [14,15,16,17,58] |
| 2,4-dichlorobenzoic acid | Symphyonema bifilamentata97.28 | [15] | ||
| Tjipanazoles | Symphyonema bifilamentata97.28 Tolypothrix tjipanasensis | indolo[2,3-a]carbazoles | Antifungal | [18,55,59] |
| Aranazoles | Fischerella sp. PCC 9339 | Halogenated NRPS/PKS | [49] | |
| Fischerellins | Fischerella muscicola | Aminoacylpolyketide | Antifungal, herbicidal | [46,47,60,61] |
| Westiellopsis sp. SAG 20.93 | ||||
| Ambiguines | Fischerella sp. Fischerella ambigua Fischerella ambigua UTEX 1903 Hapalosiphon sp. Hapalosiphon delicatulus Hapalosiphon hibemicus BZ-3-1 Westiellopsis prolifica EN-3-1 | Halogenated Indole alkaloid | Antimicrobial, phytotoxic | [62,63,64,65,66,67] |
| Fischerindoles | Fischerella sp. SAG 46.79 Fischerella muscicola Fischerella muscicola UTEX 1829 | Halogenated Indole alkaloids | Antifungal | [60,68] |
| Hapalindoles | Fischerella ambigua UTEX 1903 Fischerella sp. Fischerella sp. CENA 19 Fischerella muscicola UTEX LB1829 Westiellopsis sp. SAG 20.93 | Halogenated Indole alkaloids | Insecticidal | [61,66,69,70,71,72] |
| Welwitindolinones | Fischerella muscicola HG-39-5 Fischerella major HX-7-4 Hapalosiphon welwitschii IC-52-3 Westiella intricata HT-29-1 | Halogenated Indole alkaloids | [40,73] | |
| Hapalindolinones | Fischerella sp. ATCC 52558 | Indole alkaloids | [74] | |
| 13-hydroxydechlorofontonamide | Fischerella muscicola UTEX LB1829 | Indole alkaloids | [61] | |
| Mycosporine-like amino acids | Fischerella sp. PCC9339 Fischerella sp. 9431 | [43] | ||
| Microcystins | Fischerella sp. NQAIF311 Fischerella sp. CENA161 Fischerella major | NRPS/PKS | Hapatotoxin | [44,45,75,76] |
| Parsiguine | Fischerella ambigua PTCC 1635 | Antimicrobial | [48] | |
| Hapalosin | Hapalosiphon welwitschii IC-52-3 Fischerella sp. PCC9431 | NRPS/PKS | Multidrug-resistance reversing activity | [37,40,41,42] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anagnostidis, K.; Komárek, J. Modern approach to the classification system of Cyanophytes 5-Stigonematales. Archiv Für Hydrobiol. 1990, 86, 1–73. [Google Scholar]
- Gugger, M.F.; Hoffmann, L. Polyphyly of true branching cyanobacteria (Stigonematales). Int. J. Syst. Evol. Microbiol. 2004, 54, 349–357. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Bauersachs, T.; Miller, S.R.; Gugger, M.; Mudimu, O.; Friedl, T.; Schwark, L. Heterocyte glycolipids indicate polyphyly of stigonematalean cyanobacteria. Phytochemistry 2019, 166, 112059. [Google Scholar] [CrossRef] [PubMed]
- Alcorta, J.; Vergara-Barros, P.; Antonaru, L.A.; Alcamán-Arias, M.E.; Nürnberg, D.J.; Díez, B. Fischerella thermalis: A model organism to study thermophilic diazotrophy, photosynthesis and multicellularity in cyanobacteria. Extremophiles 2019, 23, 635–647. [Google Scholar] [CrossRef]
- Castenholz, R.W. The thermophilic cyanophytes of Iceland and the upper temperature limit. J. Phycol. 1969, 5, 360–368. [Google Scholar] [CrossRef]
- Kaštovský, J.; Johansen, J.R. Mastigocladus laminosus (Stigonematales, Cyanobacteria): Phylogenetic relationship of strains from thermal springs to soil-inhabiting genera of the order and taxonomic implications for the genus. Phycologia 2008, 47, 307–320. [Google Scholar] [CrossRef]
- Lamprinou, V.; Christodoulou, M.; Hernandez-Marine, M.; Parmakelis, A.; Economou-Amilli, A. Spelaeonaias gen. nov., a new true-branched cyanobacterium from Cave Vlychada (Diros, Peloponnese, Greece). Phytotaxa 2016, 282, 171–185. [Google Scholar] [CrossRef]
- Friedmann, E.I. Geitleria calcarea n. gen. et n. sp.: A new Atmophytic Lime-incrusting Blue-green Alga. Botaniska Notiser 1955, 108, 439–445. [Google Scholar]
- Asencio, A.D.; Aboal, M.; Hoffmann, L. A new cave-inhabiting blue-green alga: Symphyonema cavernicolum sp. nova (Mastigocladaceae, Stigonematales). Algol. Stud. Arch. für Hydrobiol. 1996, 83, 73–82. [Google Scholar] [CrossRef]
- McGregor, G.B. Freshwater Cyanobacteria of North-Eastern Australia: 3. Nostocales. Phytotaxa 2018, 359, 1–166. [Google Scholar] [CrossRef]
- Jao, C.-C. Studies on the fresh-water algae of China, XIII New Myxophyceae from Kwangsi. Sinensia 1944, 15, 75–90. [Google Scholar]
- Sarthou, C.; Thérézien, Y.; Couté, A. Cyanophycées de l’inselberg des Nouragues (Guyane française). Nova Hedwig. 1995, 61, 85–109. [Google Scholar]
- Falch, B.S.; Koenig, G.M.; Wright, A.D.; Sticher, O.; Ruegger, H.; Bernardinelli, G. Ambigol A and B: New biologically active polychlorinated aromatic compounds from the terrestrial blue-green alga Fischerella ambigua. J. Org. Chem. 1993, 58, 6570–6575. [Google Scholar] [CrossRef]
- Wright, A.D.; Papendorf, O.; König, G.M. Ambigol C and 2, 4-Dichlorobenzoic Acid, Natural Products Produced by the Terrestrial Cyanobacterium Fischerella ambigua. J. Nat. Prod. 2005, 68, 459–461. [Google Scholar] [CrossRef]
- Chilczuk, T.; Monson, R.; Schmieder, P.; Christov, V.; Enke, H.; Salmond, G.; Niedermeyer, T.H.J. Ambigols from the Cyanobacterium Fischerella ambigua increase Prodigiosin Production in Serratia spp. ACS Chem. Biol. 2020, 15, 2929–2936. [Google Scholar] [CrossRef]
- Duell, E.R.; Milzarek, T.M.; El Omari, M.; Linares-Otoya, L.J.; Schäberle, T.F.; König, G.M.; Gulder, T.A. Identification, cloning, expression and functional interrogation of the biosynthetic pathway of the polychlorinated triphenyls ambigol A–C from Fischerella ambigua 108b. Org. Chem. Front. 2020, 7, 3193–3201. [Google Scholar] [CrossRef]
- Chilczuk, T.; Schäberle, T.F.; Vahdati, S.; Mettal, U.; El Omari, M.; Enke, H.; Wiese, M.; König, G.M.; Niedermeyer, T.H.J. Halogenation-guided chemical screening provides insight into tjipanazole biosynthesis by the cyanobacterium Fischerella ambigua. ChemBioChem 2020, 21, 2170–2177. [Google Scholar] [CrossRef]
- Hyde, K.D.; Zhang, Y. Epitypification: Should we epitypify? J. Zheijang Univ. Sci. B 2008, 9, 842–846. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (Order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Williams, L.; Jung, P.; Zheng, L.J.; Maier, S.; Peer, T.; Grube, M.; Weber, B.; Büdel, B. Assessing recovery of biological soil crusts across a latitudinal gradient in Western Europe. Rest. Ecol. 2017, 26, 543–554. [Google Scholar] [CrossRef]
- Marin, B.; Nowack, E.; Melkonian, M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist 2005, 156, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Rudi, K.; Skulberg, O.M.; Jakobsen, K.S. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J. Bact. 1998, 180, 3453–3461. [Google Scholar] [CrossRef] [PubMed]
- Wilmotte, A.; Van der Auwera, G.; De Wachter, R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (“Mastigocladus laminosus HTF”) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef]
- Mikhailyuk, T.I.; Vinogradova, O.N.; Glaser, K.; Karsten, U. New taxa for the flora of Ukraine, in the context of modern approaches to taxonomy of Cyanoprokaryota/Cyanobacteria. Int. J. Algae 2016, 18, 301–320. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Wilde, S.B.; Johansen, J.R.; Wilde, H.D.; Jiang, P.; Bartelme, B.; Haynie, R.S. Aetokthonos hydrillicola gen. et sp. nov.: Epiphytic cyanobacteria on invasive aquatic plants implicated in Avian Vacuolar Myelinopathy. Phytotaxa 2014, 181, 243–260. [Google Scholar] [CrossRef]
- Romanenko, P.A.; Vinogradova, O.N.; Romanenko, E.A.; Mikhailyuk, T.I.; Babenko, L.M.; Ivannikov, R.; Scherbak, N.N. Morphological and Molecular Characterization of the Representative of Brasilonema (Scytonemataceae, Cyanoprokaryota) from the Tropical Greenhouse in Kiev (Ukraine). Int. J. Algae 2020, 22, 103–122. [Google Scholar] [CrossRef]
- Johansen, J.R.; Mareš, J.; Pietrasiak, N.; Bohunická, M.; Zima, J., Jr.; Štenclová, L.; Hauer, T. Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria). PLoS ONE 2017, 12, e0186393. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.H.; Li, D.Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Regnum Vegetabile 159; Koeltz Botanical Books: Glashütten, Germany, 2018; p. 159. [Google Scholar]
- Singh, P.; Fatma, A.; Mishra, A.K. Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann. Microbiol. 2015, 65, 799–807. [Google Scholar] [CrossRef]
- Gomont, M.A. Note sur le Scytonema ambiguum Kützing. J. Bot. 1895, 9, 49–52. [Google Scholar]
- Thurston, E.L.; Ingram, L.O. Morphology and fine structure of Fischerella ambigua. J. Phycol. 1971, 7, 203–210. [Google Scholar] [CrossRef]
- Moore, R.E.; Cheuk, C.; Patterson, G.M. Hapalindoles: New alkaloids from the blue-green alga Hapalosiphon fontinalis. J. Am. Chem. Soc. 1984, 106, 6456–6457. [Google Scholar] [CrossRef]
- Micallef, M.L.; D’Agostino, P.M.; Sharma, D.; Viswanathan, R.; Moffitt, M.C. Genome mining for natural product biosynthetic gene clusters in the Subsection V cyanobacteria. BMC Genom. 2015, 16, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Naumann, K. Influence of chlorine substituents on biological activity of chemicals: A review. Pest Manag. Sci. Former. Pestic. Sci. 2000, 56, 3–21. [Google Scholar] [CrossRef]
- Bhat, V.; Dave, A.; MacKay, J.A.; Rawal, V.H. The chemistry of hapalindoles, fischerindoles, ambiguines, and welwitindolinones. Alkaloids Chem. Biol. 2014, 73, 65–160. [Google Scholar] [PubMed]
- Stratmann, K.; Burgoyne, D.L.; Moore, R.E.; Patterson, G.M.; Smith, C.D. Hapalosin, a cyanobacterial cyclic depsipeptide with multidrug-resistance reversing activity. J. Org. Chem. 1994, 59, 7219–7226. [Google Scholar] [CrossRef]
- D’Agostino, P.M.; Gulder, T.A. Direct pathway cloning combined with sequence-and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression. ACS Synth. Biol. 2018, 7, 1702–1708. [Google Scholar] [CrossRef]
- Pereira, J.O.; de Souza, A.Q.L.; de Souza, A.D.L.; de Castro França, S.; de Oliveira, L.A. Overview on biodiversity, chemistry, and biotechnological potential of microorganisms from the Brazilian Amazon. In Diversity and Benefits of Microorganisms from the Tropics; Springer: Cham, Switzerland, 2017; pp. 71–103. [Google Scholar]
- Yang, G.; Cozad, M.A.; Holland, D.A.; Zhang, Y.; Luesch, H.; Ding, Y. Photosynthetic production of sunscreen shinorine using an engineered cyanobacterium. ACS Synth. Biol. 2018, 7, 664–671. [Google Scholar] [CrossRef]
- Cirés, S.; Alvarez-Roa, C.; Wood, S.A.; Puddick, J.; Loza, V.; Heimann, K. First report of microcystin-producing Fischerella sp. (Stigonematales, Cyanobacteria) in tropical Australia. Toxicon 2014, 88, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Fiore, M.F.; Genuário, D.B.; da Silva, C.S.P.; Shishido, T.K.; Moraes, L.A.B.; Neto, R.C.; Silva-Stenico, M.E. Microcystin production by a freshwater spring cyanobacterium of the genus Fischerella. Toxicon 2009, 53, 754–761. [Google Scholar] [CrossRef]
- Hagmann, L.; Jüttner, F. Fischerellin A, a novel photosystem-II-inhibiting allelochemical of the cyanobacterium Fischerella muscicola with antifungal and herbicidal activity. Tetrahedron Lett. 1996, 37, 6539–6542. [Google Scholar] [CrossRef]
- Papke, U.; Gross, E.M.; Francke, W. Isolation, identification and determination of the absolute configuration of Fischerellin, B. A new algicide from the freshwater cyanobacterium Fischerella muscicola (Thuret). Tetrahedron Lett. 1997, 38, 379–382. [Google Scholar] [CrossRef]
- Ghasemi, Y.; Yazdi, M.T.; Shafiee, A.; Amini, M.; Shokravi, S.; Zarrini, G. Parsiguine, a novel antimicrobial substance from Fischerella ambigua. Pharm. Biol. 2004, 42, 318–322. [Google Scholar] [CrossRef]
- Moosmann, P.; Ueoka, R.; Gugger, M.; Piel, J. Aranazoles: Extensively Chlorinated Nonribosomal Peptide–Polyketide Hybrids from the Cyanobacterium Fischerella sp. PCC 9339. Org. Lett. 2018, 20, 5238–5241. [Google Scholar] [CrossRef]
- Wagner, C.; Molitor, I.M.; König, G.M. Critical view on the monochlorodimedone assay utilized to detect haloperoxidase activity. Phytochemistry 2008, 69, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Milzarek, T.M.; Gulder, T.A. Total Synthesis of the Ambigols: A Cyanobacterial Class of Polyhalogenated Natural Products. Org. Lett. 2021, 23, 102–106. [Google Scholar] [CrossRef]
- Agarwal, V.; Blanton, J.M.; Podell, S.; Taton, A.; Schorn, M.A.; Busch, J.; Moore, B.S. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol. 2017, 13, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; El Gamal, A.A.; Yamanaka, K.; Poth, D.; Kersten, R.D.; Schorn, M.; Moore, B.S. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol. 2014, 10, 640–647. [Google Scholar] [CrossRef]
- Onaka, H.; Tanifguchi, S.I.; Igarashi, Y.; Furumai, T. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J. Antib. 2002, 55, 1063–1071. [Google Scholar] [CrossRef]
- Bonjouklian, R.; Smitka, T.A.; Doolin, L.E.; Molloy, R.M.; Debono, M.; Shaffer, S.A.; Patterson, G.M. Tjipanazoles, new antifungal agents from the blue-green alga Tolypothrix Tjipanasensis. Tetrahedron 1991, 47, 7739–7750. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Booth, T.J.; Chooi, Y.H.H. cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. bioRxiv 2020, 11, 370601. [Google Scholar]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Müller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Falch, B.S.; König, G.M.; Sticher, O.; Wright, A.D. Studies on the glycolipid content of the cyanobacterium Fischerella ambigua. Planta Med. 1995, 61, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Falch, B.S.; König, G.M.; Wright, A.D.; Sticher, O.; Angerhofer, C.K.; Pezzuto, J.M.; Bachmann, H. Biological activities of cyanobacteria: Evaluation of extracts and pure compounds. Planta Med. 1995, 61, 321–328. [Google Scholar] [CrossRef]
- Kim, H.; Krunic, A.; Lantvit, D.; Shen, Q.; Kroll, D.J.; Swanson, S.M.; Orjala, J. Nitrile-containing fischerindoles from the cultured cyanobacterium Fischerella sp. Tetrahedron 2012, 68, 3205–3209. [Google Scholar] [CrossRef]
- Kim, H.; Lantvit, D.; Hwang, C.H.; Kroll, D.J.; Swanson, S.M.; Franzblau, S.G.; Orjala, J. Indole alkaloids from two cultured cyanobacteria, Westiellopsis sp. and Fischerella muscicola. Bioorg. Med. Chem. 2012, 20, 5290–5295. [Google Scholar] [CrossRef]
- Smitka, T.A.; Bonjouklian, R.; Doolin, L.; Jones, N.D.; Deeter, J.B.; Yoshida, W.Y.; Patterson, G.M. Ambiguine isonitriles, fungicidal hapalindole-type alkaloids from three genera of blue-green algae belonging to the Stigonemataceae. J. Org. Chem. 1992, 57, 857–861. [Google Scholar] [CrossRef]
- Huber, U.; Moore, R.E.; Patterson, G.M. Isolation of a nitrile-containing indole alkaloid from the terrestrial blue-green alga Hapalosiphon delicatulus. J. Nat. Prod. 1998, 61, 1304–1306. [Google Scholar] [CrossRef]
- Raveh, A.; Carmeli, S. Antimicrobial ambiguines from the cyanobacterium Fischerella sp. collected in Israel. J. Nat. Prod. 2007, 70, 196–201. [Google Scholar] [CrossRef]
- Mo, S.; Krunic, A.; Chlipala, G.; Orjala, J. Antimicrobial ambiguine isonitriles from the cyanobacterium Fischerella ambigua. J. Nat. Prod. 2009, 72, 894–899. [Google Scholar] [CrossRef]
- Mo, S.; Krunic, A.; Santarsiero, B.D.; Franzblau, S.G.; Orjala, J. Hapalindole-related alkaloids from the cultured cyanobacterium Fischerella ambigua. Phytochem 2010, 71, 2116–2123. [Google Scholar] [CrossRef]
- Hillwig, M.L.; Zhu, Q.; Liu, X. Biosynthesis of ambiguine indole alkaloids in cyanobacterium Fischerella ambigua. ACS Chem. Biol. 2014, 9, 372–377. [Google Scholar] [CrossRef]
- Park, A.; Moore, R.E.; Patterson, G.M. Fischerindole L, a new isonitrile from the terrestrial blue-green alga Fischerella muscicola. Tetrahedron Lett. 1992, 33, 3257–3260. [Google Scholar] [CrossRef]
- Etchegaray, A.; Rabello, E.; Dieckmann, R.; Moon, D.H.; Fiore, M.F.; Von Döhren, H.; Neilan, B.A. Algicide production by the filamentous cyanobacterium Fischerella sp. CENA 19. J. Appl. Phycol. 2004, 16, 237–243. [Google Scholar] [CrossRef]
- Asthana, R.K.; Srivastava, A.; Singh, A.P.; Singh, S.P.; Nath, G.; Srivastava, R.; Srivastava, B.S. Identification of an antimicrobial entity from the cyanobacterium Fischerella sp. isolated from bark of Azadirachta indica (Neem) tree. J. Appl. Phycol. 2006, 18, 33–39. [Google Scholar] [CrossRef]
- Cagide, E.; Becher, P.G.; Louzao, M.C.; Espina, B.; Vieytes, M.R.; Juttner, F.; Botana, L.M. Hapalindoles from the cyanobacterium Fischerella: Potential sodium channel modulators. Chem. Res. Toxicol. 2014, 27, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Knoot, C.J.; Khatri, Y.; Hohlman, R.M.; Sherman, D.H.; Pakrasi, H.B. Engineered production of hapalindole alkaloids in the cyanobacterium Synechococcus sp. UTEX 2973. ACS Synth. Biol. 2019, 8, 1941–1951. [Google Scholar] [CrossRef]
- Jimenez, J.I.; Huber, U.; Moore, R.E.; Patterson, G.M. Oxidized welwitindolinones from terrestrial Fischerella spp. J. Nat. Prod. 1999, 62, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.E.; Hirsch, C.F.; Springer, J.P.; Pettibone, D.J.; Zink, D.L. Unusual cyclopropane-containing hapalindolinones from a cultured cyanobacterium. J. Org. Chem. 1987, 52, 3704–3706. [Google Scholar] [CrossRef]
- Heck, K.; Alvarenga, D.O.; Shishido, T.K.; Varani, A.M.; Dörr, F.A.; Pinto, E.; Rouhiainen, L.; Jokela, J.; Sivonen, K.; Fiore, M.F. Biosynthesis of microcystin hepatotoxins in the cyanobacterial genus Fischerella. Toxicon 2018, 141, 43–50. [Google Scholar] [CrossRef]
- Batsalova, T.; Basheva, D.; Bardarov, K.; Bardarov, V.; Dzhambazov, B.; Teneva, I. Assessment of the cytotoxicity, antioxidant activity and chemical composition of extracts from the cyanobacterium Fischerella major Gomont. Chemosphere 2019, 218, 93–103. [Google Scholar] [CrossRef] [PubMed]
| Symphyonema sinense C.-C. Jao | Symphyonema sinense var. minor C.-C. Jao | Symphyonema cavernicolum Asencio, Aboal et Hoffmann | Symphyonema kaboorum G. B. McGregor | Symphyonema bifilamentata sp. nov. P. Jung, B. Buedel et M. Lakatos | Fischerella ambigua Kützing ex Bornet et Flahault) Gomont | Symphyonema sp. 1269-1 | Symphyonema sp. 1517 | |
|---|---|---|---|---|---|---|---|---|
| Origin | Aerophytic, wet calcareous rocks, Kwangsi, China; Jao 1944 | Epilithic (subaerial), granite inselberg French Guiana; crest of Montagnes Balenfois (N 4°5′, W 52°42′) 1989; Sarthou et al., 1995 | Epilithic/chasmoendolithic, limestone cave, karstic region, Los Almadenes gorge, Spain; Asencio et al., 1996 | Aquatic; surface of large, submerged Logs/woody debris in the littoral zone of an acidic, coastal oligotrophic lake, Naree Budjong Djara National Park, Stradbroke Island, Queensland, Australia; McGregor 2018 | Edaphic, shallow, hollow, wet soil, Mellingen, Switzerland, isolated 1965; this work | Edaphic, wet, unpolluted soil, among mosses; Zurich; Gomont 1895 | Edaphic, soil, Papua New Guinea, isolated 1986; Gugger and Hoffmann, 2004 | Edaphic, soil, Papua New Guinea, isolated 1986; Gugger and Hoffmann, 2004 |
| Thallus | Pulvinate, later expanded, up to 1.5 cm thick, greyish-blue; filaments parallel arranged, mostly erect, | Greyish, interwoven filaments | Pulvinate, formed by more or less parallel arranged filaments | Pulvinate, up to 1 cm thick, dark green, wooly; interwoven filaments, on agar mainly creeping, parallel arranged rarely slightly erect; dyes liquid medium transparent-brownish | Prostrate, dark brown, or only in in solitary filaments | Creeping filaments | Creeping and erect filaments | |
| Branching Type | False-branching (scytonematoid), usually solitary, less frequently geminate | False branching, true-branching, reverse-type true branching | Mainly Y-type, but also T-type and false-branching (scytonematoid) | T- and Y-type true branching, false-branching (scytonematoid) | T-type true-branching towards all sides of the basal section, false-branching (scytonematoid) | T- and Y-type true branching, false-branching (scytonematoid) | Mainly Y-type but also T-type true-branching and false-branching (scytonematoid) | Mainly Y-type but also T-type true-branching and false-branching (scytonematoid) |
| Basal/main section | Filaments 9–12 µm wide, flexuose, irregularly ramified, densely intricate; trichomes 7–10 µm wide, commonly not constricted at cross walls; cells cylindrical, 15–38 µm long, up to 1.5, up to 3× longer than wide; blue green; apical cells rounded | Filaments 8–13 µm without ramification, simple; cell content homogeneous, greenish, cells cylindrical (6)8–14 µm × 2–4(5) µm | No differentiation; trichomes uniseriate, constricted at cross-walls, often tapering towards ends; cells irregular barrel shaped or cylindrical, isodiametric or longer than wide, 4–9.6 µm × 4–8 µm; pale blue green to violet, with scattered cyanophycin granules | Filaments 11–17 μm wide, erect, straight to irregularly flexuous, not tapered towards the ends, irregularly ramified, densely intricate, not constricted at the cross walls; Vegetative cells isodiametric, or up to 4 × longer than broad, shorter than broad towards the apices, (8.5–) 12.5–18.5 (–26) μm long × 4.8–10.5 μm wide, with fine granular contents, often vacuolate; apical cells rounded | Strictly divaricated from lateral branches; trichomes 3.3–6.6 µm wide, with necridia, mostly uniseriate, bloated but tapered/waisted towards heterocytes; cells barrel-shaped, rounded, squeezed, constricted at cross-walls, wider than long, 2.4–3.3 (6) µm × 1.3–1.7 µm, coarse granulated, with at least one big vacuole-like structure in each cell, blue-green | Creeping, irregular, coiled or flexuouse, 3–7 µm wide, cylindrical, not or slightly narrowed towards ends, sometimes fasciculated; monoseriate but sometimes biseriate, cells clearly constricted at cross walls, barrel shped, up to spherical 3–4 µm wide | not evaluated | not evaluated |
| Branches | Solitary, divaricated from basal sections | Slightly narrower than main filament | Strictly divaricated from basal sections, up to 200 µm long, uniseriate, without necridia; filaments straight, parallel; regular, not tapered towards the ends, fine granulated, fine vacuolated, cells regular, quadratic or slightly longer than wide, 2–2.3 µm × 2–2.8 µm, very slightly constricted at cross-walls, blue-green; apical tip rounded to slightly conical, yellowish, 2.8 µm × 1.8 µm; | Cells cylindrical, 2–3 µm wide and up to 4× longer than wide, terminal cells rounded | not evaluated | not evaluated | ||
| Heterocyst | Solitary, rare, 12–15 µm × 8–10, ±rectangular, rarely subglobose | Intercalary, cylindrical, 7–13 µm × 4–6 µm | Very rare, intercalary, cylindrical, 8.2 × 5.8 µm | solitary, intercalary, rarely located at the point of branching, spherical to elongated cylindrical, up to 3.5 × longer than broad, 9.0–18.5 (–26) μm × 5.5–13.8 μm | Solitary, rarely in pairs of two, thick-walled, yellow, intercalary; rectangular in branches, 2.3 µm × 3.6–5 (8.5) µm; subglobose, squeezed and boalated in basal section, 2–2.2 µm × 3.6–3.9 µm | Intercalary, cylindrical | Intercalary, rarely lateral-sessile | Intercalary, rarely lateral-sessile |
| Sheath | Slightly widened, not or partly slightly lamellated, initially hyaline, brownish with age | Yellow-brownish, 3–4 µm thick, stratified, marked | Hyaline, often lamellated, heavily lime-encrusted | Firm, hyaline mucilage, up to 25 μm wide, lamellated closer to the trichome | Hyaline, colorless, firm, limited, not lamellated, very tight, ruptured by the apical cells of lateral branches; wider and prominent in mature cultures | Colorless, up to yellow brown, lamellated in old filaments | not evaluated | not evaluated |
| Multiplication | Hormogonia, 30–35 µm × 9–10 µm | Apical hormogonia, 44–50 × 7 µm | Terminal hormogonia | not evaluated | Fragmentation of lateral branches | Long hormogonia | Hormogonia | Hormogonia |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, P.; D’Agostino, P.M.; Büdel, B.; Lakatos, M. Symphyonema bifilamentata sp. nov., the Right Fischerella ambigua 108b: Half a Decade of Research on Taxonomy and Bioactive Compounds in New Light. Microorganisms 2021, 9, 745. https://doi.org/10.3390/microorganisms9040745
Jung P, D’Agostino PM, Büdel B, Lakatos M. Symphyonema bifilamentata sp. nov., the Right Fischerella ambigua 108b: Half a Decade of Research on Taxonomy and Bioactive Compounds in New Light. Microorganisms. 2021; 9(4):745. https://doi.org/10.3390/microorganisms9040745
Chicago/Turabian StyleJung, Patrick, Paul M. D’Agostino, Burkhard Büdel, and Michael Lakatos. 2021. "Symphyonema bifilamentata sp. nov., the Right Fischerella ambigua 108b: Half a Decade of Research on Taxonomy and Bioactive Compounds in New Light" Microorganisms 9, no. 4: 745. https://doi.org/10.3390/microorganisms9040745
APA StyleJung, P., D’Agostino, P. M., Büdel, B., & Lakatos, M. (2021). Symphyonema bifilamentata sp. nov., the Right Fischerella ambigua 108b: Half a Decade of Research on Taxonomy and Bioactive Compounds in New Light. Microorganisms, 9(4), 745. https://doi.org/10.3390/microorganisms9040745