Functional and Phylogenetic Diversity of BSH and PVA Enzymes
Abstract
1. Introduction
2. An Overview of Enzyme Function and Distribution
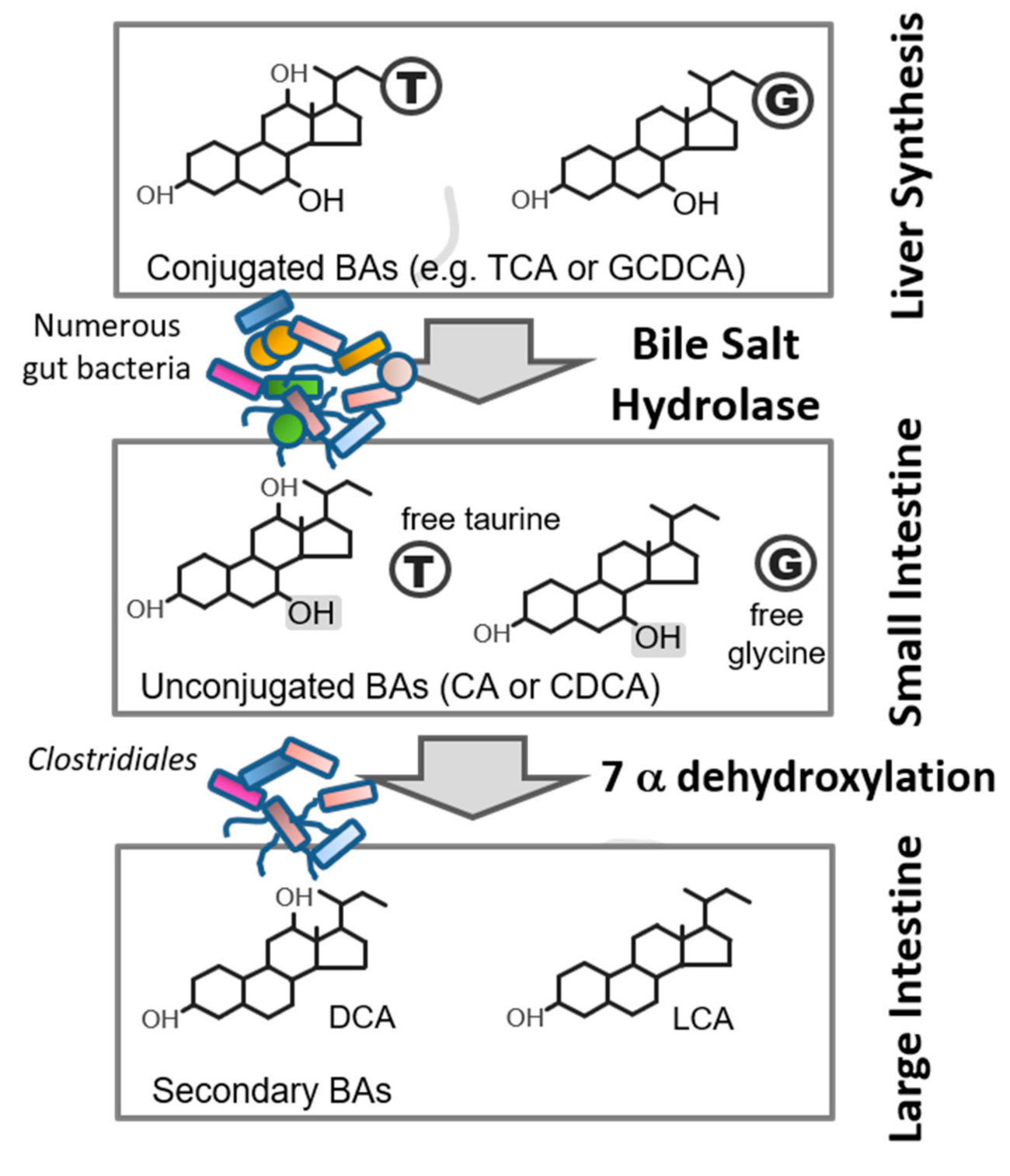
2.1. BSH Function and Distribution
2.2. PVA Enzyme Activities and Distribution
3. Biochemistry of BSH and PVA Enzymes
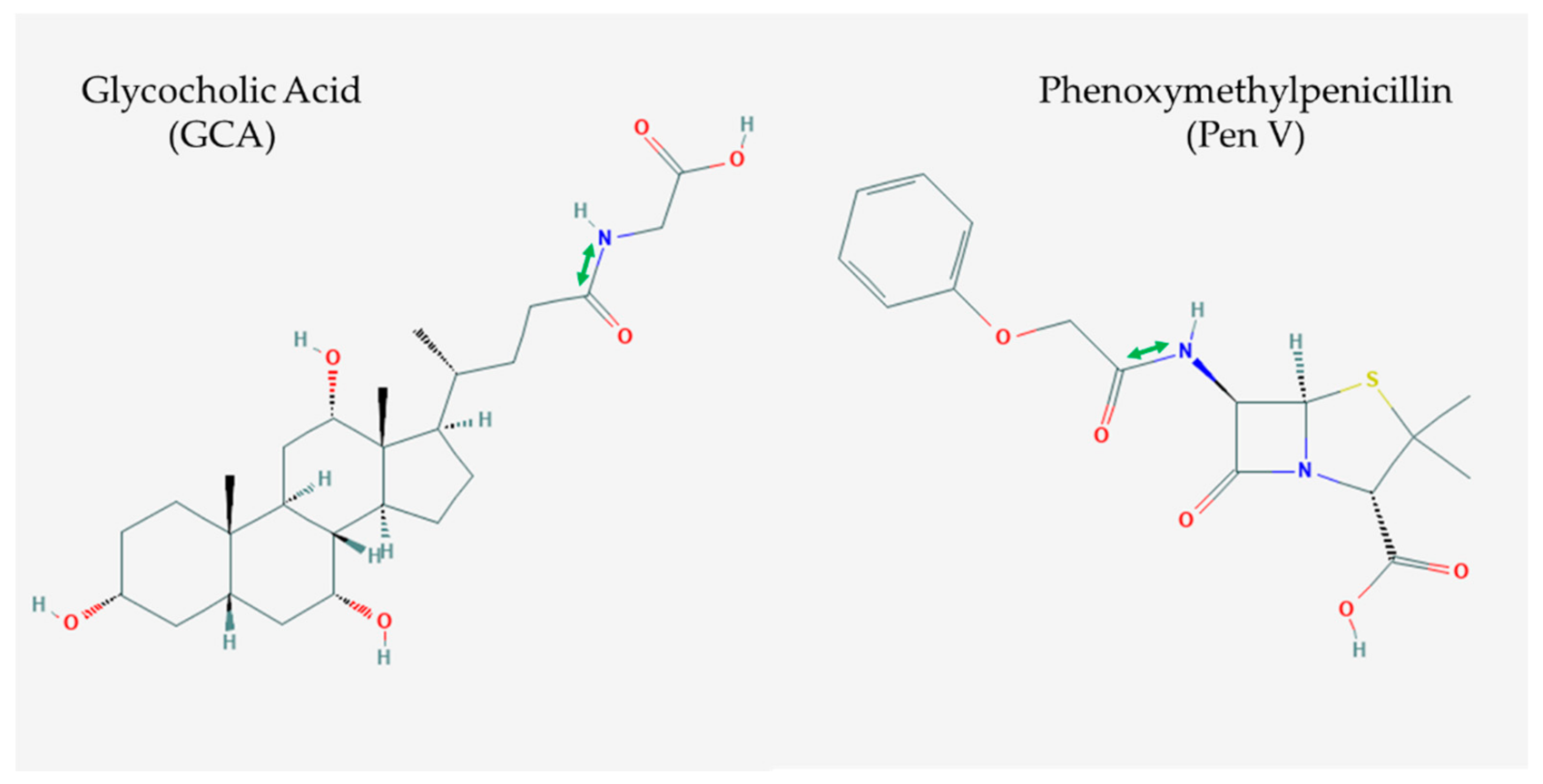
3.1. Biochemical Overview of BSH and PVA Enzymes
3.2. Catalytic Mechanism
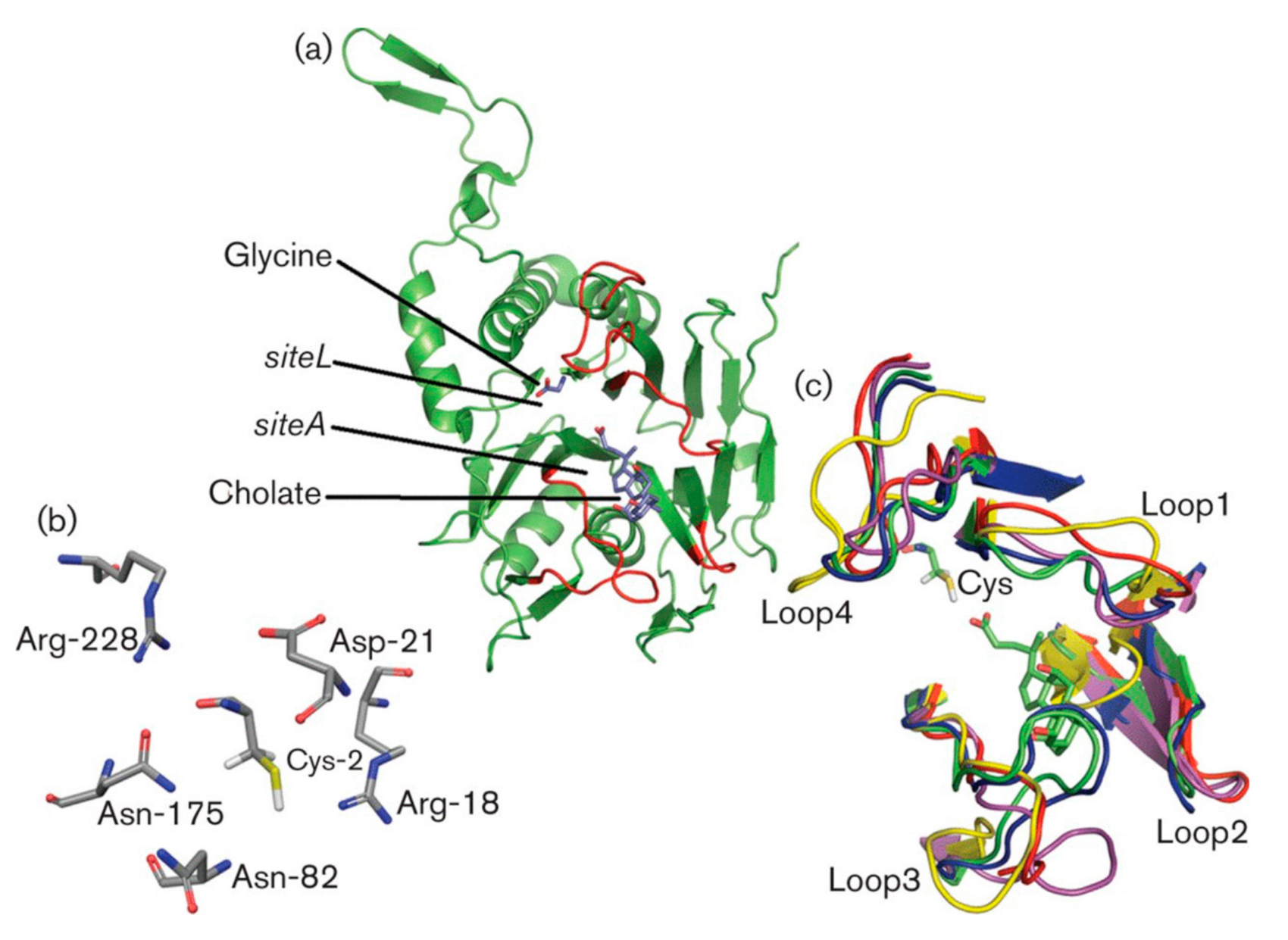
3.3. Substrate Binding and Specificity
3.4. Key Residues Involved in Activity
4. In Silico Differentiation and Functional Characterisation
5. Potential Role of BSH and PVA in Environmental Survival, Host Interaction and Metabolism
5.1. Role of BSH in Host Physiology and Metabolism
5.2. Role of PVA in Environmental Modulation and Survival
6. Conclusions
Funding
Conflicts of Interest
References
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Oinonen, C.; Rouvinen, J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000, 9, 2329–2337. [Google Scholar] [CrossRef] [PubMed]
- Rossocha, M.; Schultz-Heienbrok, R.; von Moeller, H.; Coleman, J.P.; Saenger, W. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 2005, 44, 5739–5748. [Google Scholar] [CrossRef]
- Brannigan, J.A.; Dodson, G.; Duggleby, H.J.; Moody, P.C.; Smith, J.L.; Tomchick, D.R.; Murzin, A.G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 1995, 378, 416–419. [Google Scholar] [CrossRef] [PubMed]
- Philem, P.D.; Yadav, Y.; Vellore Sunder, A.; Ghosh, D.; Prabhune, A.; Ramasamy, S. Structural and enzymatic analysis of a dimeric cholylglycine hydrolase like acylase active on N-acyl homoserine lactones. Biochimie 2020, 177, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Chand, D.; Avinash, V.S.; Yadav, Y.; Pundle, A.V.; Suresh, C.G.; Ramasamy, S. Molecular features of bile salt hydrolases and relevance in human health. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2981–2991. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.M.; Siezen, R.J.; de Vos, W.M.; Kleerebezem, M. Improved annotation of conjugated bile acid hydrolase superfamily members in Gram-positive bacteria. Microbiology 2008, 154, 2492–2500. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Briner Crawley, A.; Theriot, C.M.; Barrangou, R. The Lactobacillus Bile Salt Hydrolase Repertoire Reveals Niche-Specific Adaptation. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Panigrahi, P.; Sule, M.; Sharma, R.; Ramasamy, S.; Suresh, C.G. An improved method for specificity annotation shows a distinct evolutionary divergence among the microbial enzymes of the cholylglycine hydrolase family. Microbiology 2014, 160, 1162–1174. [Google Scholar] [CrossRef]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 2017, 56, 54–65. [Google Scholar] [CrossRef]
- Winston, J.A.; Theriot, C.M. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 2020, 11, 158–171. [Google Scholar] [CrossRef]
- Bustos, A.Y.; Font de Valdez, G.; Fadda, S.; Taranto, M.P. New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res. Int. 2018, 112, 250–262. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef]
- Sjovall, J. Dietary glycine and taurine on bile acid conjugation in man; bile acids and steroids 75. Proc. Soc. Exp. Biol. Med. 1959, 100, 676–678. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acid Biology, Pathophysiology, and Therapeutics. Clin. Liver Dis. 2020, 15, 91–94. [Google Scholar] [CrossRef]
- Dong, Z.; Lee, B.H. Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci. 2018, 27, 1742–1754. [Google Scholar] [CrossRef]
- Liang, L.; Yi, Y.; Lv, Y.; Qian, J.; Lei, X.; Zhang, G. A Comprehensive Genome Survey Provides Novel Insights into Bile Salt Hydrolase (BSH) in Lactobacillaceae. Molecules 2018, 23, 1157. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Kalavagunta, P.K.; Liao, J.; Jin, L.; Shang, J.; et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 2019, 7, 9. [Google Scholar] [CrossRef]
- Jia, B.; Park, D.; Hahn, Y.; Jeon, C.O. Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes 2020, 11, 1300–1313. [Google Scholar] [CrossRef]
- Elkins, C.A.; Moser, S.A.; Savage, D.C. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 2001, 147, 3403–3412. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Grover, S.; Kaushik, J.K.; Batish, V.K. IS30-related transposon mediated insertional inactivation of bile salt hydrolase (bsh1) gene of Lactobacillus plantarum strain Lp20. Microbiol. Res. 2014, 169, 553–560. [Google Scholar] [CrossRef]
- McAuliffe, O.; Cano, R.J.; Klaenhammer, T.R. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2005, 71, 4925–4929. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.M.; Bongers, R.S.; de Vos, W.M.; Kleerebezem, M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 2008, 74, 4719–4726. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gui, W.; Koo, I.; Smith, P.B.; Allman, E.L.; Nichols, R.G.; Rimal, B.; Cai, J.; Liu, Q.; Patterson, A.D. The microbiome modulating activity of bile acids. Gut Microbes 2020, 11, 979–996. [Google Scholar] [CrossRef]
- Begley, M.; Sleator, R.D.; Gahan, C.G.; Hill, C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 2005, 73, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Allen, G.; Rivera, A.J.; Stewart, A.K.; Barrangou, R.; Theriot, C.M. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Avinash, V.S.; Chauhan, P.D.; Gaikwad, S.; Pundle, A. Biotransformation of penicillin V to 6-aminopenicillanic acid using immobilized whole cells of E. coli expressing a highly active penicillin V acylase. Prep. Biochem. Biotechnol. 2017, 47, 52–57. [Google Scholar] [CrossRef]
- Avinash, V.S.; Pundle, A.V.; Ramasamy, S.; Suresh, C.G. Penicillin acylases revisited: Importance beyond their industrial utility. Crit. Rev. Biotechnol. 2016, 36, 303–316. [Google Scholar] [CrossRef]
- Xu, G.; Zhao, Q.; Huang, B.; Zhou, J.; Cao, F. Directed evolution of a penicillin V acylase from Bacillus sphaericus to improve its catalytic efficiency for 6-APA production. Enzyme Microb. Technol. 2018, 119, 65–70. [Google Scholar] [CrossRef]
- Arroyo, M.; de la Mata, I.; Acebal, C.; Castillón, M.P. Biotechnological applications of penicillin acylases: State-of-the-art. Appl. Microbiol. Biotechnol. 2003, 60, 507–514. [Google Scholar] [CrossRef]
- Maresova, H.; Plackova, M.; Grulich, M.; Kyslik, P. Current state and perspectives of penicillin G acylase-based biocatalyses. Appl. Microbiol. Biotechnol. 2014, 98, 2867–2879. [Google Scholar] [CrossRef]
- Illanes, A.; Valencia, P. 13—Industrial and Therapeutic Enzymes: Penicillin Acylase. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Negi, S., Soccol, C.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 267–305. [Google Scholar] [CrossRef]
- Kumar, R.S.; Brannigan, J.A.; Prabhune, A.A.; Pundle, A.V.; Dodson, G.G.; Dodson, E.J.; Suresh, C.G. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 2006, 281, 32516–32525. [Google Scholar] [CrossRef]
- Srirangan, K.; Orr, V.; Akawi, L.; Westbrook, A.; Moo-Young, M.; Chou, C.P. Biotechnological advances on penicillin G acylase: Pharmaceutical implications, unique expression mechanism and production strategies. Biotechnol. Adv. 2013, 31, 1319–1332. [Google Scholar] [CrossRef]
- Valle, F.; Balba’s, P.; Merino, E.; Bollvar, F. The role of penicillin amidases in nature and in industry. Trends Biochem. Sci. 1991, 16, 36–40. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Mukherji, R.; Varshney, N.K.; Panigrahi, P.; Suresh, C.G.; Prabhune, A. A new role for penicillin acylases: Degradation of acyl homoserine lactone quorum sensing signals by Kluyvera citrophila penicillin G acylase. Enzyme Microb. Technol. 2014, 56, 1–7. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Cui, X.; Zhang, X.H. A novel stress response mechanism, triggered by indole, involved in quorum quenching enzyme MomL and iron-sulfur cluster in Muricauda olearia Th120. Sci. Rep. 2017, 7, 4252. [Google Scholar] [CrossRef]
- Velasco-Bucheli, R.; Hormigo, D.; Fernández-Lucas, J.; Torres-Ayuso, P.; Alfaro-Ureña, Y.; Saborido, A.I.; Serrano-Aguirre, L.; García, J.L.; Ramón, F.; Acebal, C.; et al. Penicillin Acylase from Streptomyces lavendulae and Aculeacin A Acylase from Actinoplanes utahensis: Two Versatile Enzymes as Useful Tools for Quorum Quenching Processes. Catalysts 2020, 10, 730. [Google Scholar] [CrossRef]
- Sunder, A.V.; Utari, P.D.; Ramasamy, S.; van Merkerk, R.; Quax, W.; Pundle, A. Penicillin V acylases from gram-negative bacteria degrade N-acylhomoserine lactones and attenuate virulence in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2017, 101, 2383–2395. [Google Scholar] [CrossRef]
- Kusada, H.; Tamaki, H.; Kamagata, Y.; Hanada, S.; Kimura, N. A Novel Quorum-Quenching N-Acylhomoserine Lactone Acylase from Acidovorax sp. Strain MR-S7 Mediates Antibiotic Resistance. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Chand, D.; Panigrahi, P.; Varshney, N.; Ramasamy, S.; Suresh, C.G. Structure and function of a highly active Bile Salt Hydrolase (BSH) from Enterococcus faecalis and post-translational processing of BSH enzymes. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 507–518. [Google Scholar] [CrossRef]
- Avinash, V.S.; Ramasamy, S.; Suresh, C.G.; Pundle, A. Penicillin V acylase from Pectobacterium atrosepticum exhibits high specific activity and unique kinetics. Int. J. Biol. Macromol. 2015, 79, 1–7. [Google Scholar] [CrossRef]
- Koreishi, M.; Zhang, D.; Imanaka, H.; Imamura, K.; Adachi, S.; Matsuno, R.; Nakanishi, K. A Novel Acylase from Streptomyces mobaraensis that Efficiently Catalyzes Hydrolysis/Synthesis of Capsaicins as Well as N-Acyl-l-amino Acids and N-Acyl-peptides. J. Agric. Food Chem. 2006, 54, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Torres-Bacete, J.; Hormigo, D.; Torres-Guzman, R.; Arroyo, M.; Castillon, M.P.; Garcia, L.; Acebal, C.; de la Mata, I. Overexpression of penicillin V acylase from Streptomyces lavendulae and elucidation of its catalytic residues. Appl. Environ. Microbiol. 2015, 81, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Rathinaswamy, P.; Gaikwad, S.M.; Suresh, C.G.; Prabhune, A.A.; Brannigan, J.A.; Dodson, G.G.; Pundle, A.V. Purification and characterization of YxeI, a penicillin acylase from Bacillus subtilis. Int. J. Biol. Macromol. 2012, 50, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Gopal-Srivastava, R.; Hylemon, P.B. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 1988, 29, 1079–1085. [Google Scholar] [CrossRef]
- Tanaka, H.; Hashiba, H.; Kok, J.; Mierau, I. Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 2000, 66, 2502–2512. [Google Scholar] [CrossRef]
- Wang, Z.; Zeng, X.; Mo, Y.; Smith, K.; Guo, Y.; Lin, J. Identification and characterization of a bile salt hydrolase from Lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters. Appl. Environ. Microbiol. 2012, 78, 8795–8802. [Google Scholar] [CrossRef]
- Pundle, A.; SivaRaman, H. Bacillus sphaericus penicillin V acylase: Purification, substrate specificity, and active-site characterization. Curr. Microbiol. 1997, 34, 144–148. [Google Scholar] [CrossRef]
- Suresh, C.G.; Pundle, A.V.; SivaRaman, H.; Rao, K.N.; Brannigan, J.A.; McVey, C.E.; Verma, C.S.; Dauter, Z.; Dodson, E.J.; Dodson, G.G. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat. Struct. Biol. 1999, 6, 414–416. [Google Scholar] [CrossRef]
- Rathinaswamy, P.; Pundle, A.V.; Prabhune, A.A.; Sivaraman, H.; Brannigan, J.A.; Dodson, G.G.; Suresh, C.G. Cloning, purification, crystallization and preliminary structural studies of penicillin V acylase from Bacillus subtilis. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2005, 61, 680–683. [Google Scholar] [CrossRef]
- Avinash, V.S.; Panigrahi, P.; Suresh, C.G.; Pundle, A.V.; Ramasamy, S. Structural modelling of substrate binding and inhibition in penicillin V acylase from Pectobacterium atrosepticum. Biochem. Biophys. Res. Commun. 2013, 437, 538–543. [Google Scholar] [CrossRef]
- Avinash, V.S.; Panigrahi, P.; Chand, D.; Pundle, A.; Suresh, C.G.; Ramasamy, S. Structural analysis of a penicillin V acylase from Pectobacterium atrosepticum confirms the importance of two Trp residues for activity and specificity. J. Struct. Biol. 2016, 193, 85–94. [Google Scholar] [CrossRef]
- Rani, R.P.; Anandharaj, M.; Ravindran, A.D. Characterization of Bile Salt Hydrolase from Lactobacillus gasseri FR4 and Demonstration of Its Substrate Specificity and Inhibitory Mechanism Using Molecular Docking Analysis. Front. Microbiol. 2017, 8, 1004. [Google Scholar] [CrossRef]
- Chae, J.P.; Valeriano, V.D.; Kim, G.-B.; Kang, D.-K. Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J. Appl. Microbiol. 2013, 114, 121–133. [Google Scholar] [CrossRef]
- Ozturk, M.; Onal, C. Asparagine 79 is an important amino acid for catalytic activity and substrate specificity of bile salt hydrolase (BSH). Mol. Biol. Rep. 2019, 46, 4361–4368. [Google Scholar] [CrossRef]
- Lodola, A.; Branduardi, D.; De Vivo, M.; Capoferri, L.; Mor, M.; Piomelli, D.; Cavalli, A. A catalytic mechanism for cysteine N-terminal nucleophile hydrolases, as revealed by free energy simulations. PLoS ONE 2012, 7, e32397. [Google Scholar] [CrossRef]
- Xu, F.; Guo, F.; Hu, X.J.; Lin, J. Crystal structure of bile salt hydrolase from Lactobacillus salivarius. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 376–381. [Google Scholar] [CrossRef]
- Xu, F.; Hu, X.J.; Singh, W.; Geng, W.; Tikhonova, I.G.; Lin, J. The complex structure of bile salt hydrolase from Lactobacillus salivarius reveals the structural basis of substrate specificity. Sci. Rep. 2019, 9, 12438. [Google Scholar] [CrossRef]
- Hu, P.L.; Yuan, Y.H.; Yue, T.L.; Guo, C.F. A new method for the in vitro determination of the bile tolerance of potentially probiotic lactobacilli. Appl. Microbiol. Biotechnol. 2018, 102, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Li, Y.; Bumann, M.; Raftis, E.J.; Casey, P.G.; Cooney, J.C.; Walsh, M.A.; O’Toole, P.W. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J. Bacteriol. 2009, 191, 5743–5757. [Google Scholar] [CrossRef]
- Ren, J.; Sun, K.; Wu, Z.; Yao, J.; Guo, B. All 4 bile salt hydrolase proteins are responsible for the hydrolysis activity in Lactobacillus plantarum ST-III. J. Food Sci. 2011, 76, M622–M628. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Seaton, S.C.; Ndousse-Fetter, S.; Adhikari, A.A.; DiBenedetto, N.; Mina, A.I.; Banks, A.S.; Bry, L.; Devlin, A.S. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Smet, I.D.; Hoorde, L.V.; Saeyer, N.D.; Woestyne, M.V.; Verstraete, W. In Vitro Study of Bile Salt Hydrolase (BSH) Activity of BSH Isogenic Lactobacillus plantarum 80 Strains and Estimation of Cholesterol Lowering through Enhanced BSH Activity. Microbial. Ecol. Health Dis. 1994, 7, 315–329. [Google Scholar] [CrossRef]
- Joyce, S.A.; Gahan, C.G. Bile Acid Modifications at the Microbe-Host Interface: Potential for Nutraceutical and Pharmaceutical Interventions in Host Health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef]
- Joyce, S.A.; Kamil, A.; Fleige, L.; Gahan, C.G.M. The Cholesterol-Lowering Effect of Oats and Oat Beta Glucan: Modes of Action and Potential Role of Bile Acids and the Microbiome. Front. Nutr. 2019, 6, 171. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu. Rev. Nutr. 2019, 39, 175–200. [Google Scholar] [CrossRef]
- Ikpa, P.T.; Doktorova, M.; Meijsen, K.F.; Nieuwenhuijze, N.D.A.; Verkade, H.J.; Jonker, J.W.; de Jonge, H.R.; Bijvelds, M.J.C. Impaired Intestinal Farnesoid X Receptor Signaling in Cystic Fibrosis Mice. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 47–60. [Google Scholar] [CrossRef]
- Jones, H.; Alpini, G.; Francis, H. Bile acid signaling and biliary functions. Acta Pharm. Sin. B 2015, 5, 123–128. [Google Scholar] [CrossRef]
- Duboc, H.; Tache, Y.; Hofmann, A.F. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig. Liver Dis. 2014, 46, 302–312. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef]
- Malhi, H.; Camilleri, M. Modulating bile acid pathways and TGR5 receptors for treating liver and GI diseases. Curr. Opin. Pharmacol. 2017, 37, 80–86. [Google Scholar] [CrossRef]
- Watanabe, M.; Houten, S.M.; Mataki, C.; Christoffolete, M.A.; Kim, B.W.; Sato, H.; Messaddeq, N.; Harney, J.W.; Ezaki, O.; Kodama, T.; et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006, 439, 484–489. [Google Scholar] [CrossRef]
- Degirolamo, C.; Rainaldi, S.; Bovenga, F.; Murzilli, S.; Moschetta, A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014, 7, 12–18. [Google Scholar] [CrossRef]
- Qu, T.; Yang, L.; Wang, Y.; Jiang, B.; Shen, M.; Ren, D. Reduction of serum cholesterol and its mechanism by Lactobacillus plantarum H6 screened from local fermented food products. Food Funct. 2020, 11, 1397–1409. [Google Scholar] [CrossRef]
- Joyce, S.A.; Shanahan, F.; Hill, C.; Gahan, C.G. Bacterial bile salt hydrolase in host metabolism: Potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 2014, 5, 669–674. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; de Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef]
- Guban, J.; Korver, D.R.; Allison, G.E.; Tannock, G.W. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult. Sci. 2006, 85, 2186–2194. [Google Scholar] [CrossRef]
- Gonzalez, F.J.; Jiang, C.; Patterson, A.D. An Intestinal Microbiota-Farnesoid X Receptor Axis Modulates Metabolic Disease. Gastroenterology 2016, 151, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Jiang, C.; Krausz, K.W.; Li, Y.; Albert, I.; Hao, H.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.D.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013, 4, 2384. [Google Scholar] [CrossRef] [PubMed]
- Drissi, F.; Raoult, D.; Merhej, V. Metabolic role of lactobacilli in weight modification in humans and animals. Microb. Pathog. 2017, 106, 182–194. [Google Scholar] [CrossRef] [PubMed]
- DiMarzio, M.; Rusconi, B.; Yennawar, N.H.; Eppinger, M.; Patterson, A.D.; Dudley, E.G. Identification of a mouse Lactobacillus johnsonii strain with deconjugase activity against the FXR antagonist T-beta-MCA. PLoS ONE 2017, 12, e0183564. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; Gahan, C.G. Disease-Associated Changes in Bile Acid Profiles and Links to Altered Gut Microbiota. Dig. Dis. 2017, 35, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; McDonald, J.A.K.; Pechlivanis, A.; Allegretti, J.R.; Kao, D.; Barker, G.F.; Kapila, D.; Petrof, E.O.; Joyce, S.A.; Gahan, C.G.M.; et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 2019, 68, 1791–1800. [Google Scholar] [CrossRef]
- Grandclement, C.; Tannieres, M.; Morera, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86. [Google Scholar] [CrossRef]
- Enright, E.F.; Griffin, B.T.; Gahan, C.G.M.; Joyce, S.A. Microbiome-mediated bile acid modification: Role in intestinal drug absorption and metabolism. Pharmacol. Res. 2018, 133, 170–186. [Google Scholar] [CrossRef]

| Organism | Enzyme | pH | Temp. | Form | aa | Activity in Descending Order | Other Activity | References |
|---|---|---|---|---|---|---|---|---|
| Clostridium perfringens | CpBSH | Optimum 5.8–6.4 | - | Homotetramer | 328 | GCA, GCDCA, GDCA, TCA, TDCA | Positive Penicillin V | [48] |
| Bifidobacterium longum | BlBSH | Optimum 5–7 | 40 °C | Homotetramer | 315 | GCDCA, GDCA, GCA, TCDCA, TCA, TDCA | Negative Penicillin V | [34,49] |
| Lactobacillus salivarius | LsBSH | Optimum 5–6 (5.4) | 41 °C | Tetramer and dimer | 324/325 | GCDCA, TDCA, TCDCA, GCA, TCA, GDCA | - | [50] |
| Enterococcus fecalis | EfBSH | Optimum 5 | 50 °C | Homotetramer | 324 | GCA, GDCA, TDCA, TCA, GCDCA, TCDCA | - | [43] |
| Lyinibacillus sphaericus | BspPVA | Optimum 6 | 60 °C | Homotetramer | 335 | Penicillin V | Positive TCA | [51,52] |
| Bacillus subtilis | BsuPVA | Active 5.5–9 Optimal 6.6–7.4 | 40 °C | Homotetramer | 328 | Penicillin V | - | [47,53] |
| Pectobacterium atrosepticum | PaPVA | Active 3–6 Optimum 5 | 40 °C | Homotetramer | 355 | Penicillin V | Negative GCA and TCA | [44,54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daly, J.W.; Keely, S.J.; Gahan, C.G.M. Functional and Phylogenetic Diversity of BSH and PVA Enzymes. Microorganisms 2021, 9, 732. https://doi.org/10.3390/microorganisms9040732
Daly JW, Keely SJ, Gahan CGM. Functional and Phylogenetic Diversity of BSH and PVA Enzymes. Microorganisms. 2021; 9(4):732. https://doi.org/10.3390/microorganisms9040732
Chicago/Turabian StyleDaly, Jack W., Stephen J. Keely, and Cormac G. M. Gahan. 2021. "Functional and Phylogenetic Diversity of BSH and PVA Enzymes" Microorganisms 9, no. 4: 732. https://doi.org/10.3390/microorganisms9040732
APA StyleDaly, J. W., Keely, S. J., & Gahan, C. G. M. (2021). Functional and Phylogenetic Diversity of BSH and PVA Enzymes. Microorganisms, 9(4), 732. https://doi.org/10.3390/microorganisms9040732






