Identifying a Novel Bile Salt Hydrolase from the Keystone Gut Bacterium Christensenella minuta
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Growth and Bile Acid Tolerance Assay
2.2. Cloning of C. minuta DSM 33407 BSH Gene
2.3. Expression and of Recombinant BSH
2.4. Enzymatic Assays
2.5. Protein Sequence and In Silico Structural Analysis
2.6. BSH Alignment, Phylogenetic Analysis and Clustering
3. Results
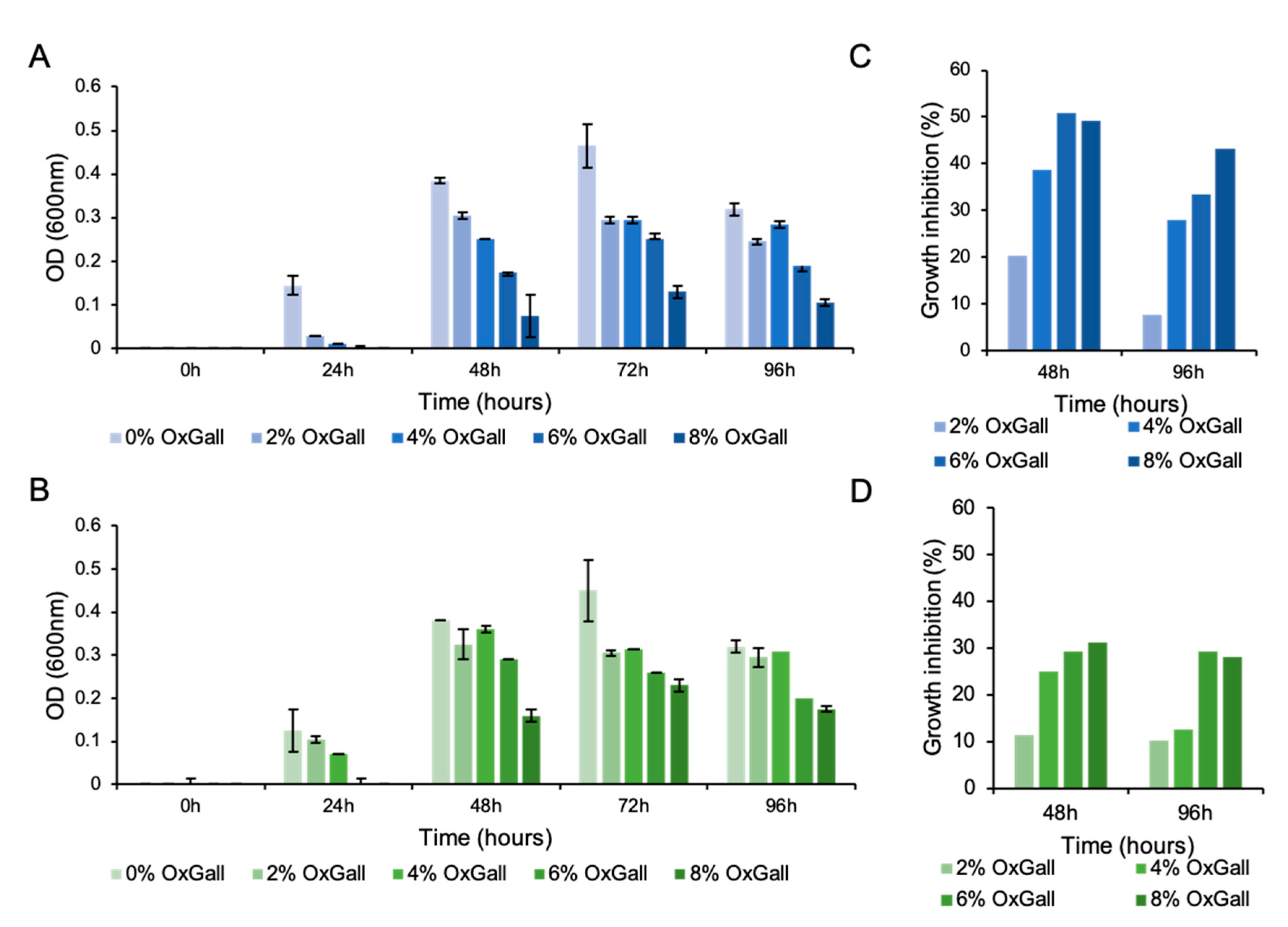
3.1. Christensenella minuta DSM 33407 Displays High Resistance to Bile Acids
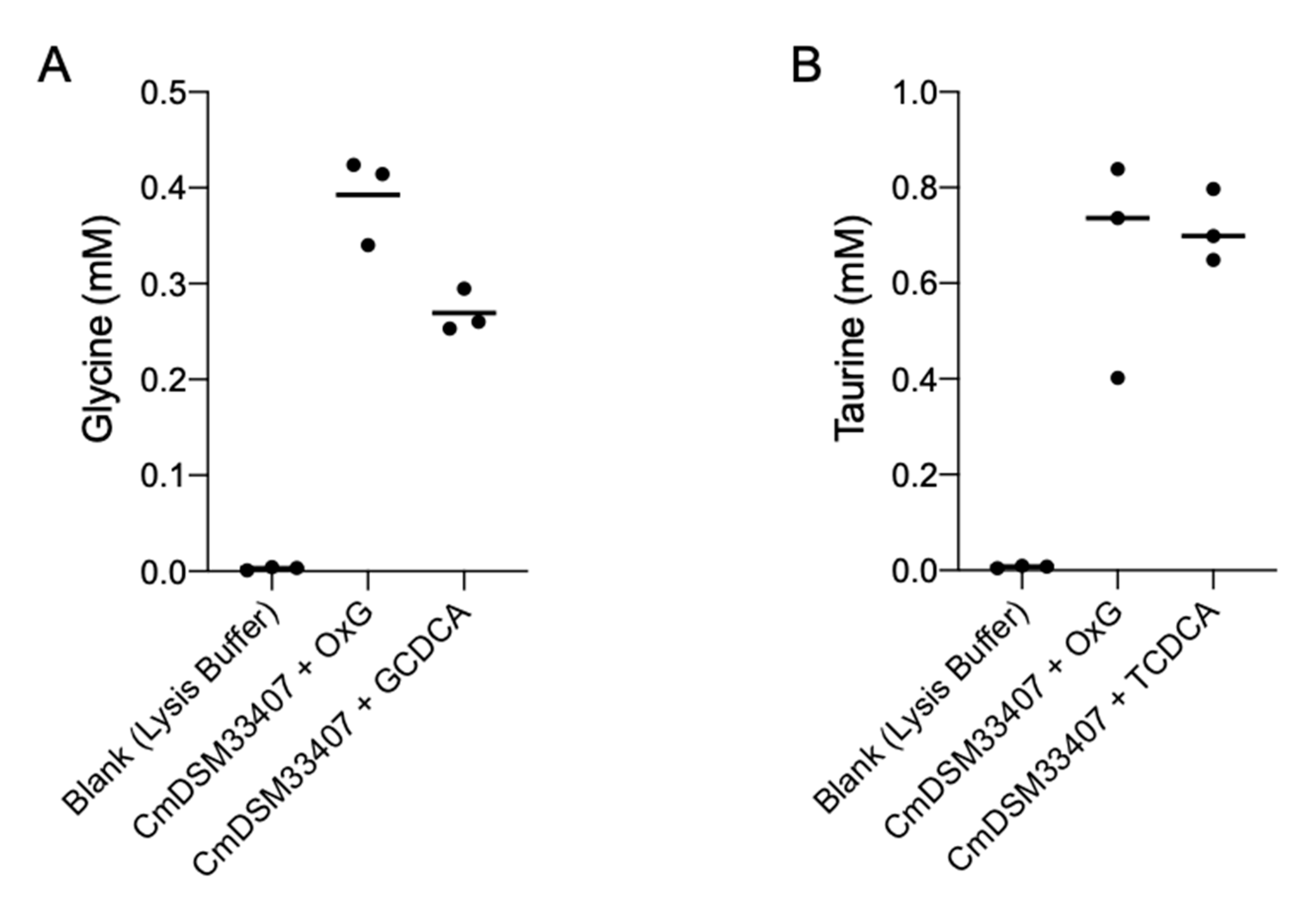
3.2. Christensenella minuta DSM 33407 and DSM 22607 Carry an Active Bile Salt Hydrolase Enzyme
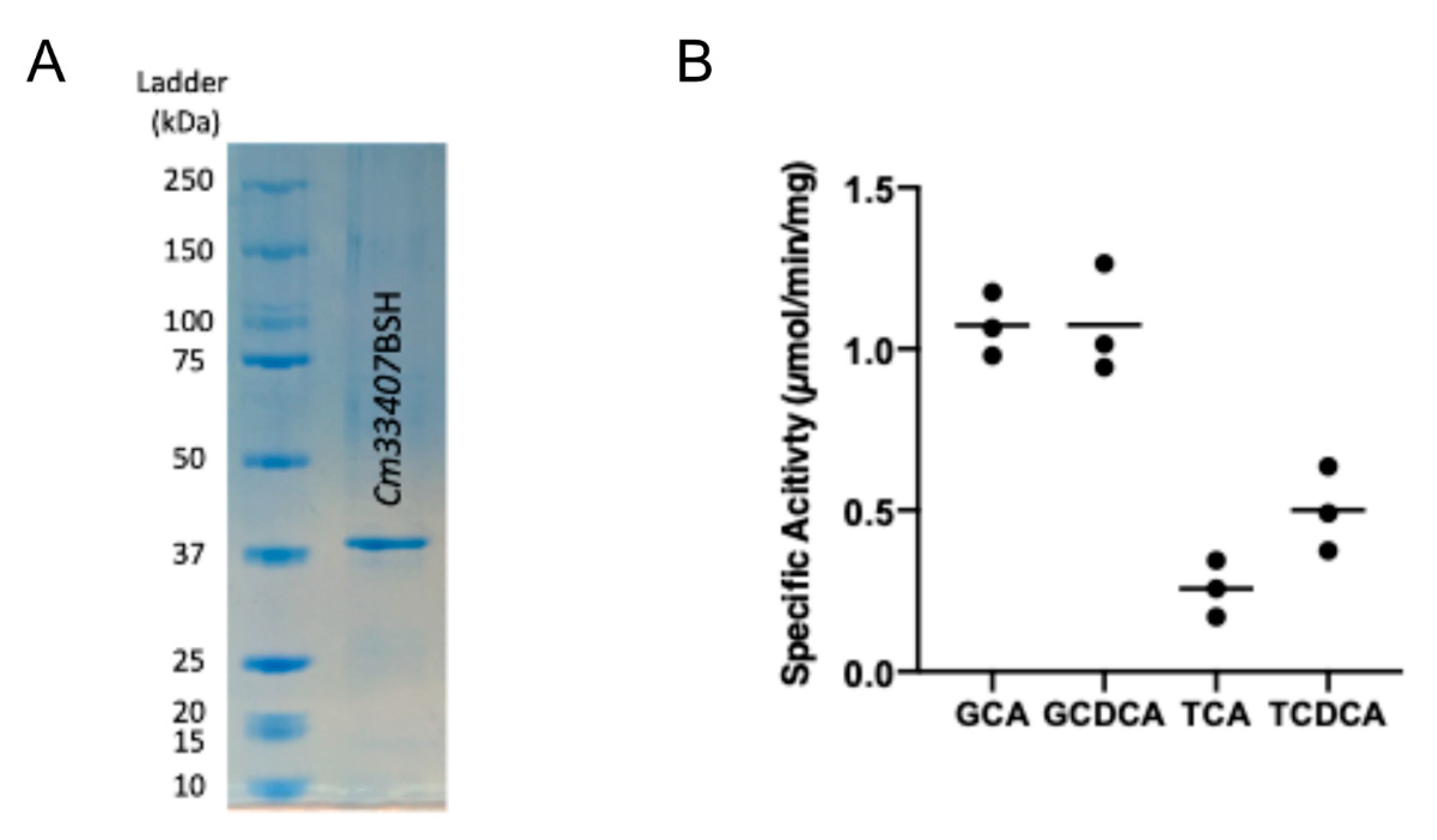
3.3. Cm33407BSH Carries a Potent BSH Activity in C. minuta DSM33407
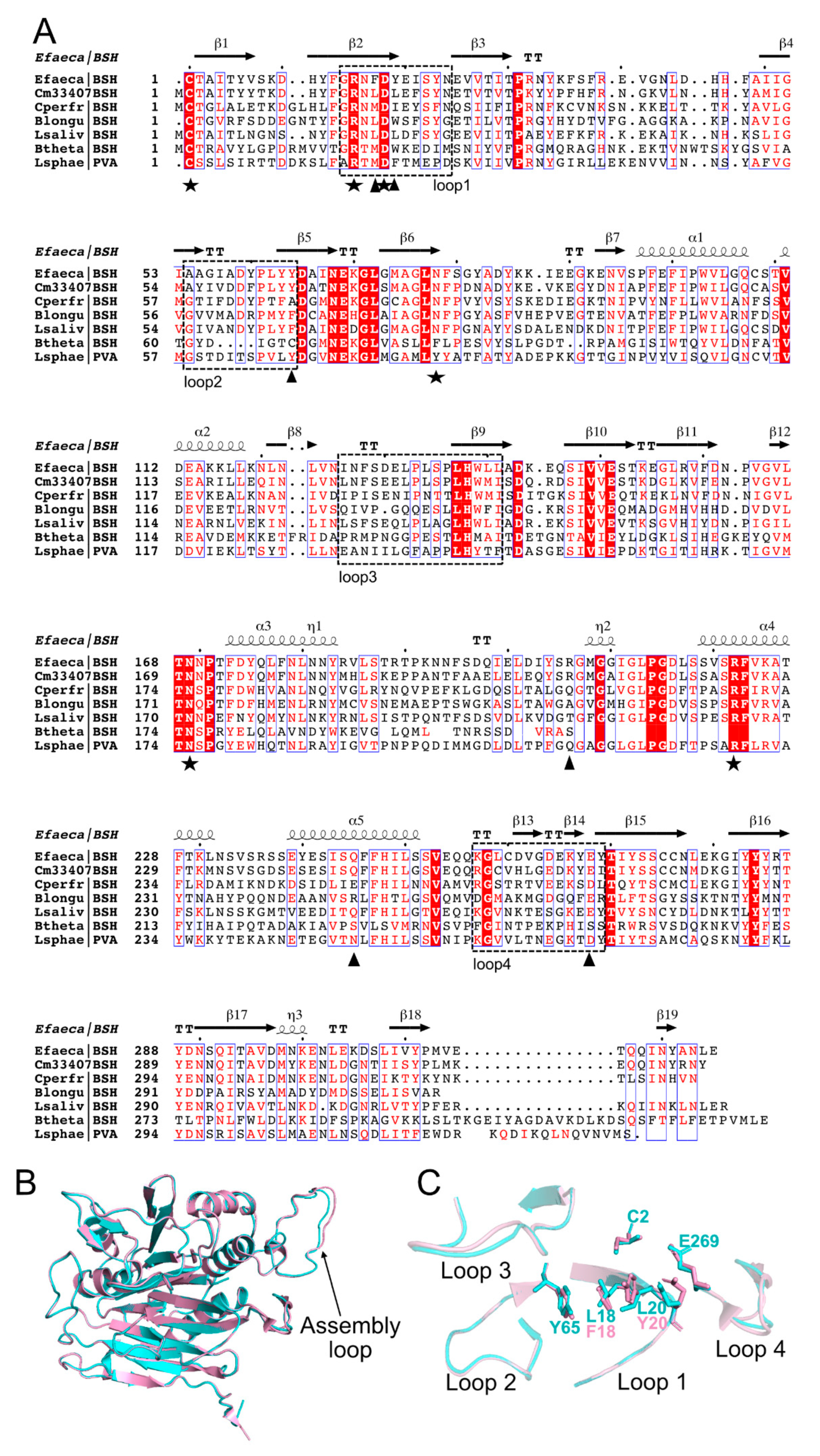
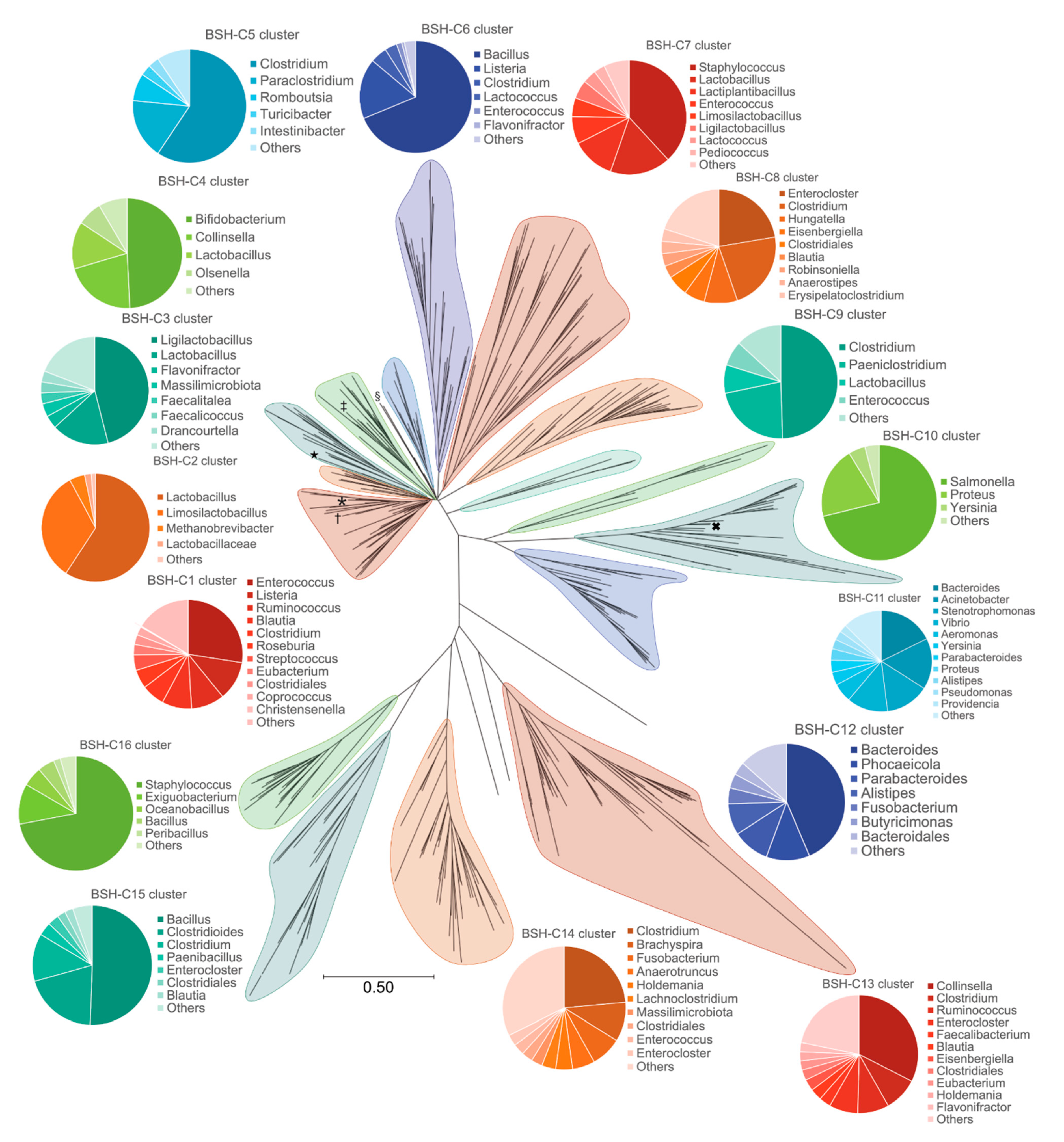
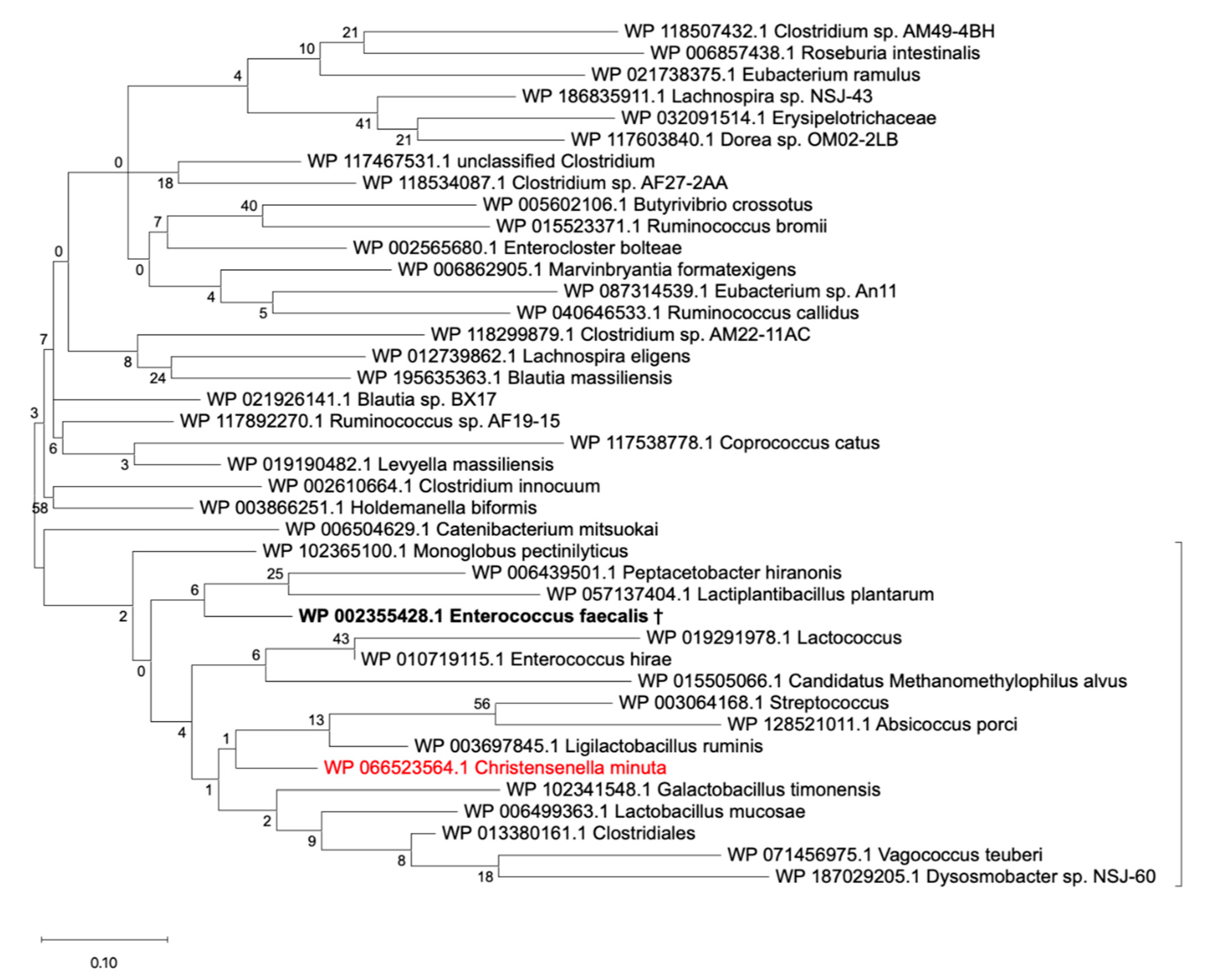
3.4. The BSH Protein Carried by C. minuta sp. Clusters away from all Identified Putative BSH Sequences in the Human Gut Microbiome
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019, 17, 641. [Google Scholar] [CrossRef]
- Lim, M.Y.; You, H.J.; Yoon, H.S.; Kwon, B.; Lee, J.Y.; Lee, S.; Song, Y.-M.; Lee, K.; Sung, J.; Ko, G. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 2017, 66, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Shapiro, J.A.; Church, T.R.; Miller, G.; Trinh-Shevrin, C.; Yuen, E.; Friedlander, C.; Hayes, R.B.; Ahn, J. A taxonomic signature of obesity in a large study of American adults. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Alemán, J.O.; Bokulich, N.A.; Swann, J.R.; Walker, J.M.; De Rosa, J.C.; Battaglia, T.; Costabile, A.; Pechlivanis, A.; Liang, Y.; Breslow, J.L.; et al. Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J. Transl. Med. 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; He, Y.; Li, P.; Zhou, H.; Zeng, N. Regional distribution of Christensenellaceae and its associations with metabolic syndrome based on a population-level analysis. PeerJ 2020, 8, e9591. [Google Scholar] [CrossRef] [PubMed]
- Morotomi, M.; Nagai, F.; Watanabe, Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 144–149. [Google Scholar] [CrossRef]
- Mazier, W.; Le Corf, K.; Martinez, C.; Tudela, H.; Kissi, D.; Kropp, C.; Coubard, C.; Soto, M.; Elustondo, F.; Rawadi, G.; et al. A new strain of Christensenella minuta as a potential biotherapy for obesity and associated metabolic diseases. Cells 2021, 10, 823. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Backhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Chávez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 2017, 152, 1679–1694.e3. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.-J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G.M. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Harris, S.C.; Ridlon, J.M. Metabolism of hydrogen gases and bile acids in the gut microbiome. FEBS Lett. 2018, 592, 2070–2082. [Google Scholar] [CrossRef] [PubMed]
- Kliewer, S.A.; Mangelsdorf, D.J. Bile Acids as Hormones: The FXR-FGF15/19 Pathway. Dig. Dis. 2015, 33, 327–331. [Google Scholar] [CrossRef]
- Kim, G.B.; Yi, S.H.; Lee, B.H. Purification and characterization of three different types of bile salt hydrolases from Bifidobacterium strains. J. Dairy Sci. 2004, 87, 258–266. [Google Scholar] [CrossRef]
- Kumar, R.S.; Brannigan, J.A.; Prabhune, A.A.; Pundle, A.V.; Dodson, G.G.; Dodson, E.J.; Suresh, C.G. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 2006, 281, 32516–32525. [Google Scholar] [CrossRef]
- Foley, M.H.; O’Flaherty, S.; Allen, G.; Rivera, A.J.; Stewart, A.K.; Barrangou, R.; Theriot, C.M. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Crawley, A.B.; Theriot, C.M.; Barrangou, R. The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Chand, D.; Panigrahi, P.; Varshney, N.; Ramasamy, S.; Suresh, C.G. Structure and function of a highly active Bile Salt Hydrolase (BSH) from Enterococcus faecalis and post-translational processing of BSH enzymes. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.M.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef]
- Yao, L.; Seaton, S.C.; Ndousse-Fetter, S.; Adhikari, A.A.; DiBenedetto, N.; Mina, A.I.; Banks, A.S.; Bry, L.; Devlin, A.S. A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 2018, 7, 675. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Kalavagunta, P.K.; Liao, J.; Jin, L.; Shang, J.; et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 2019, 7, 1–16. [Google Scholar] [CrossRef]
- Bustos, A.Y.; Font de Valdez, G.; Fadda, S.; Taranto, M.P. New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res. Int. 2018, 112, 250–262. [Google Scholar] [CrossRef]
- Moser, S.A.; Savage, D.C. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 2001, 67, 3476. [Google Scholar] [CrossRef] [PubMed]
- Déjean, G.; Tamura, K.; Cabrera, A.; Jain, N.; Pudlo, N.A.; Pereira, G.; Viborg, A.H.; Van Petegem, F.; Martens, E.C.; Brumer, H. Synergy between cell surface glycosidases and glycan-binding proteins dictates the utilization of specific beta (1, 3)-glucans by human gut Bacteroides. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Eschenfeldt, W.H.; Lucy, S.; Millard, C.S.; Joachimiak, A.; Mark, I.D. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 2009, 498, 105–115. [Google Scholar] [PubMed]
- Tanaka, H.; Hashiba, H.; Kok, J.; Mierau, I. Bile salt hydrolase of Bifidobacterium longum—Biochemical and genetic characterization. Appl. Environ. Microbiol. 2000, 66, 2502–2512. [Google Scholar] [CrossRef] [PubMed]
- Juncker, A.S.; Willenbrock, H.; Von Heijne, G.; Brunak, S.; Nielsen, H.; Krogh, A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003, 12, 1652–1662. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting secretory proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A web-compatible database search or multiple alignment viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Nayfach, S.; Boland, M.; Strozzi, F.; Beracochea, M.; Shi, Z.J.; Pollard, K.S.; Sakharova, E.; Parks, D.H.; Hugenholtz, P.; et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nature Biotechnol. 2021, 39, 105–114. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207. [Google Scholar]
- Human, M.P.C. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.W.; Keely, S.J.; Gahan, C.G.M. Functional and Phylogenetic Diversity of BSH and PVA Enzymes. Microorganisms 2021, 9, 732. [Google Scholar] [CrossRef]
- Dong, Z.; Lee, B.H. Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci. 2018, 27, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Núñez, G. Pathogen Colonization Resistance in the Gut and Its Manipulation for Improved Health. Am. J. Pathol. 2019, 189, 1300–1310. [Google Scholar] [CrossRef]
- Gopal-Srivastava, R.; Hylemon, P.B. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 1988, 29, 1079–1085. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.-U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.F.; Pye, G.; Bramley, R.; Clark, A.G.; Dyson, T.J.; Hardcastle, J.D. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 1988, 29, 1035–1041. [Google Scholar] [CrossRef]
- Fallingborg, J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999, 46, 183–196. [Google Scholar] [PubMed]
- Begley, M.; Gahan, C.G.M.; Hill, C. Bile stress response in Listeria monocytogenes LO28: Adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 2002, 68, 6005. [Google Scholar] [CrossRef] [PubMed]
- Chand, D.; Avinash, V.S.; Yadav, Y.; Pundle, A.V.; Suresh, C.G.; Ramasamy, S. Molecular features of bile salt hydrolases and relevance in human health. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2981–2991. [Google Scholar] [CrossRef] [PubMed]
- Grill, J.P.; Cayuela, C.; Antoine, J.M.; Schneider, F. Isolation and characterization of a Lactobacillus amylovorus mutant depleted in conjugated bile salt hydrolase activity: Relation between activity and bile salt resistance. J. Appl. Microbiol. 2000, 89, 553–563. [Google Scholar] [CrossRef]
- De Smet, I.; Van Hoorde, L.; Vande Woestyne, M.; Christiaens, H.; Verstrate, W. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 1995, 79, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Hu, X.J.; Singh, W.; Geng, W.; Tikhonova, I.G.; Lin, J. The complex structure of bile salt hydrolase from Lactobacillus salivarius reveals the structural basis of substrate specificity. Sci. Rep. 2019, 9, 12438. [Google Scholar] [CrossRef]
- Xu, F.; Guo, F.; Hu, X.J.; Lin, J. Crystal structure of bile salt hydrolase from Lactobacillus salivarius. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Rossocha, M.; Schultz-Heienbrok, R.; Moeller, H.V.; Coleman, J.P.; Saenger, W. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 2005, 44, 5739–5748. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, A.A.; Seegar, T.C.M.; Ficarro, S.B.; McCurry, M.D.; Ramachandran, D.; Yao, L.; Chaudhari, S.N.; Ndousse-Fetter, S.; Banks, A.S.; Marto, J.A.; et al. Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat. Chem. Biol. 2020, 16. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Foley, M.H.; O’Flaherty, S.; Barrangou, R.; Theriot, C.M. Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019, 15, e1007581. [Google Scholar] [CrossRef]
- Ogilvie, L.A.; Jones, B.V. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: A mechanism and marker of disease. Gut 2012, 61, 1642–1643. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Zhang, X.; Xiao, F.; Hu, H.; Li, X.; Dong, F.; Sun, M.; Xiao, Y.; Ge, T.; et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn’s disease. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Khodakivskyi, P.V.; Lauber, C.L.; Yevtodiyenko, A.; Bazhin, A.A.; Bruce, S.; Ringel-Kulka, T.; Ringel, Y.; Bétrisey, B.; Torres, J.; Hu, J.; et al. Noninvasive imaging and quantification of bile salt hydrolase activity: From bacteria to humans. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; McDonald, J.A.K.; Pechlivanis, A.; Allegretti, J.R.; Kao, D.; Barker, G.F.; Kapila, D.; Petrof, E.O.; Joyce, S.A.; Gahan, C.G.M.; et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 2019, 68, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Déjean, G.; Tudela, H.; Bruno, L.; Kissi, D.; Rawadi, G.; Claus, S.P. Identifying a Novel Bile Salt Hydrolase from the Keystone Gut Bacterium Christensenella minuta. Microorganisms 2021, 9, 1252. https://doi.org/10.3390/microorganisms9061252
Déjean G, Tudela H, Bruno L, Kissi D, Rawadi G, Claus SP. Identifying a Novel Bile Salt Hydrolase from the Keystone Gut Bacterium Christensenella minuta. Microorganisms. 2021; 9(6):1252. https://doi.org/10.3390/microorganisms9061252
Chicago/Turabian StyleDéjean, Guillaume, Héloïse Tudela, Lisa Bruno, Déborah Kissi, Georges Rawadi, and Sandrine P. Claus. 2021. "Identifying a Novel Bile Salt Hydrolase from the Keystone Gut Bacterium Christensenella minuta" Microorganisms 9, no. 6: 1252. https://doi.org/10.3390/microorganisms9061252
APA StyleDéjean, G., Tudela, H., Bruno, L., Kissi, D., Rawadi, G., & Claus, S. P. (2021). Identifying a Novel Bile Salt Hydrolase from the Keystone Gut Bacterium Christensenella minuta. Microorganisms, 9(6), 1252. https://doi.org/10.3390/microorganisms9061252





