Clinically Relevant Escherichia coli Isolates from Process Waters and Wastewater of Poultry and Pig Slaughterhouses in Germany
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sample Preparation
2.2. Isolation and Identification of Target E. coli
2.3. Antimicrobial Susceptibility Testing (AST) and Molecular Typing
2.4. DNA Preparation, Whole-Genome Sequencing and Bioinformatics Analysis
3. Results
3.1. Isolation and Selection of target E. coli
3.2. Phenotypic Antimicrobial Resistance
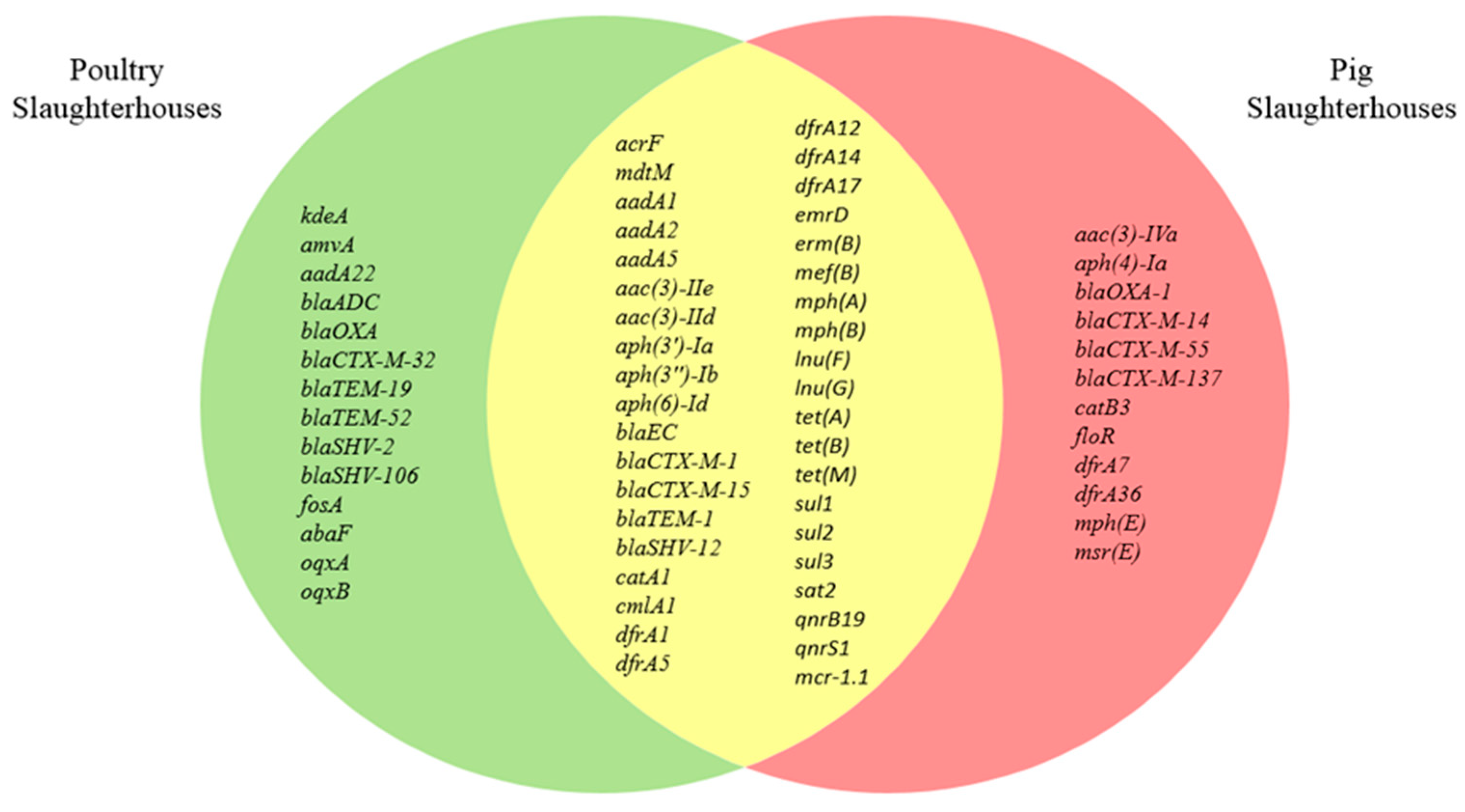
3.3. Characterization of Antimicrobial Resistance Genes
3.4. Distribution of Phylogenetic Groups and MLST Sequence Types
3.5. Characterization of Virulence Genes
3.6. Heavy Metal and Biocide Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Federal Ministry of Food and Agriculture. Report of the Federal Ministry of Food and Agriculture on the Evaluation of the Antibiotics Minimisation Concept introduced with the 16th Act to Amend the Medicinal Products Act (16th AMG Amendment): Evaluation based on section 58g of the Medicinal Products Act. Available online: https://www.bmel.de/SharedDocs/Downloads/EN/_Animals/Report-16thAMGAmendment.pdf?__blob=publicationFile&v=4 (accessed on 23 February 2021).
- EMEA. Antibiotic Resistance in the European Union Associated with Therapeutic Use of Veterinary Medicines: Report and Quantitative Risk Assessment by the Committee for Veterinary Medicinal Products. Available online: https://www.ema.europa.eu/en/documents/report/antibiotic-resistance-european-union-associated-therapeutic-use-veterinary-medicines-report_en-0.pdf (accessed on 23 February 2021).
- European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef] [Green Version]
- Ghodousi, A.; Bonura, C.; Di Noto, A.M.; Mammina, C. Extended-Spectrum ß-Lactamase, AmpC-Producing, and Fluoroquinolone-Resistant Escherichia coli in Retail Broiler Chicken Meat, Italy. Foodborne Pathog. Dis. 2015, 12, 619–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaak, H.; Van Hoek, A.H.A.M.; Hamidjaja, R.A.; van der Plaats, R.Q.; Kerkhof-de Heer, L.; de Roda Husman, A.M.; Schets, F.M. Distribution, Numbers, and Diversity of ESBL-Producing E. coli in the Poultry Farm Environment. PLoS ONE 2015, 10, e0135402. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE Bacteria and Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [Green Version]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020, 727, 138788. [Google Scholar] [CrossRef]
- Schechter, L.M.; Creely, D.P.; Garner, C.D.; Shortridge, D.; Nguyen, H.; Chen, L.; Hanson, B.M.; Sodergren, E.; Weinstock, G.M.; Dunne, W.M.; et al. Extensive Gene Amplification as a Mechanism for Piperacillin-Tazobactam Resistance in Escherichia coli. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livermore, D.M.; Day, M.; Cleary, P.; Hopkins, K.L.; Toleman, M.A.; Wareham, D.W.; Wiuff, C.; Doumith, M.; Woodford, N. OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J. Antimicrob. Chemother. 2019, 74, 326–333. [Google Scholar] [CrossRef] [Green Version]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [Green Version]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Janssen, T.; Wieler, L.H. Aviäre pathogene Escherichia coli (APEC). Berl. Munch. Tierarztl. Wochenschr. 2003, 116, 381–395. [Google Scholar]
- Maluta, R.P.; Logue, C.M.; Casas, M.R.T.; Meng, T.; Guastalli, E.A.L.; Rojas, T.C.G.; Montelli, A.C.; Sadatsune, T.; de Carvalho Ramos, M.; Nolan, L.K.; et al. Overlapped Sequence Types (STs) and Serogroups of Avian Pathogenic (APEC) and Human Extra-Intestinal Pathogenic (ExPEC) Escherichia coli Isolated in Brazil. PLoS ONE 2014, 9, e105016. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Johnson, J.R.; Johnston, B.; Curtiss, R.; Mellata, M. Zoonotic Potential of Escherichia coli Isolates from Retail Chicken Meat Products and Eggs. Appl. Environ. Microbiol. 2015, 81, 1177–1187. [Google Scholar] [CrossRef] [Green Version]
- Dohmen, W.; Van Gompel, L.; Schmitt, H.; Liakopoulos, A.; Heres, L.; Urlings, B.A.; Mevius, D.; Bonten, M.J.M.; Heederik, D.J.J. ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol. Infect. 2017, 145, 2003–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nde, C.W.; McEvoy, J.M.; Sherwood, J.S.; Logue, C.M. Cross Contamination of Turkey Carcasses by Salmonella Species during Defeathering. Poult. Sci. 2007, 86, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Bagutti, C.; Brodmann, P.; Alt, M.; Schulze, J.; Fanning, S.; Stephan, R.; Nüesch-Inderbinen, M. Wastewater is a reservoir for clinically relevant carbapenemase- and 16s rRNA methylase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2017, 50, 436–440. [Google Scholar] [CrossRef] [Green Version]
- KRINKO. Hygienemaßnahmen bei Infektionen oder Besiedlung mit multiresistenten gramnegativen Stäbchen. Empfehlung der Kommission für Kranken-haushygiene und Infektionsprävention (KRINKO) beim Robert Koch-Institut (RKI). Bundesgesundh. Gesundh. Gesundh. 2012, 55, 1311–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Johnson, J.R.; Gajewski, A.; Lesse, A.J.; Russo, T.A. Extraintestinal Pathogenic Escherichia coli as a Cause of Invasive Nonurinary Infections. J. Clin. Microbiol. 2003, 41, 5798–5802. [Google Scholar] [CrossRef] [Green Version]
- Spurbeck, R.R.; Dinh, P.C.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L.T. Escherichia coli Isolates That Carry vat, fyuA, chuA, and yfcV Efficiently Colonize the Urinary Tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Using the NCBI AMRFinder Tool to Determine Antimicrobial Resistance Genotype-Phenotype Correlations Within a Collection of NARMS Isolates. BioRxiv 2019, 550707. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit. BVL-Report 13.7. Bericht zur Resistenzmonitoringstudie 2017: Resistenzsituation bei Klinisch Wichtigen Tierpathogenen Bakterien. Available online: https://www.bvl.bund.de/SharedDocs/Berichte/07_Resistenzmonitoringstudie/Bericht_Resistenzmonitoring_2017.html (accessed on 5 March 2021).
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit. BVL-Report 15.2. Berichte zur Lebensmittelsicherheit: Zoonosen-Monitoring 2019. Available online: https://www.bvl.bund.de/SharedDocs/Downloads/01_Lebensmittel/04_Zoonosen_Monitoring/Zoonosen_Monitoring_Bericht_2019.pdf?__blob=publicationFile&v=5 (accessed on 5 March 2021).
- World Organisation for Animal Health. Oie List of Antimicrobial Agents of Veterinary Importance. Available online: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/A_OIE_List_antimicrobials_May2018.pdf (accessed on 23 February 2021).
- World Health Organisation. Critically Important Antimicrobials for Human Medicine: 6th Revision 2018. Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance due to Non-human Use. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf?ua=1 (accessed on 23 February 2021).
- European Medicines Agency. Reflection Paper on Use of Aminoglycosides in Animals in the European Union: Development of Resistance and Impact on Human and Animal Health. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/draft-reflection-paper-use-aminoglycosides-animals-european-union-development-resistance-impact_en.pdf (accessed on 23 February 2021).
- Hidalgo-Grass, C.; Strahilevitz, J. High-Resolution Melt Curve Analysis for Identification of Single Nucleotide Mutations in the Quinolone Resistance Gene aac(6′)-Ib-cr. Antimicrob. Agents Chemother. 2010, 54, 3509–3511. [Google Scholar] [CrossRef] [Green Version]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suetens, C.; Latour, K.; Kärki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikäinen, O.; et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: Results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance 2018, 23, 1800516. [Google Scholar] [CrossRef] [Green Version]
- Zhou, K.; Tao, Y.; Han, L.; Ni, Y.; Sun, J. Piperacillin-Tazobactam (TZP) Resistance in Escherichia coli Due to Hyperproduction of TEM-1 β-Lactamase Mediated by the Promoter Pa/Pb. Front. Microbiol. 2019, 10, 833. [Google Scholar] [CrossRef]
- Roer, L.; Overballe-Petersen, S.; Hansen, F.; Schønning, K.; Wang, M.; Røder, B.L.; Hansen, D.S.; Justesen, U.S.; Andersen, L.P.; Fulgsang-Damgaard, D.; et al. Escherichia coliSequence Type 410 Is Causing New International High-Risk Clones. mSphere 2018, 3, e00337-18. [Google Scholar] [CrossRef] [Green Version]
- Pietsch, M.; Eller, C.; Wendt, C.; Holfelder, M.; Falgenhauer, L.; Fruth, A.; Grössl, T.; Leistner, R.; Valenza, G.; Werner, G.; et al. Molecular characterisation of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet. Microbiol. 2017, 200, 130–137. [Google Scholar] [CrossRef]
- Schaekel, F.; May, T.; Seiler, J.; Hartmann, M.; Kreienbrock, L. Antibiotic drug usage in pigs in Germany—Are the class profiles changing? PLoS ONE 2017, 12, e0182661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosato, A.; Vicarini, H.; Leclercq, R. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 1999, 43, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Meng, J.; McDermott, P.F.; Wang, F.; Yang, Q.; Cao, G.; Hoffmann, M.; Zhao, S. Presence of disinfectant resistance genes in Escherichia coli isolated from retail meats in the USA. J. Antimicrob. Chemother. 2014, 69, 2644–2649. [Google Scholar] [CrossRef] [Green Version]
- Leverstein-van Hall, M.A.; Dierikx, C.M.; Cohen Stuart, J.; Voets, G.M.; van den Munckhof, M.P.; van Essen-Zandbergen, A.; Platteel, T.; Fluit, A.C.; van de Sande-Bruinsma, N.; Scharinga, J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, C.A.; Tarlton, N.J.; Riley, L.W. Escherichia coli from Commercial Broiler and Backyard Chickens Share Sequence Types, Antimicrobial Resistance Profiles, and Resistance Genes with Human Extraintestinal Pathogenic Escherichia coli. Foodborne Pathog. Dis. 2019, 16, 813–822. [Google Scholar] [CrossRef]
- Büdel, T.; Kuenzli, E.; Campos-Madueno, E.I.; Mohammed, A.H.; Hassan, N.K.; Zinsstag, J.; Hatz, C.; Endimiani, A. On the island of Zanzibar people in the community are frequently colonized with the same MDR Enterobacterales found in poultry and retailed chicken meat. J. Antimicrob. Chemother. 2020, 75, 2432–2441. [Google Scholar] [CrossRef]
- Blaak, H.; de Kruijf, P.; Hamidjaja, R.A.; van Hoek, A.H.A.M.; de Roda Husman, A.M.; Schets, F.M. Prevalence and characteristics of ESBL-producing E. coli in Dutch recreational waters influenced by wastewater treatment plants. Vet. Microbiol. 2014, 171, 448–459. [Google Scholar] [CrossRef]
- Paulshus, E.; Thorell, K.; Guzman-Otazo, J.; Joffre, E.; Colque, P.; Kühn, I.; Möllby, R.; Sørum, H.; Sjöling, Å. Repeated Isolation of Extended-Spectrum-β-Lactamase-Positive Escherichia coli Sequence Types 648 and 131 from Community Wastewater Indicates that Sewage Systems Are Important Sources of Emerging Clones of Antibiotic-Resistant Bacteria. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Bleichenbacher, S.; Stevens, M.J.A.; Zurfluh, K.; Perreten, V.; Endimiani, A.; Stephan, R.; Nüesch-Inderbinen, M. Environmental dissemination of carbapenemase-producing Enterobacteriaceae in rivers in Switzerland. Environ. Pollut. 2020, 265, 115081. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Johnson, J.R. Food-Borne Origins of Escherichia coli Causing Extraintestinal Infections. Clin. Infect. Dis. 2012, 55, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Aslam, M.; Toufeer, M.; Narvaez Bravo, C.; Lai, V.; Rempel, H.; Manges, A.; Diarra, M.S. Characterization of Extraintestinal Pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. Int. J. Food Microbiol. 2014, 177, 49–56. [Google Scholar] [CrossRef]
- Müller, A.; Stephan, R.; Nüesch-Inderbinen, M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 2016, 541, 667–672. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.B.; Zou, G.; Cheng, Y.-T.; Xiao, R.; Li, L.; Wu, B.; Zhou, R. Phylogenetic grouping and distribution of virulence genes in Escherichia coli along the production and supply chain of pork around Hubei, China. J. Microbiol. Immunol. Infect. 2017, 50, 382–385. [Google Scholar] [CrossRef] [Green Version]
- Singer, R.S. Urinary tract infections attributed to diverse ExPEC strains in food animals: Evidence and data gaps. Front. Microbiol. 2015, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Poole, N.M.; Green, S.I.; Rajan, A.; Vela, L.E.; Zeng, X.-L.; Estes, M.K.; Maresso, A.W. Role for FimH in Extraintestinal Pathogenic Escherichia coli Invasion and Translocation through the Intestinal Epithelium. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [Green Version]
- Tseng, C.-C.; Lin, W.-H.; Wu, A.-B.; Wang, M.-C.; Teng, C.-H.; Wu, J.-J. Escherichia coli FimH adhesins act synergistically with PapG II adhesins for enhancing establishment and maintenance of kidney infection. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Maluta, R.P.; Leite, J.L.; Rojas, T.C.G.; Scaletsky, I.C.A.; Guastalli, E.A.L.; Ramos, M.D.C.; Dias da Silveira, W. Variants of ast A gene among extra-intestinal Escherichia coli of human and avian origin. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatsuyanagi, J.; Saito, S.; Miyajima, Y.; Amano, K.-I.; Enomoto, K. Characterization of Atypical Enteropathogenic Escherichia coli Strains Harboring the astA Gene That Were Associated with a Waterborne Outbreak of Diarrhea in Japan. J. Clin. Microbiol. 2003, 41, 2033–2039. [Google Scholar] [CrossRef] [Green Version]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manges, A.R.; Dietrich, P.S.; Riley, L.W. Multidrug-ResistantEscherichia coliClonal Groups Causing Community-Acquired Pyelonephritis. Clin. Infect. Dis. 2004, 38, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Zhi, S.; Stothard, P.; Banting, G.; Scott, C.; Huntley, K.; Ryu, K.; Otto, S.; Ashbolt, N.; Checkley, S.; Dong, T.; et al. Characterization of water treatment-resistant and multidrug-resistant urinary pathogenic Escherichia coli in treated wastewater. Water Res. 2020, 182, 115827. [Google Scholar] [CrossRef]
- Zhang, M.; Wan, K.; Zeng, J.; Lin, W.; Ye, C.; Yu, X. Co-selection and stability of bacterial antibiotic resistance by arsenic pollution accidents in source water. Environ. Int. 2020, 135, 105351. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Kempf, I.; Chidaine, B.; Cariolet, R.; Ersbøll, A.K.; Houe, H.; Bruun Hansen, H.C.; Aarestrup, F.M. Copper Resistance in Enterococcus faecium, Mediated by the tcrB Gene, Is Selected by Supplementation of Pig Feed with Copper Sulfate. Appl. Environ. Microbiol. 2006, 72, 5784–5789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazdankhah, S.; Rudi, K.; Bernhoft, A. Zinc and copper in animal feed—Development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb. Ecol. Health Dis. 2014, 25. [Google Scholar] [CrossRef] [Green Version]
- Medardus, J.J.; Molla, B.Z.; Nicol, M.; Morrow, W.M.; Rajala-Schultz, P.J.; Kazwala, R.; Gebreyes, W.A. In-Feed Use of Heavy Metal Micronutrients in U.S. Swine Production Systems and Its Role in Persistence of Multidrug-Resistant Salmonellae. Appl. Environ. Microbiol. 2014, 80, 2317–2325. [Google Scholar] [CrossRef] [Green Version]


| Poultry Slaughterhouses, n = 35 | Pig Slaughterhouses, n = 36 | |||
|---|---|---|---|---|
| Genes | Percentage | Genes | Percentage | |
| Aminoglycosides | aadA1, aadA2 | 22.9 | aadA1 | 8.3 |
| aadA2 | 14.3 | aadA1, aadA2 | 8.3 | |
| aadA1, aph(3″)-Ib, aph(6)-Id | 11.4 | aac(3)-IIe, aph(3″)-Ib, aph(6)-Id | 5.6 | |
| aadA22 | 11.4 | aadA1, aadA2, aph(3″)-Ib, aph(6)-Id | 5.6 | |
| aac(3)-IIe, aadA1 | 5.7 | aadA1, aph(3″)-Ib, aph(6)-Id | 5.6 | |
| aadA, aadA5, aph(3″)-Ib, aph(6)-Id | 5.7 | aac(3)-IId, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | 2.8 | |
| aac(3)-IId, aadA2 | 2.9 | aac(3)-IIe, aadA1 | 2.8 | |
| aadA1, aadA2, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | 2.9 | aac(3)-IIe, aadA1, aph(3′)-Ia | 2.8 | |
| aadA1, aadA5, aph(3″)-Ib, aph(6)-Id | 2.9 | aac(3)-IIe, aadA5, aph(3″)-Ib, aph(6)-Id | 2.8 | |
| aadA2, aph(3″)-Ib, aph(6)-Id | 2.9 | aac(3)-IIe, aph(3″)-Ib, aph(6)-Id | 2.8 | |
| aadA22, aph(3″)-Ib;aph(3′)-Ia, aph(6)-Id | 2.9 | aac(3)-IVa, aadA1, aph(3″)-Ib, aph(3′)-Ia, aph(4)-Ia, aph(6)-Id | 2.8 | |
| aadA5, aph(3″)-Ib, aph(6)-Id | 2.9 | aadA1, aadA5 | 2.8 | |
| aadA1, aadA5, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | 2.8 | |||
| aadA5 | 2.8 | |||
| aadA5, aph(3″)-Ib, aph(6)-Id | 2.8 | |||
| aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | 2.8 | |||
| aph(3″)-Ib, aph(6)-Id | 2.8 | |||
| Overall | 88.6 | 66.7 | ||
| Aminoglycosides and Fluoroquinolones | aac(6’)-Ib-cr5 | 16.7 | ||
| Overall | 16.7 | |||
| β-lactams | blaEC, blaSHV-12 | 20.0 | blaCTX-M-1, blaEC | 25.0 |
| blaEC, blaTEM-52 | 17.1 | blaCTX-M-1, blaEC, blaTEM-1 | 22.2 | |
| blaEC, blaTEM-1 | 11.4 | blaCTX-M-15, blaEC, blaOXA-1 | 13.9 | |
| blaCTX-M-1, blaEC, blaTEM-1 | 8.6 | blaCTX-M-55, blaEC | 8.3 | |
| blaCTX-M-32, blaEC, blaTEM-1 | 8.6 | blaEC, blaOXA-1, blaTEM-1 | 8.3 | |
| blaEC, blaSHV-12, blaTEM-1 | 8.6 | blaCTX-M-15, blaEC, blaTEM-1 | 5.6 | |
| blaCTX-M-1, blaEC | 5.7 | blaEC, blaSHV-12 | 5.6 | |
| blaCTX-M-15, blaEC, blaTEM-1 | 5.7 | blaCTX-M-1, blaEC, blaOXA-1 | 2.8 | |
| blaEC, blaSHV-2, blaTEM-1 | 5.7 | blaCTX-M-137, blaEC, blaTEM-1 | 2.8 | |
| blaADC, blaOXA | 2.9 | blaCTX-M-14, blaEC, blaTEM-1 | 2.8 | |
| blaCTX-M-1, blaEC, blaSHV-106, blaTEM-1 | 2.9 | blaCTX-M-15, blaEC, blaOXA-1, blaTEM-1 | 2.8 | |
| blaEC, blaTEM-1, blaTEM-19 | 2.9 | |||
| Overall | 100 | 100 | ||
| Phenicols | cmlA1 | 17.1 | catB3 | 13.9 |
| catA1 | 11.4 | floR | 11.1 | |
| catA1, cmlA1 | 5.7 | catA1, floR | 8.3 | |
| cmlA1, floR | 8.3 | |||
| catB3, cmlA1 | 2.8 | |||
| cmlA1 | 2.8 | |||
| Overall | 34.3 | 47.2 | ||
| Diaminopyrimidines | dfrA1 | 14.3 | dfrA17 | 13.9 |
| dfrA12 | 8.6 | dfrA12, dfrA36 | 8.3 | |
| dfrA17 | 5.7 | dfrA1 | 2.8 | |
| dfrA1, dfrA12 | 2.9 | dfrA1, dfrA17 | 2.8 | |
| dfrA1, dfrA14 | 2.9 | dfrA12 | 2.8 | |
| dfrA1, dfrA17 | 2.9 | dfrA12, dfrA5 | 2.8 | |
| dfrA12, dfrA17 | 2.9 | dfrA14 | 2.8 | |
| dfrA14 | 2.9 | dfrA17, dfrA7 | 2.8 | |
| dfrA5 | 2.9 | dfrA5 | 2.8 | |
| Overall | 45.7 | 41.7 | ||
| Macrolides | emrD | 77.1 | emrD | 61.1 |
| emrD, mph(B) | 8.6 | emrD, mph(A) | 30.6 | |
| emrD, mph(A) | 5.7 | emrD, erm(B) | 2.8 | |
| emrD, erm(B) | 2.9 | emrD, mph(A), mph(B) | 2.8 | |
| emrD, mef(B) | 2.9 | emrD, mphE, mef(B),msrE | 2.8 | |
| Overall | 97.1 | 100 | ||
| Lincosamides | lnu(F) | 48.6 | lnu(G) | 11.1 |
| lnu(F), lnu(G) | 2.9 | lnu(F) | 2.8 | |
| Overall | 51.4 | 13.9 | ||
| Phosphonic Acid (Fosfomycin) | fosA | 2.9 | ||
| abaF | 2.9 | |||
| Overall | 5.8 | |||
| Tetracyclines | tet(A) | 48.6 | tet(A) | 36.1 |
| tet(B) | 8.6 | tet(B) | 13.9 | |
| tet(A), tet(M) | 2.9 | tet(A), tet(M) | 8.3 | |
| tet(A), tet(B) | 2.8 | |||
| Overall | 60.0 | 61.1 | ||
| Sulfonamides | sul2 | 28.6 | sul2 | 25.0 |
| sul1, sul2 | 14.3 | sul1, sul2, sul3 | 11.1 | |
| sul3 | 14.3 | sul1 | 8.3 | |
| sul1 | 5.7 | sul1, sul2 | 8.3 | |
| sul2, sul3 | 5.7 | sul3 | 8.3 | |
| sul1, sul3 | 2.9 | sul2, sul3 | 2.8 | |
| Overall | 71.4 | 63.9 | ||
| Streptothricin (Nucleosides) | sat2 | 5.7 | sat2 | 8.3 |
| Overall | 5.7 | 8.3 | ||
| Fluoroquinolones | qnrS1 | 17.1 | qnrS1 | 11.1 |
| qnrB19 | 11.4 | qnrB19 | 5.6 | |
| oqxA, oqxB | 2.9 | |||
| Overall | 31.4 | 16.7 | ||
| Cyclic Polypeptides (Colistin) | mcr-1.1 | 11.4 | mcr-1.1 | 8.3 |
| Overall | 11.4 | 8.3 | ||
| E. coli, n = 35 Poultry Slaughterhouses | E. coli, n = 36 Pig Slaughterhouses | ||||
|---|---|---|---|---|---|
| Sequence Type | n | % | Sequence Type | n | % |
| ST117 | 3 | 8.6 | ST410 | 5 | 13.9 |
| ST10 | 2 | 5.7 | ST10 | 4 | 11.1 |
| ST224 | 2 | 5.7 | ST359 | 3 | 8.3 |
| ST533 | 2 | 5.7 | ST744 | 3 | 8.3 |
| ST648 | 2 | 5.7 | ST58 | 2 | 5.6 |
| ST1011 | 2 | 5.7 | ST88 | 2 | 5.6 |
| ST1730 | 2 | 5.7 | ST117 | 2 | 5.6 |
| ST3995 | 2 | 5.7 | ST101 | 1 | 2.8 |
| ST4994 | 2 | 5.7 | ST131 | 1 | 2.8 |
| ST34 | 1 | 2.9 | ST156 | 1 | 2.8 |
| ST58 | 1 | 2.9 | ST167 | 1 | 2.8 |
| ST101 | 1 | 2.9 | ST224 | 1 | 2.8 |
| ST135 | 1 | 2.9 | ST398 | 1 | 2.8 |
| ST155 | 1 | 2.9 | ST542 | 1 | 2.8 |
| ST162 | 1 | 2.9 | ST617 | 1 | 2.8 |
| ST297 | 1 | 2.9 | ST641 | 1 | 2.8 |
| ST361 | 1 | 2.9 | ST1170 | 1 | 2.8 |
| ST457 | 1 | 2.9 | ST1284 | 1 | 2.8 |
| ST515 | 1 | 2.9 | ST1431 | 1 | 2.8 |
| ST711 | 1 | 2.9 | ST3595 | 1 | 2.8 |
| ST1485 | 1 | 2.9 | |||
| ST6617 | 1 | 2.9 | |||
| unknown STs | 3 | 8.6 | unknown STs | 2 | 5.6 |
| E. coli (n = 35), % Poultry Slaughterhouses | E. coli (n = 36), % Pig Slaughterhouses | ||
|---|---|---|---|
| Adhesins | |||
| fimH | Type 1 fimbriae | 94.3 | 91.7 |
| papC | Genes of P fimbriae operon | 5.7 | 11.1 |
| papEFG | Genes of P fimbriae operon | 5.7 | 8.3 |
| sfa/foc | S or F1C fimbriae | 5.7 | 11.1 |
| focG | F1C fimbriae adhesin | 0 | 0 |
| iha | Adhesin siderophore | 0 | 0 |
| F10 papA | P fimbriae subunit variant | 0 | 0 |
| tsh | Temperature sensitive hemagglutinin | 0 | 0 |
| hra | Heat-resistant agglutinin | 0 | 0 |
| afa/draBC | Dr-binding adhesins | 0 | 0 |
| Toxins | |||
| astA | Enteroaggregative E. coli toxin | 40.0 | 16.7 |
| vat | Vacuolating toxin | 8.6 | 5.6 |
| pic | Serine protease | 8.6 | 2.8 |
| hlyD | Alpha-hemolysin | 0 | 2.8 |
| cnf1 | Cytotoxic necrotizing factor | 0 | 2.8 |
| sat | Secreted autotransporter toxin | 0 | 0 |
| Siderophores | |||
| iutA | Aerobactin receptor | 51.4 | 58.3 |
| iroN | Salmochelin receptor | 48.6 | 30.6 |
| fyuA | Yersiniabactin receptor | 37.1 | 27.8 |
| ireA | Siderophore receptor | 0 | 0 |
| Capsule | |||
| kpsM II | kpsM II group 2 capsule | 22.9 | 5.6 |
| K1 | K1 group 2 capsule variants | 0 | 0 |
| K2 | K2 group 2 capsule variants | 0 | 0 |
| K5 | K5 group 2 capsule variants | 0 | 0 |
| kpsMT III | Group 3 capsule | 0 | 0 |
| Metal | E. coli (n = 35), % Poultry Slaughterhouses | E. coli (n = 36), % Pig Slaughterhouses |
|---|---|---|
| Arsenic | 97.1 | 100.0 |
| Copper | 5.7 | 13.9 |
| Copper/silver | 5.7 | 27.8 |
| Mercury | 25.7 | 38.9 |
| Silver | 5.7 | 27.8 |
| Tellurium | 2.9 | 2.8 |
| Nickel | 2.9 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savin, M.; Bierbaum, G.; Kreyenschmidt, J.; Schmithausen, R.M.; Sib, E.; Schmoger, S.; Käsbohrer, A.; Hammerl, J.A. Clinically Relevant Escherichia coli Isolates from Process Waters and Wastewater of Poultry and Pig Slaughterhouses in Germany. Microorganisms 2021, 9, 698. https://doi.org/10.3390/microorganisms9040698
Savin M, Bierbaum G, Kreyenschmidt J, Schmithausen RM, Sib E, Schmoger S, Käsbohrer A, Hammerl JA. Clinically Relevant Escherichia coli Isolates from Process Waters and Wastewater of Poultry and Pig Slaughterhouses in Germany. Microorganisms. 2021; 9(4):698. https://doi.org/10.3390/microorganisms9040698
Chicago/Turabian StyleSavin, Mykhailo, Gabriele Bierbaum, Judith Kreyenschmidt, Ricarda Maria Schmithausen, Esther Sib, Silvia Schmoger, Annemarie Käsbohrer, and Jens Andre Hammerl. 2021. "Clinically Relevant Escherichia coli Isolates from Process Waters and Wastewater of Poultry and Pig Slaughterhouses in Germany" Microorganisms 9, no. 4: 698. https://doi.org/10.3390/microorganisms9040698
APA StyleSavin, M., Bierbaum, G., Kreyenschmidt, J., Schmithausen, R. M., Sib, E., Schmoger, S., Käsbohrer, A., & Hammerl, J. A. (2021). Clinically Relevant Escherichia coli Isolates from Process Waters and Wastewater of Poultry and Pig Slaughterhouses in Germany. Microorganisms, 9(4), 698. https://doi.org/10.3390/microorganisms9040698






