Abstract
Cotton leaf curl Multan virus (CLCuMuV) is one of the major casual agents of cotton leaf curl disease. Previous studies show that two indigenous whitefly species of the Bemisia tabaci complex, Asia II 1 and Asia II 7, are able to transmit CLCuMuV, but the molecular mechanisms underlying the transmission are poorly known. In this study, we attempted to identify the whitefly proteins involved in CLCuMuV transmission. First, using a yeast two-hybrid system, we identified 54 candidate proteins of Asia II 1 that putatively can interact with the coat protein of CLCuMuV. Second, we examined interactions between the CLCuMuV coat protein and several whitefly proteins, including vacuolar protein sorting-associated protein (Vps) twenty associated 1 (Vta1). Third, using RNA interference, we found that Vta1 positively regulated CLCuMuV acquisition and transmission by the Asia II 1 whitefly. In addition, we showed that the interaction between the CLCuMuV coat protein and Vta1 from the whitefly Middle East-Asia Minor (MEAM1), a poor vector of CLCuMuV, was much weaker than that between Asia II 1 Vta1 and the CLCuMuV coat protein. Silencing of Vta1 in MEAM1 did not affect the quantity of CLCuMuV acquired by the whitefly. Taken together, our results suggest that Vta1 may play an important role in the transmission of CLCuMuV by the whitefly.
1. Introduction
Plant viruses pose considerable threats to the production of many crops in modern agriculture [1]. The majority of plant viruses are transmitted by insect vectors (vector borne) [2]. In the past decades, geminiviruses, a subgroup of vector-borne plant viruses, have caused extensive epidemics in many crops, most notably in developing countries [3]. Among the nine genera in the family Geminiviridae, Begomovirus is the largest genus, containing over 400 species [4,5]. Begomoviruses are transmitted by whiteflies of the Bemisia tabaci complex, which comprises over 40 cryptic species, in a persistent circulative manner [2,3,6].
So far, studies concerning whitefly transmission of begomoviruses have been mostly conducted with tomato yellow leaf curl viruses (TYLCV). As learned from these studies, once TYLCV is acquired by the whitefly, the virus goes through the food canal to reach the filter chamber, from where it crosses the midgut wall and reaches the whitefly hemolymph; the virus then infects whitefly primary salivary glands and is secreted with saliva during feeding [7]. During this process, TYLCV hijacks clathrin-mediated endocytosis and the endosomal network to cross the midgut barrier of the whitefly vector [8,9]. Moreover, the coat protein (CP) of TYLCV may interact with many whitefly proteins, thereby facilitating virus transmission [3,10,11,12]. Two recent reviews on whitefly transmission of begomoviruses indicate that the transmission efficiency of a given virus may vary with different whitefly species, and different viruses may be transmitted with disparate efficiencies by a given whitefly species [3,10]. These variations indicate that the transmission mechanisms among different whitefly–begomovirus combinations may vary, highlighting the need for unravelling transmission mechanisms with previously unexplored whitefly species or begomoviruses.
Cotton leaf curl Multan virus (CLCuMuV) is one of the major casual agents of cotton leaf curl disease, one of the most significant constraints in cotton production in South Asia [13]. CLCuMuV was the major virus causing cotton leaf curl disease in South Asia in the 1990s and seemed to have been displaced by the Burewala strain of the cotton leaf curl Kokhran virus (CLCuKoV-Bur) at the beginning of this century [13,14]. In recent years, however, field surveys in India have revealed the rebound of CLCuMuV and the association of the recombinant variants of this virus with the breakdown of resistance in cotton [15,16]. The field surveys also indicate that in some regions in northwest India, CLCuMuV became the dominant virus in the cotton field, a sign of displacement of CLCuKoV-Bur by CLCuMuV [17].
Laboratory studies on the transmission of CLCuMuV by different whitefly species show that the virus can be efficiently transmitted by Asia II 1 and Asia II 7, two indigenous species of whiteflies from Asia, but can be hardly transmitted by other species of whiteflies, including MEAM1, Mediterranean (MED) and Asia 1 [18,19]. In this study, first we used split-ubiquitin yeast two-hybrid assay to identify Asia II 1 whitefly proteins that putatively interact with the CP of CLCuMuV. Next, we used yeast two-hybrid and pull-down assay to detect the interaction between CLCuMuV CP and several putative whitefly proteins, including vacuolar protein sorting-associated protein (Vps) twenty associated 1 (Vta1). We then used RNA interference to investigate the role of Vta1 in the acquisition and transmission of CLCuMuV by Asia II 1. In addition, we examined the function of Vta1 in MEAM1, a poor vector of CLCuMuV, in its transmission of the virus. Our findings provide new insights into the transmission of CLCuMuV by whiteflies.
2. Material and Methods
2.1. Plants, Insects, and Viruses
For plants, cotton (Gossypium hirsutum L. cv. Zhemian 1793 and Xinhai 21) and tobacco (Nicotiana tabacum L. cv. NC89) were used. Plants were grown in insect-proof greenhouses under natural lighting at controlled temperatures of 25 ± 3 °C and 14 h light/10 h darkness. Infectious clones of CLCuMuV isolate GD37 (GenBank accession number: JN968573) with its conjugated beta-satellite (GenBank accession number: JN968574) were introduced into 3–4 true-leaf-stage tobacco plants. Next, Asia II 1 transmission was used to obtained CLCuMuV-infected tobacco plants. Infection of plants was verified by symptom inspection and PCR detection of the virus using primers CLCuMuV-PCR-F and CLCuMuV-PCR-R (Table S1).
For insects, two whitefly species, namely Asia II 1 (mtCOI GenBank accession number: DQ309077) and MEAM1 (mtCOI GenBank accession number: KM821540), were used. Cultures of the two species were originally established from whiteflies collected from the field and have been maintained on cotton plants (cv. Zhemian 1793). Maintenance of whitefly cultures and all experiments were conducted in climate chambers at 26 ± 2 °C, in 14 h light/10 h darkness, and at 60–80% relative humidity. The purity of each of the whitefly cultures was monitored every 2 months using the mtCOI PCR-RFLP technique and sequencing, as previously reported [20]. All female adult whiteflies were within 7 days post-emergence when used in experiments.
2.2. Yeast Two-Hybrid System
The split-ubiquitin yeast two-hybrid system (Dualsystems Biotech, Zurich, Switzerland) was used to identify Asia II 1 whitefly proteins that interact with the CLCuMuV CP [11]. A cDNA library of Asia II 1 whitefly was constructed in the prey plasmid, SfiI-digested pPR3-N, with the EasyClone cDNA library construction kit (Dualsystems Biotech, Zurich, Switzerland). The quality of the cDNA library was determined as per the kit manual. The titer of the cDNA library of Asia II 1 was over 2 × 106 cfu/19.5 μL, with an average insert size of over 1.0 kb, meeting the requirements of a standard cDNA library. The CLCuMuV CP gene was ligated into the bait plasmid pDHB1 using primers CLCuMuV-CP-pDHB1-infusion-F and CLCuMuV-CP-pDHB1-infusion-R (Table S1). The recombinant plasmid pDHB1-CLCuMuV CP was introduced into yeast strain NMY51, and the expression of the CLCuMuV CP in yeast was verified by Western blotting using anti-TYLCV CP mouse monoclonal antibodies (mAb) (provided by Professor Xue-Ping Zhou, Institute of Biotechnology, Zhejiang University). Next, the cDNA library was introduced into yeast cells containing the pDHB1-CLCuMuV CP. Yeast clones were selected on triple dropout (TDO) medium (S.D./-His/-Leu/-Trp) containing 2.5 mM of 3-aminotriazole (3-AT). The yeast cells were then resuspended in 0.9% NaCl solution (to OD600 = 1.0) and later restreaked on quadruple dropout (QDO) medium (S.D./-Ade/-His/-Leu/-Trp) containing 2.5 mM of 3-AT to verify interactions. In addition, a yeast beta-Gal assay kit (Thermo Scientific, Waltham, MA, USA) was used to examine the interactions by detecting beta-galactosidase activity in yeast clones. Finally, plasmids were recovered from yeast and transformed into Escherichia. coli strain DH5α and then sequenced.
For the verification of interaction, plasmids recovered from yeast clones were transformed into yeast cells containing the pDHB1-CLCuMuV CP using the method described above. The full length of whitefly genes was cloned into the plasmid pPR3-N with primers Vta1-pPR3-N-infusion-F, Vta1-pPR3-N-infusion-R, pPR3-N-infusion-F, and pPR3-N-infusion-R (Table S1) and analyzed using procedures as described above.
2.3. Bioinformatics Analysis
The whitefly genes whose coding proteins were verified to interact with the CLCuMuV CP were annotated using BLAST (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi). Gene Ontology (GO) enrichment analysis was then performed using the OmicShare tools (http://www.omicshare.com/tools).
2.4. Cloning of Vta1 in Asia II 1 and MEAM1
The predicted full length of MEAM1 Vta1 was found on the NCBI database (Genbank accession code: LOC109029924). Therefore, we cloned MEAM1 Vta1 and submitted it to the NCBI database under accession number NW380743. The full length of Vta1 in the Asia II 1 whitefly was amplified by the SMARTer RACE 5′/3′ kit (Clontech, Kyoto Japan), as per the manufacturer’s protocol, with primers 5′ race-Vta1, 5′ race-Vta1, and 3′ race-Vta1-CS1 (Table S1). Total RNAs extracted from the Asia II 1 whitefly using TRIzol reagent (Ambion, Waltham, MA, USA) were used in RACE. Next, Asia II 1 Vta1 was cloned and submitted to the NCBI database under accession number MW346674.
2.5. Pull-Down Assay
The full length of the CLCuMuV CP gene was cloned into pGEX-6p-1 for fusion with glutathione S-transferase (GST) using primers CLCuMuV-CP-pGEX-6p-1-F and CLCuMuV-CP-pGEX-6p-1-R (Table S1). The full length of Vta1 was cloned into pMAL-c5x for fusion with maltose-binding protein (MBP) using primers Vta1-pMAL-c5x-F and Vta1-pMAL-c5x-R (Table S1). Recombinant proteins were expressed in E. coli strain BL21. After purification, the GST-CLCuMuV CP and GST (control) were allowed to bind to glutathione agarose beads (GE Healthcare, Boston, MA, USA) for 2 h at 4 °C. The beads were then washed and incubated with MBP-Vta1 or MBP (control) at 4 °C for 4 h. Next, the beads were washed and boiled, and the bead-bound proteins were separated using SDS-PAGE and detected using Western blotting with anti-MBP rabbit polyclonal antibodies (pAb) (Abcam, Cambridge, UK).
2.6. Double Strand RNA (DsRNA) Synthesis and Membrane Feeding
DNA templates for dsRNA synthesis were amplified by PCR using primers with the T7 promoter at both ends, namely Vta1 (Asia II 1)-T7-F, Vta1 (Asia II 1)-T7-R, Vta1 (MEAM1)-T7-F, and Vta1 (MEAM1)-T7-R (Table S1). DsRNA synthesis was conducted with a T7 high-yield RNA transcription kit (Vazyme, Nanjing, China). Next, dsRNA was purified, and the quality and concentration were determined using agarose gel electrophoresis and Nanodrop (Thermo Fisher, Waltham, MA, USA). For membrane feeding, dsRNA-targeting Vta1 or GFP (control) was added to 15% sucrose solution to make the final concentration 200 ng/μL. Whiteflies were collected and released into artificial diet feeding chambers, as described before [8]. The duration of membrane feeding was 48 h.
2.7. Analysis of Gene Expression Level
Total RNAs of the whitefly were extracted using TRIzol reagent as per the manufacturer’s instructions. cDNA was synthesized from 1 μg of RNA using the PrimeScript RT reagent kit (Takara, Kyoto, Japan). qPCR was performed on the CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with SYBR Premix Ex Taq II (TaKaRa, Kyoto, Japan). Primers β-actin-qPCR-F and β-actin-qPCR-R were used as internal controls. Vta1 (Asia II 1)-qPCR-F and Vta1 (Asia II 1)-qPCR-R were used for Asia II 1 Vta1, and Vta1 (MEAM1)-qPCR-F and Vta1 (MEAM1)-qPCR-R were used for MEAM1 Vta1. All primes are listed in Table S1.
2.8. Virus Acquisition and Quantification of CLCuMuV in the Whitefly
For virus acquisition, whiteflies were collected and allowed to feed on CLCuMuV-infected tobacco plants. Forty-eight hours later, virus quantification was performed. For the whitefly whole body, whitefly adults were collected as groups of 15 and lysed in lysis buffer (50 mM KCl, 10 mM Tris, 0.45% Tween 20, 0.2% gelatin, 0.45% NP40, 60 mg/mL proteinase K, with pH 8.4). As for organs, 4 midguts or primary salivary glands were collected as one sample, and hemolymph from 4 whiteflies was treated as one sample. The collection and preparation of whitefly organs were conducted, as described before [18]. qPCR was conducted, as described above, with primers β-actin-qPCR-F, β-actin-qPCR-R, CLCuMuV-qPCR-F, and CLCuMuV-qPCR-R (Table S1).
2.9. Virus Transmission
Plants of tobacco (cv. NC89) and cotton (cv. Xinhai 21) were used. When tobacco plants were used, female adult whiteflies that had fed on virus-infected plants for 48 h were collected as groups of 10 and then transferred to be placed on a 3–4 true-leaf-stage tobacco seedling using clip cages [21] to feed for 48 h. Three replicates were conducted, with each containing 6–10 plants. The whiteflies were then removed, and the plants were sprayed with imidacloprid (20 mg/L) to kill all the eggs. The infection status of test plants was examined by symptom inspection and PCR detection of viral DNAs, as described above, 30 days post-inoculation.
When cotton plants were used, female adult whiteflies that had fed on virus-infected plants for 72 h were collected as groups of 10 and then transferred to feed on a 1–2 true-leaf-stage cotton seedling (enclosed in leaf-clip cages) for 72 h. Three replicates were conducted, with each containing 6–10 plants of both tobacco and cotton. After that, observations were conducted using the same procedure as that for tobacco except that the infection status of the plants was conducted 70 days post-inoculation.
2.10. Statistical Analysis
All qPCR data were calculated using 2−△Ct as normalized to whitefly actin. For the comparison of gene expression level and quantity of viruses, an independent t-test was used. For the comparison of transmission efficiency, percentage data were arcsine-square-root-transformed for statistical analysis using an independent t-test and back-transformed for presentation. All statistical analyses were conducted using SPSS 20.0 Statistics (IBM, Armonk, NY, USA) and Microsoft Excel (version 2016, Microsoft, Redmond, WA, USA).
3. Results
3.1. Identification of Asia II 1 Whitefly Proteins That Interact with the CLCuMuV CP
We used the split-ubiquitin yeast two-hybrid system to identify proteins in the Asia II 1 whitefly that potentially interact with the CLCuMuV CP. The expression of the bait plasmid pDHB1-CLCuMuV CP in yeast was verified using Western blotting (Figure S1), showing the functionality of the bait plasmid pDHB1-CLCuMuV CP. After screening of the Asia II 1 whitefly cDNA library using this yeast two-hybrid system, more than 300 positive clones were isolated and the sequencing results indicated that plasmids in these positive clones encode 200 unique proteins. To examine the interactions between the CLCuMuV CP and whitefly proteins encoded by the plasmids recovered from positive clones, we chose 70 proteins to examine their interaction with the CLCuMuV CP using the yeast two-hybrid system. Of these proteins, 54 were found to interact with the CLCuMuV CP. A BLAST search of the NCBI database was then conducted for the 54 proteins, and the names of the protein sequences that share the highest homology were obtained (Table 1).

Table 1.
List of Asia II 1 proteins that putatively interact with the CLCuMuV CP as identified by the yeast two-hybrid system.
3.2. Bioinformatics Analysis of Whitefly Proteins That Interact with the CLCuMuV CP
Using GO analysis, the whitefly proteins that putatively interact with the CLCuMuV CP were assigned to 13 biological processes (BPs) or seven molecular functions (MFs) or two cellular components (CCs) (Figure 1). Specifically, of the 13 BPs, cellular process contained the majority of proteins (19). Further analysis of these proteins indicated that some proteins can be further classified into vesicle-mediated transport (GO: 0016192), including calmodulin (CALM), vesicle transport protein SEC22 (SEC22), vacuolar protein sorting-associated protein (Vps) twenty associated 1 (Vta1), Ras-related protein Rab-6A (RAB6A), and vesicle-associated membrane protein 7 (VAMP7).
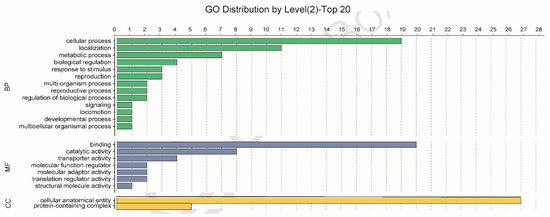
Figure 1.
Gene Ontology (GO) analysis of the 54 Asia II 1 whitefly proteins that putatively interact with the CLCuMuV CP, as indicated by the yeast two-hybrid system. Different colors represent different GO categories (BP: biological process; MF: molecular function; CC: cellular components). GO annotation was conducted using Blast2GO software, and the figure was generated using OmicShare tools (http://www.omicshare.com/tools).
3.3. Verification of Interaction Using Yeast Two-Hybrid and Pull-Down Systems
For verification of the interaction between the CLCuMuV CP and prey proteins, we selected the five proteins in the GO category vesicle-mediated transport (GO:0016192) for further analysis using the yeast two-hybrid system. The full-length open reading frames of the five genes were amplified and ligated into pPR3-N, and then these prey plasmids and the bait plasmid pDHB1-CLCuMuV CP were co-transformed into yeast cells. Among the five proteins, the full lengths of calmodulin (CALM), vesicle transport protein SEC22 (SEC22), and vacuolar protein sorting-associated protein (Vps) twenty associated 1 (Vta1) were found to interact with the CLCuMuV CP (Figure 2A). Since Vta1 has been reported to regulate virus–host interactions, we subjected it to further analysis [22,23]. Analysis of beta-gal activity confirmed the interaction between Vta1 and the CLCuMuV CP (Figure 2B). And in the pull-down assay, when the fusion protein GST-CLCuMuV CP was used as a bait protein and the fusion protein MBP-Vta1 used as the prey protein, the prey protein could co-elute with the GST-fused CLCuMuV CP (Figure 2C). These results suggest that Vta1 from Asia II 1 whitefly can interact with the CLCuMuV CP both in vivo and in vitro.
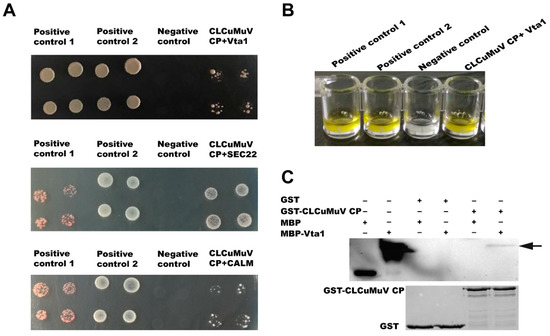
Figure 2.
Verification of interaction between the CLCuMuV CP and whitefly proteins. The interaction between the full length of calmodulin (CALM), vesicle transport protein SEC22 (SEC22), and vacuolar protein sorting-associated protein (Vps) twenty associated 1 (Vta1) with the CLCuMuV CP in yeast (A). Positive control 1: pDHB1-CLCuMV-CP+pOst1-NubI; positive control 2: pDHB1-large T+pDSL- p53; negative control: pDHB1-CLCuMV-CP+pPR3-N. Beta-gal activity in yeast cells (B). Pull-down assay between CLCuMuV CP and Vta1 (C).
3.4. Functional Characterization of Vta1 in Asia II 1 Transmission of CLCuMuV
To examine the function of Vta1, Asia II 1 whiteflies were fed with Vta1 dsRNA. Following dsRNA feeding, the expression of Vta1 in whiteflies was down-regulated by 27.4% as compared to controls (Figure 3A). Next, the whiteflies were transferred to feed on CLCuMuV-infected tobacco plants for 48 h for virus acquisition. Knockdown of Vta1 resulted in a significant decrease in the relative virus quantity in the whiteflies’ whole body, midgut, and hemolymph but did not cause a significant change in the relative virus quantity in primary salivary glands (Figure 3B). Further, transmission trails were performed. When tobacco plants were used as test plants, knockdown of Vta1 resulted in a significant decrease in CLCuMuV transmission, as shown by percentages of plants with viral symptoms but not by viral detection using PCR (Figure 4A,B). Similar results were found when cotton plants were used as test plants (Figure 4C,D). These results suggest that Vta1 plays an important role in CLCuMuV acquisition and transmission by Asia II 1.
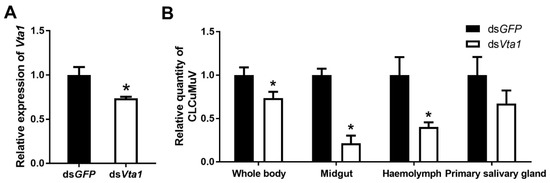
Figure 3.
Effects of Vta1 knockdown on CLCuMuV acquisition by Asia II 1. Gene expression level of Vta1 following dsRNA feeding (n = 4 for dsVta1 or dsGFP) (A). Following feeding, virus quantity in whitefly whole body and organs (n = 3–8 for whole body and midgut, 19–22 for hemolymph, and 12 for primary salivary glands) (B). * stands for significant difference (independent t-test, p < 0.05).
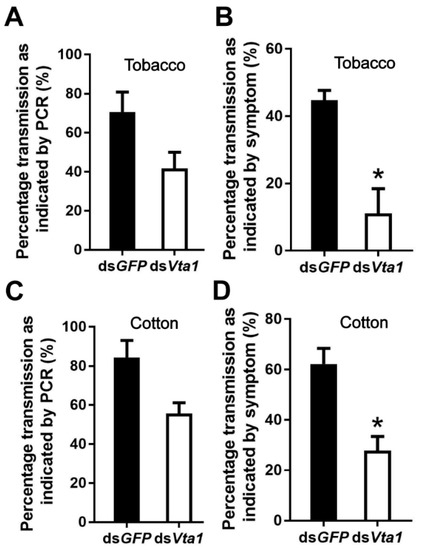
Figure 4.
Effects of Vta1 knockdown on CLCuMuV transmission by Asia II 1. Whiteflies that had acquired CLCuMuV were collected and transferred to feed on tobacco (A,B) or cotton (C,D). On each of the two test plants, 3 replicates were conducted for both dsGFP and dsVta1, with each replicate containing 6–10 plants. * stands for significant difference (independent t-test, p < 0.05).
3.5. The Role of Vta1 in CLCuMuV Transmission by MEAM1
Asia II 1 was able to readily transmit CLCuMuV, while MEAM1 can only transmit this virus with very low efficiency [18]. To explore whether Vta1 plays a role in CLCuMuV transmission by MEAM1, we first compared the amino acid sequence of MEAM1 Vta1 with that of Asia II 1 Vta1 and found that six amino acids are different between the two Vta1s (Figure 5). The yeast two-hybrid system was then used to compare Asia II 1 Vta1 and MEAM1 Vta1 in interaction with the CLCuMuV CP. Yeast cells were resuspended to certain ODs and then cultured on quadruple dropout medium (SD/-Leu/-Trp/-His/-Ade) containing 2.5 mM of 3-AT. When OD600 was 1.0, yeast cells containing the pDHB1-CLCuMuV CP and pPR3-N-MEAM1 Vta1 did not grow, but cells containing pDHB1-CLCuMuV CP and pPR3-N-Asia II 1 Vta1 grew to noticeable colonies. To further determine whether there is any detectable interaction between the CLCuMuV CP and MEAM1 Vta1, we adjusted OD600 to 2.0. Under this condition, yeast cells containing the pDHB1-CLCuMuV CP and pPR3-N-MEAM1 Vta1 grew to colonies, but they were much smaller than those containing the pDHB1-CLCuMuV CP and pPR3-N-Asia II 1 Vta1 (Figure 6A). Analysis of beta-gal activity in yeast cells containing different combination of plasmids revealed that the interaction between MEAM1 Vta1 and the CLCuMuV CP was very weak, if any, as judged by the unappreciable yellow color in the solution (Figure 6B). Knockdown of MEAM1 Vta1 resulted in significant down-regulation of the Vta1 expression level by 44.4% (Figure 6C). Following virus acquisition, knockdown of Vta1 did not change the quantity of CLCuMuV acquired by MEAM1 in two independent experiments (Figure 6D). These results suggest that Vta1 plays a minor, if any, role in CLCuMuV transmission by the MEAM1 whitefly.
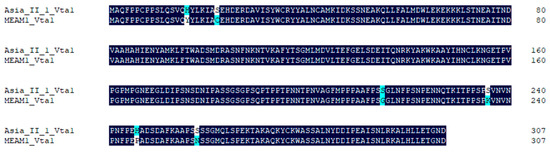
Figure 5.
Comparison of amino acid sequences of Asia II 1 Vta1 and MEAM1 Vta1. Alignments were performed using DNAMAN. Dark blue indicates consensus in amino acids between the two sequences, and light blue or white indicates divergence in amino acids.
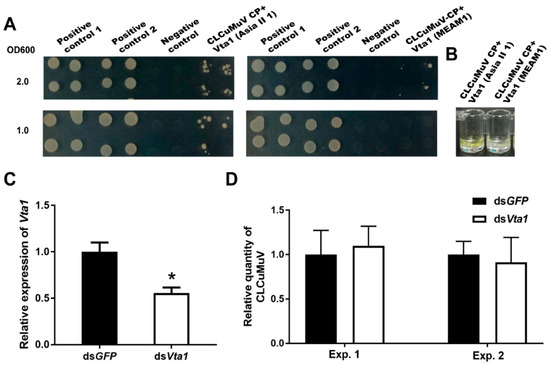
Figure 6.
The role of Vta1 in CLCuMuV transmission by MEAM1. The yeast two-hybrid assay was performed to compare the affinity of the CLCuMuV CP to Asia II 1 Vta1 and MEAM1 Vta1. Yeast cells containing different combination of plasmids were resuspended to OD600 being 1.0 and 2.0 and then cultured on quadruple dropout medium containing 2.5 mM of 3-AT (A). Positive control 1: pDHB1-CLCuMV-CP+pOst1-NubI; positive control 2: pDHB1-large T+pDSL-p53; negative control: pDHB1-CLCuMV-CP+pPR3-N. Yeast beta-gal assay kit was used to examine the interactions (B). Further, knockdown of MEAM1 Vta1 was performed, and the whiteflies were then set to acquire CLCuMuV for 48 h. MEAM1 Vta1 expression level (n = 4 for dsGFP or dsVta1) (C), and quantity of CLCuMuV in whitefly whole body in two experiments (n = 4–5 for dsGFP or dsVta1 in each experiment) (D). * stands for significant difference (independent t-test, p < 0.05 for C,D).
4. Discussion
In this study, using the yeast two-hybrid system, we identified 54 candidate Asia II 1 proteins that putatively interact with the CLCuMuV CP (Table 1). GO enrichment analysis showed that these proteins may be responsible for 13 different biological processes (Figure 1). Based on the molecular function of the identified proteins, we selected five proteins to verify their interactions with the CLCuMuV CP using the yeast two-hybrid system and confirmed the interaction of Vta1 with the CLCuMuV CP using the GST pull-down assay (Figure 2). Next, we found that RNA interference of Vta1 in Asia II 1 reduced the virus quantity in the whitefly and the efficiency of virus transmission (Figure 3 and Figure 4). It should be noted that decreases in virus quantity were significant in the whitefly midgut and hemolymph but not in primary salivary glands upon Vta1 silencing (Figure 3). This might be due to the fact that the silencing efficiency of Vta1 is lower in primary salivary glands than that in the midgut and hemolymph. Moreover, knockdown of Vta1 in Asia II 1 results in significant decreases in the virus transmission efficiency, as indicated by symptom inspection, but the decrease is not significant when examined by PCR (Figure 4). The possible reason for the discrepancy is that PCR is very sensitive, so it may amplify even trace amounts of viral DNAs in whitefly-inoculated plants. However, symptom appearance requires the accumulation of a substantial amount of viruses, which is a better indicator of successful inoculation. In addition, we showed that MEAM1 Vta1, the sequence of which is slightly different from that of Asia II 1 Vta1, exhibited much lower affinity to the CLCuMuV CP than Asia II 1 Vta1, and RNA interference of MEAM1 Vta1 did not affect the quantity of virus acquired by MEAM1 (Figure 5 and Figure 6).
In eukaryotic cells, Vta1 functions as a cofactor of vacuolar protein sorting 4 (Vps4) by impacting its oligomerization, thereby regulating the activity of Vps4 to modulate endosomal sorting complexes required for transport (ESCRT) [24]. ESCRTs include ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III and are involved in regulating the function of multivesicular bodies, which are an endosomal-membrane-trafficking and protein-sorting station [25]. In the context of virus–host or virus–vector interactions, the role of Vta1 is little known. The mammalian homologue of Vta1, LIP5, positively regulates the budding of human immunodeficiency virus type 1 in human cells [23]. In addition, deletion of MIT domains of Spodoptera frugiperda Vta1 reduces the replication of Autographa californica multiple nucleopolyhedrovirus [22]. In this study, we found that Vta1 positively regulates CLCuMuV transport across the midgut of Asia II 1 whiteflies following viral acquisition, as well as the efficiency of virus transmission. This may be due to the fact that Vta1 affects the ESCRT machinery via its action on Vps4, and the ESCRT machinery regulates vesicle trafficking, which has been shown to regulate whitefly transmission of begomoviruses [8,9]. Further, our recent findings show that the early endosome plays an important role in begomoviruses intracellular transport and an endocytic receptor can regulate vesicle transport of begomoviruses in whitefly midgut cells [9,12]. These results suggest that endosomal trafficking is important for the transport of both CLCuMuV and TYLCV across epithelial cells in the whitefly midgut. However, the unambiguous dissection of the role of endosomes and Vta1 in Asia II 1 transmission of CLCuMuV warrants further investigations.
For a given begomovirus, different whitefly species may transmit with disparate efficiencies [3,10]. Case studies have shown that this can be attributed to the differential capacity of the virus to cross barriers within the body of different whitefly species [18,26,27]. For example, the relatively high and low efficiencies in transmitting CLCuMuV by Asia II 1 and MEAM1 were found to be associated with the relatively high and low efficiencies of the virus to cross the midgut wall of the two species of whiteflies [18]. Here, we found that Vta1 from Asia II 1 positively regulates CLCuMuV transport across the midgut and transmission, and MEAM1 Vta1 does not seem to play a role in CLCuMuV transmission. Additionally, Asia II 1 Vta1 displays a stronger affinity to the CLCuMuV CP than MEAM1 Vta1. Hence, we propose that Vta1 may be a significant factor in determining the disparate efficiencies of CLCuMuV transmission by Asia II 1 and MEAM1, possibly through its different functions in facilitating the virus to cross physiological barriers such as the midgut in the vector body. Interestingly, a comparison of the amino acid sequences of MEAM1 Vta1 and Asia II 1 Vta1 showed that they differ in only six amino acids. The divergence at the six amino acids may directly contribute to the differential transmission of CLCuMuV by MEAM1 and Asia II 1. Of course, more empirical studies are needed to verify this hypothesis.
In summary, we identified 54 proteins from the Asia II 1 whitefly that putatively interact with the CLCuMuV CP. We showed that Asia II 1 Vta1 positively regulates the acquisition and transmission of CLCuMuV by the Asia II 1 whitefly. We further found that Vta1 from MEAM1, a poor vector of CLCuMuV, interacts with the CLCuMuV CP weakly and does not seem to play a role in CLCuMuV transmission by this whitefly. Taken together, our findings indicate that the protein Vta1 may be an important factor in determining the efficiency of CLCuMuV transmission by a given species of whitefly, and provide new insight into the interaction between single strand DNA (ssDNA) viruses and their insect vectors.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/2/304/s1, Figure S1: The expression of pDHB1-CLCuMuV CP in yeast. Expression of CLCuMuV CP bait fusion protein was detected by western blot using anti-TYLCV CP antibodies; Table S1: Primers used in this study.
Author Contributions
Conceptualization, L.-L.P., S.-S.L., S.M. and X.-W.W.; Data curation, Y.C. and L.-L.P.; Formal analysis, Y.C., L.-L.P. and X.-W.W.; Funding acquisition, S.-S.L. and X.-W.W.; Investigation, Y.C.; Methodology, Y.C. and L.-L.P.; Project administration, Y.C.; Resources, S.-S.L.; Supervision, S.-S.L. and X.-W.W.; Validation, Y.C.; Visualization, Y.C.; Writing-original draft, Y.C. and L.-L.P.; Writing-review & editing, L.-L.P., S.-S.L., S.M. and X.-W.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China, grant number 31925033 and 31930092.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data in this study are available from the authors upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lefeuvre, P.; Martin, D.P.; Elena, S.F.; Shepherd, D.N.; Roumagnac, P.; Varsani, A. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 2019, 17, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Ammar, E.; Whitfield, A.E.; Redinbaugh, M.G. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [PubMed]
- Wang, X.; Blanc, S. Insect transmission of plant single-stranded DNA viruses. Annu. Rev. Phytopathol. 2021, 66, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV Virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses ICTV. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/ssdna-viruses/w/geminiviridae (accessed on 25 November 2020).
- Kanakala, S.; Ghanim, M. Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of Tomato yellow leaf curl virus. Front. Plant. Sci. 2016, 7, 1702. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, M.; Morin, S.; Czosnek, H. Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 2001, 91, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.L.; Chen, Q.F.; Zhao, J.J.; Guo, T.; Wang, X.W.; Hariton-Shalev, A.; Czosnek, H.; Liu, S.S. Clathrin-mediated endocytosis is involved in Tomato yellow leaf curl virus transport across the midgut barrier of its whitefly vector. Virology 2017, 502, 152–159. [Google Scholar] [CrossRef]
- Xia, W.Q.; Liang, Y.; Chi, Y.; Pan, L.L.; Zhao, J.; Liu, S.S.; Wang, X.W. Intracellular trafficking of begomoviruses in the midgut cells of their insect vector. PLoS Pathog. 2018, 14, e1006866. [Google Scholar] [CrossRef]
- Fiallo-Olive, E.; Pan, L.L.; Liu, S.S.; Navas-Castillo, J. Transmission of begomoviruses and other whitefly-borne viruses: Dependence on the vector species. Phytopathology 2020, 110, 10–17. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, T.; Lei, T.; Zhu, J.; Wang, F.; Wang, X.W.; Liu, S.S. Proteomic analyses of whitefly-begomovirus interactions reveal the inhibitory role of tumorous imaginal discs in viral retention. Front. Immunol. 2020, 11, 1596. [Google Scholar] [CrossRef]
- Zhao, J.; Lei, T.; Zhang, X.J.; Yin, T.Y.; Wang, X.W.; Liu, S.S. A vector whitefly endocytic receptor facilitates the entry of begomoviruses into its midgut cells via binding to virion capsid proteins. PLoS Pathog. 2020, 16, e1009053. [Google Scholar] [CrossRef] [PubMed]
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease—An emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W. Cotton leaf curl disease, a multicomponent begomovirus complex. Mol. Plant. Pathol. 2003, 4, 427–434. [Google Scholar] [CrossRef]
- Datta, S.; Budhauliya, R.; Das, B.; Gopalakrishnan, R.; Sharma, S.; Chatterjee, S.; Vanlalhmuaka; Raju, P.S.; Veer, V. Rebound of Cotton leaf curl Multan virus and its exclusive detection in cotton leaf curl disease outbreak, Punjab (India), 2015. Sci. Rep. 2017, 7, 17361. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, P.K.; Kumar, P.; Kalbande, B.B.; Chavhan, R.L.; Koundal, V.; Monga, D.; Pappu, H.R.; Roy, A.; Mandal, B. Recombinant variants of cotton leaf curl Multan virus is associated with the breakdown of leaf curl resistance in cotton in northwestern India. Virus Dis. 2020, 31, 45–55. [Google Scholar] [CrossRef]
- Biswas, K.K.; Bhattacharyya, U.K.; Palchoudhury, S.; Balram, N.; Kumar, A.; Arora, R.; Sain, S.K.; Kumar, P.; Khetarpal, R.K.; Sanyal, A.; et al. Dominance of recombinant cotton leaf curl Multan-Rajasthan virus associated with cotton leaf curl disease outbreak in northwest India. PLoS ONE 2020, 15, e231886. [Google Scholar] [CrossRef]
- Pan, L.L.; Cui, X.Y.; Chen, Q.F.; Wang, X.W.; Liu, S.S. Cotton leaf curl disease: Which whitefly is the vector? Phytopathology 2018, 108, 1172–1183. [Google Scholar] [CrossRef]
- Chen, T.; Saeed, Q.; He, Z.; Lu, L. Transmission efficiency of Cotton leaf curl Multan virus by three cryptic species of Bemisia tabaci complex in cotton cultivars. Peer J. 2019, 7, e7788. [Google Scholar] [CrossRef]
- Qin, L.; Wang, J.; Bing, X.L.; Liu, S.S. Identification of nine cryptic species of Bemisia tabaci (Hemiptera: Aleyrodidae) from China by using the mtCOI PCR-RFLP technique. Acta Entomol. Sin. 2013, 56, 186–194, (In Chinese with English abstract). [Google Scholar]
- Ruan, Y.M.; Luan, J.B.; Zang, L.S.; Liu, S.S. Observing and recording copulation events of whiteflies on plants using a video camera. Entomol. Exp. Appl. 2007, 124, 229–233. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Wang, S.; Yu, Q.; Li, Z. Effects of deletion of MIT domains of host Vta1 on replication of Autographa californica multiple nucleopolyhedrovirus. Acta Microbiol. Sin. 2019, 59, 247–257, (In Chinese with English abstract). [Google Scholar]
- Ward, D.M.; Vaughn, M.B.; Shiflett, S.L.; White, P.L.; Pollock, A.L.; Hill, J.; Schnegelberger, R.; Sundquist, W.I.; Kaplan, J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 2005, 280, 10548–10555. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H. The ESCRT complexes. Crit. Rev. Biochem. Mol. 2010, 45, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.I.; Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Bi. 2012, 28, 337–362. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhao, J.J.; Zhang, T.; Li, F.F.; Ghanim, M.; Zhou, X.P.; Ye, G.Y.; Liu, S.S.; Wang, X.W. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. J. Virol. 2014, 88, 13460–13468. [Google Scholar] [CrossRef]
- Guo, T.; Zhao, J.; Pan, L.L.; Geng, L.; Lei, T.; Wang, X.W.; Liu, S.S. The level of midgut penetration of two begomoviruses affects their acquisition and transmission by two species of Bemisia tabaci. Virology 2018, 515, 66–73. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).