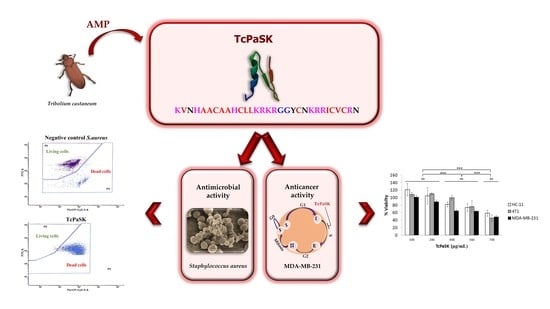
Dual Antimicrobial and Antiproliferative Activity of TcPaSK Peptide Derived from a Tribolium castaneum Insect Defensin
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
- VNHAACAAHCLLKRKRGGYCNKRRICVCR;
- TcPaSK: KVNHAACAAHCLLKRKRGGYCNKRRICVCRN;
- hBD-3: GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK
2.2. Reagents and Chemicals
2.3. Cell Culture Conditions
2.4. Minimal Inhibitory Concentration (MIC)
2.5. Hemolytic Activity
2.6. Antimicrobial Activity
2.7. Scanning Electron Microscopy (SEM)
2.8. Transmission Electron Microscopy (TEM)
2.9. Cell Viability Assay
2.10. Cell Proliferation Assay
2.11. Cell Cycle Analysis
2.12. Sample Preparation for Proteomic Analyses
2.13. Library LC-MS/MS Analysis
2.14. SWATH LC-MS/MS Analysis
2.15. SWATH Data Analysis and Protein Quantitation
3. Results
3.1. TcPaSK Is a Cationic Peptide that Shares the Common Structural Features of AMPs and May Form an Amphipathic α-Helix
3.2. TcPaSK Peptide Exhibits Potent Antibacterial Activity against S. aureus
3.3. TcPaSK Peptide Induces Morphological Alteration of S. aureus Cells
3.4. TcPaSK Peptide Alters Cell Membrane Integrity and Impairs Cell Division in S. aureus
3.5. TcPaSK Peptide Interacts with Mammalian Cells
3.6. TcPaSK Peptide Inhibits MDA-MB-231 TNBC Cell Proliferation at a Subcytotoxic Concentration by Blocking G1-S Cell Cycle Progression
3.7. TcPaSK Affects MDA-MB-231 TNBC Cell Expression of Proteins Involved in Cell Growth and Tumor Progression
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Stork, N.E. How many species of insects and other terrestrial arthropods are there on earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Uvell, H.; Engström, Y. A multilayered defense against infection: Combinatorial control of insect immune genes. Trends Genet. 2007, 23, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Phil. Trans. R. Soc. B 2016, 371, 20150290. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Patočka, J.; Kuča, K. Insect antimicrobial peptides, a mini review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Tonk, M.; Vilcinskas, A. The medical potential of antimicrobial peptides from insects. Curr. Top. Med. Chem. 2017, 17, 554–575. [Google Scholar] [CrossRef]
- Ratcliffe, N.A.; Mello, C.B.; Garcia, E.S.; Butt, T.M.; Azambuja, P. Insect natural products and processes: New treatments for human disease. Insect Biochem. Mol. Biol. 2011, 41, 747–769. [Google Scholar] [CrossRef]
- Ratcliffe, N.; Azambuja, P.; Mello, C.B. Recent advances in developing insect natural products as potential modern day medicines. Evid. Based Complement. Alternat. Med. 2014, 2014, 904958. [Google Scholar] [CrossRef]
- Koehbach, J. Structure-activity relationships of insect defensins. Front. Chem. 2017, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed]
- Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with dual antimicrobial and anticancer activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Tonk, M.; Vilcinskas, A.; Rahnamaeian, M. Insect antimicrobial peptides: Potential tools for the prevention of skin cancer. Appl. Microbiol. Biotechnol. 2016, 100, 7397–7405. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Ishibashi, J.; Tanaka, H.; Sato, M.; Asaoka, A.; Taylor, D.M.; Yamakawa, M. Selective cancer cell cytotoxicity of enantiomeric 9-mer peptides derived from beetle defensins depends on negatively charged phosphatidylserine on the cell Surface. Peptides 2009, 30, 660–668. [Google Scholar] [CrossRef]
- Kim, I.W.; Kim, S.J.; Kwon, Y.N.; Yun, E.Y.; Ahn, M.Y.; Kang, D.C.; Hwang, J.S. Effects of the synthetic coprisin analog peptide, CopA3 in pathogenic microorganisms and mammalian cancer cells. J. Microbiol. Biotechnol. 2012, 22, 156–158. [Google Scholar] [CrossRef]
- Kim, I.W.; Lee, J.H.; Kwon, Y.N.; Yun, E.Y.; Nam, S.H.; Ahn, M.Y.; Kang, D.C.; Hwang, J.S. Anticancer activity of a synthetic peptide derived from harmoniasin, an antibacterial peptide from the ladybug Harmonia axyridis. Int. J. Oncol. 2013, 43, 622–628. [Google Scholar] [CrossRef]
- Contreras, E.; Benito-Jardon, M.; Lopez-Galiano, M.J.; Real, M.D.; Rausell, C. Tribolium castaneum immune defense genes are differentially expressed in response to Bacillus thuringiensis toxins sharing common receptor molecules and exhibiting disparate toxicity. Dev. Comp. Immunol. 2015, 50, 139–145. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides. Methods Mol. Biol. 2017, 1548, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Landon, C.; Barbault, F.; Legrain, M.; Guenneugues, M.; Vovelle, F. Rational design of peptides active against the gram positive bacteria. Staphylococcus aureus. Proteins 2008, 72, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Z.; Wang, C.; Lang, L.; Zhou, Y.; Wang, H.; Shang, D.J. Design of potent, non-toxic anticancer peptides based on the structure of the antimicrobial peptide, temporin-1CEa. Arch. Pharm. Res. 2013, 36, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Milburn, D.; Laskowski, R.A.; Thornton, J.M. Sequences annotated by structure: A tool to facilitate the use of structural information in sequence analysis. Prot. Eng. 1998, 11, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Takashima, H.; Tamaoki, H.; Kyogoku, Y.; Lambert, P.; Kuroda, H.; Chino, N.; Watanabe, T.X.; Kimura, T.; Sakakibara, S.; et al. The cysteine-stabilized α-helix: A common structural motif of ion-channel blocking neurotoxic peptides. Biopolymers 1991, 31, 1213–1220. [Google Scholar] [CrossRef]
- Tamaoki, H.; Miura, R.; Kusunoki, M.; Kyogoku, Y.; Kobayashi, Y.; Moroder, L. Folding motifs induced and stabilized by distinct cysteine frameworks. Protein Eng. 1998, 11, 649–659. [Google Scholar] [CrossRef]
- Cornet, B.; Bonmatin, J.-M.; Hetru, C.; Hoffmann, J.A.; Ptak, M.; Vovelle, F. Refined three-dimensional solution structure of insect defensin A. Structure 1995, 3, 435–448. [Google Scholar] [CrossRef]
- Patil, A.A.; Cai, Y.; Sang, Y.; Blecha, F.; Zhang, G. Cross-species analysis of themammalian beta-defensin gene family: Presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol. Genom. 2005, 23, 5–17. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.; Wang, H.; Xi, X.; Ma, C.; Liu, Y.; Zhou, M.; Du, Q.; Burrows, J.F.; Wei, M.; Chen, T.; et al. Design of N-terminal derivatives from a novel Dermaseptin exhibiting broad-spectrum antimicrobial activity against isolates from cystic fibrosis patients. Biomolecules 2019, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Hilchie, A.L.; Doucette, C.D.; Pinto, D.M.; Patrzykat, A.; Dougla, S.; Hoskin, D.W. Pleurocidin-family cationic antimicrobial peptides are cytolytic for breast carcinoma cells and prevent growth of tumor xenografts. Breast Cancer Res. 2011, 13, R102. [Google Scholar] [CrossRef] [PubMed]
- Thundimadathil, J. Cancer treatment using peptides: Current therapies and future prospects. J. Amino Acids 2012, 2012. [Google Scholar] [CrossRef]
- Lawrence, R.T.; Perez, E.M.; Hernández, D.; Miller, C.P.; Haas, K.M.; Irie, H.Y.; Lee, S.I.; Blau, C.A.; Villén, J. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015, 11, 1–15. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Mura, M.; Hopkins, T.G.; Michael, T.; Abd-Latip, N.; Weir, J.; Aboagye, E.; Mauri, F.; Jameson, C.; Sturge, J.; Gabra, H.; et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 2015, 34, 5025–5036. [Google Scholar] [CrossRef]
- Wang, D.; Wang, L.; Ren, C.; Zhang, P.; Wang, M.; Zhang, S. High expression of density-regulated re-initiation and release factor drives tumourigenesis and affects clinical outcome. Oncol. Lett. 2019, 17, 141–148. [Google Scholar] [CrossRef]
- Jiménez-Vacas, J.M.; Herrero-Aguayo, V.; Montero-Hidalgo, A.J.; Gómez-Gómeza, E.; Fuentes-Fayos, A.C.; León-González, A.J.; Sáez-Martínez, P.; Alors-Pérez, E.; Pedraza-Arévalo, S.; González-Serrano, T.; et al. Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cáncer. EBioMedicine 2020, 51, 102547. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Pu, Y.; Amina, Q.; Wang, Q.; Zhang, M.; Song, J.; Guo, J.; Mardan, M. Prognostic and therapeutic potential of Adenylate kinase 2 in lung adenocarcinoma. Sci. Rep. 2019, 9, 17757. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Tan, Q.; Li, J.; Yang, W.; Lian, B.; Mo, Q.; Wei, C. Elevated expression of POLD1 is associated with poor prognosis in breast cancer. Oncol. Lett. 2018, 16, 5591–5598. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, T.; Watt, K.; Truesdell, P.; Meens, J.; Schneider, M.M.; Sengupta, S.K.; Craig, A.W. Endophilin A2 promotes TNBC cell invasion and tumor metastasis. Mol. Cancer Res. 2015, 13, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Peng, Y.; Mao, B.; Qian, K. Thioredoxin reductase: A novel, independent prognostic marker in patients with hepatocellular carcinoma. Oncotarget 2015, 6, 17792–17804. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Cai, F.; Ding, Z.; Gao, L. MMP14 predicts a poor prognosis in patients with colorectal cancer. Hum. Pathol. 2019, 83, 36–42. [Google Scholar] [CrossRef]
- Lee, J.R.; Roh, J.-L.; Lee, S.M.; Park, Y.; Cho, K.J.; Choi, S.-H.; Nam, S.Y.; Kim, S.Y. Overexpression of glutathione peroxidase 1 predicts poor prognosis in oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 2257–2265. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Andreeva, A.; Howorth, D.; Chothia, C.; Kulesha, E.; Murzin, A. SCOP2 prototype: A new approach to protein structure mining. Nucleic Acid Res. 2014, 42, 10–14. [Google Scholar] [CrossRef]
- De Oliveira Dias, R.; Franco, O.L. Cysteine-stabilized αβdefensins: From a common fold to antibacterial activity. Peptides 2015, 72, 64–72. [Google Scholar] [CrossRef]
- Tarr, D.E.K. Establishing a reference array for the CS-αβ superfamily of defensive peptides. BMC Res. Notes 2016, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.C.; Hancock, R.E.W.; Yu, P.L. Antimicrobial activity and bacterial membrane interaction of ovine-derived cathelicidins. Antimicrob. Agents Chemother. 2004, 48, 673–676. [Google Scholar] [CrossRef] [PubMed]
- Che, C.H.; Lu, T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef]
- Hemmingsson, O.; Zhang, Y.; Still, M.; Naredi, P. ASNA1, an ATPase targeting tail-anchored proteins, regulates melanoma cell growth and sensitivity to cisplatin and arsenite. Cancer Chemother. Pharmacol. 2009, 63, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Hemmingsson, O.; Nojd, M.; Kao, G.; Naredi, P. Increased sensitivity to platinating agents and arsenite in human ovarian cancer by downregulation of ASNA1. Oncol. Rep. 2009, 22, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Ito, E.; Yue, S.; Moriyama, E.H.; Hui, A.B.; Kim, I.; Shi, W.; Alaje, N.M.; Bhogal, N.; Li, G.H.; Datti, A.; et al. Uroporphyrinogen decarboxylase is a radiosensitizing target for head and neck cáncer. Sci. Transl. Med. 2011, 3, 67ra7. [Google Scholar] [CrossRef] [PubMed]
- Booy, E.P.; Howard, R.; Marushchak, O.; Ariy, E.O.; Meie, M.; Novakowski, S.K.; De, S.R.; Dzananovic, E.; Stetefeld, J.; McKenna, S.A. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res. 2014, 42, 3346–3361. [Google Scholar] [CrossRef]
- Kato, M.; Goto, Y.; Matsushita, R.; Kurozumi, A.; Fukumoto, I.; Nishikawa, R.; Sakamoto, S.; Enokida, H.; Nakagawa, M.; Ichikawa, T.; et al. MicroRNA-26a/b directly regulate La-related protein 1 and inhibit cancer cell invasion in prostate cancer. Int. J. Oncol. 2015, 47, 710–718. [Google Scholar] [CrossRef]
- Ye, L.; Lin, S.-T.; Mi, Y.-S.; Liu, Y.; Ma, Y.; Sun, H.-M.; Peng, Z.-A.; Fan, J.-W. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumor Biol. 2016, 37, 14585–14594. [Google Scholar] [CrossRef]
- Hopkins, T.G.; Mura, M.; Al-Ashtal, H.A.; Lahr, R.M.; Abd-Latip, N.; Sweeney, K.; Lu, H.; Weir, J.; El-Bahrawy, M.; Steel, J.H.; et al. The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cáncer. Nucleic Acids Res. 2016, 44, 1227–1246. [Google Scholar] [CrossRef]
- Zhang, P.; Feng, S.; Liu, G.; Wang, H.; Fu, A.; Zhu, H.; Ren, Q.; Wang, B.; Xu, X.; Bai, H.; et al. CD82 suppresses CD44 alternative splicing-dependent melanoma metastasis by mediating U2AF2 ubiquitination and degradation. Oncogene 2016, 35, 5056–5069. [Google Scholar] [CrossRef] [PubMed]
- Bossi, D.; Cicalese, A.; Dellino, G.A.; Luzi, L.; Riva, L.; D’Alesio, C.; Diaferia, G.R.; Carugo, A.; Cavallaro, E.; Piccioni, R.; et al. In vivo genetic screens of patient-derived tumors revealed unexpected frailty of the transformed phenotype. Cancer Dicov. 2016, 6, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, F.; Borghi, M.; Marzoli, F.; Azzarito, T.; Matarrese, P.; Iessi, E.; Venturi, G.; Meschini, S.; Canitano, A.; Bona, R.; et al. TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene 2015, 34, 5163–5174. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Jiang, H.; Liang, S.; Wang, Y.; Jiang, W.; Zhu, C. Ribosomal protein L15 is involved in colon carcinogenesis. Int. J. Med. Sci. 2019, 16, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Schneider, M.; Kölle, P.; Kuhlencordt, P.; Förster, H.; Beck, H.; Bornkamm, G.W.; Conrad, M. Loss of thioredoxin reductase 1 renders tumors highly susceptible to pharmacologic glutathione deprivation. Cancer Res. 2010, 70, 9505–9514. [Google Scholar] [CrossRef] [PubMed]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Sign. 2012, 15, 1957–1997. [Google Scholar] [CrossRef]
- Gonzalez-Molina, J.; Gramolelli, S.; Liao, Z.; Carlson, J.W.; Ojal, P.V.; Lehti, K. MMP14 in sarcoma: A regulator of tumor microenvironment communication in connective tissues. Cells 2019, 8, 991. [Google Scholar] [CrossRef]
- Le, C.-F.; Fang, C.-M.; Sekaranc, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef]
- De la Fuente-Núñez, C.; Silva, O.N.; Lu, T.K.; Franco, O.L. Antimicrobial peptides: Role in human disease and potential as immunotherapies. Pharmacol. Ther. 2017, 178, 132–140. [Google Scholar] [CrossRef]







| TcPaSK | Tcdef3 | |
|---|---|---|
| Sequence | KVNHAACAAHCLLKRKRGGYCNKRRICVCRN 31 residues:Hydrophobic residues are in red, P and G are in blue, positively charged are in magenta | VNHAACAAHCLLKRKRGGYCNKRRICVCR 29 residues:Hydrophobic residues are in red, P and G are in blue, positively charged are in magenta |
| MW (daltons) * | 3543.30 | 3301.02 |
| NC * | +9 | +8 |
| pI * | 10.6 | 10.5 |
| HR ** | 45% | 48% |
| H ** | 0.225 | 0.296 |
| GRAVY ** | −0.545 | −0.328 |
| APD Theoretical predictions ** |
|
|
| Peak Name | Group | t-Value | p-Value | Fold Change C/T |
|---|---|---|---|---|
| sp|O43681|ASNA_HUMAN | ATPase ASNA1 | 3.92852 | 0.02483 | 4.81 |
| sp|P06132|DCUP_HUMAN | Uroporphyrinogen decarboxylase | 4.99399 | 0.00485 | 4.66 |
| sp|Q9H2U1|DHX36_HUMAN | ATP-dependent RNA helicase DHX36 | 2.90488 | 0.02776 | 3.69 |
| sp|Q6PKG0|LARP1_HUMAN | La-related protein 1 | 3.78106 | 0.00944 | 3.63 |
| sp|Q9NR46|SHLB2_HUMAN | Endophilin-B2 | 4.20419 | 0.00903 | 3.44 |
| sp|O43583|DENR_HUMAN | Density-regulated protein | 2.88173 | 0.02873 | 3.22 |
| sp|P26368|U2AF2_HUMAN | Splicing factor U2AF 65 kDa subunit | 2.82794 | 0.03412 | 3.20 |
| sp|Q9UIG0|BAZ1B_HUMAN | Tyrosine-protein kinase BAZ1B | 4.12155 | 0.00622 | 3.19 |
| sp|Q92544|TM9S4_HUMAN | Transmembrane 9 superfamily member 4 | 4.36180 | 0.01640 | 2.72 |
| sp|P28340|DPOD1_HUMAN | DNA polymerase delta catalytic subunit | 2.99222 | 0.04858 | 2.01 |
| sp|P54819|KAD2_HUMAN | Adenylate kinase 2, mitochondrial | 2.88071 | 0.02808 | 1.98 |
| sp|P46777|RL5_HUMAN | 60S ribosomal protein L5 | 2.54086 | 0.04416 | 1.66 |
| sp|Q99961|SH3G1_HUMAN | Endophilin-A2 | 3.82975 | 0.01530 | 1.58 |
| sp|P55145|MANF_HUMAN | Mesencephalic astrocyte-derived neurotrophic factor | 2.87810 | 0.04330 | 1.58 |
| sp|Q05682|CALD1_HUMAN | Caldesmon | 2.76783 | 0.03276 | 1.40 |
| sp|P67936|TPM4_HUMAN | Tropomyosin alpha-4 chain | 2.82893 | 0.03159 | 1.36 |
| sp|P37802|TAGL2_HUMAN | Transgelin-2 | 3.76778 | 0.00985 | 1.34 |
| sp|P51114|FXR1_HUMAN | Fragile X mental retardation syndrome-related protein 1 | 2.57835 | 0.04190 | 1.31 |
| sp|P62277|RS13_HUMAN | 40S ribosomal protein S13 | 2.98365 | 0.02454 | 1.22 |
| sp|P17812|PYRG1_HUMAN | CTP synthase 1 | 3.22684 | 0.03034 | 1.22 |
| sp|Q9H3H3|CK068_HUMAN | UPF0696 protein C11orf68 | 5.03897 | 0.00243 | 1.20 |
| sp|P55884|EIF3B_HUMAN | Eukaryotic translation initiation factor 3 subunit B | 2.84937 | 0.03000 | 1.19 |
| sp|Q92841|DDX17_HUMAN | Probable ATP-dependent RNA helicase DDX17 | 3.09782 | 0.04655 | 1.19 |
| sp|Q01105|SET_HUMAN | Protein SET | 2.81192 | 0.04233 | 1.11 |
| sp|Q9H5V8|CDCP1_HUMAN | CUB domain-containing protein 1 | −3.09560 | 0.03259 | 0.82 |
| sp|Q9Y6N5|SQRD_HUMAN | Sulfide:quinone oxidoreductase, mitochondrial | −2.72138 | 0.03583 | 0.77 |
| sp|O00560|SDCB1_HUMAN | Syntenin-1 | −2.95845 | 0.03792 | 0.70 |
| sp|P15153|RAC2_HUMAN | Ras-related C3 botulinum toxin substrate 2 | −3.38378 | 0.01906 | 0.69 |
| sp|Q16881|TRXR1_HUMAN | Thioredoxin reductase 1, cytoplasmic | −2.83327 | 0.03590 | 0.63 |
| sp|P50281|MMP14_HUMAN | Matrix metalloproteinase-14 | −3.22740 | 0.03678 | 0.52 |
| sp|P07203|GPX1_HUMAN | Glutathione peroxidase 1 | −5.48171 | 0.00695 | 0.17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robles-Fort, A.; García-Robles, I.; Fernando, W.; Hoskin, D.W.; Rausell, C.; Real, M.D. Dual Antimicrobial and Antiproliferative Activity of TcPaSK Peptide Derived from a Tribolium castaneum Insect Defensin. Microorganisms 2021, 9, 222. https://doi.org/10.3390/microorganisms9020222
Robles-Fort A, García-Robles I, Fernando W, Hoskin DW, Rausell C, Real MD. Dual Antimicrobial and Antiproliferative Activity of TcPaSK Peptide Derived from a Tribolium castaneum Insect Defensin. Microorganisms. 2021; 9(2):222. https://doi.org/10.3390/microorganisms9020222
Chicago/Turabian StyleRobles-Fort, Aida, Inmaculada García-Robles, Wasundara Fernando, David W. Hoskin, Carolina Rausell, and María Dolores Real. 2021. "Dual Antimicrobial and Antiproliferative Activity of TcPaSK Peptide Derived from a Tribolium castaneum Insect Defensin" Microorganisms 9, no. 2: 222. https://doi.org/10.3390/microorganisms9020222
APA StyleRobles-Fort, A., García-Robles, I., Fernando, W., Hoskin, D. W., Rausell, C., & Real, M. D. (2021). Dual Antimicrobial and Antiproliferative Activity of TcPaSK Peptide Derived from a Tribolium castaneum Insect Defensin. Microorganisms, 9(2), 222. https://doi.org/10.3390/microorganisms9020222