Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Conditions
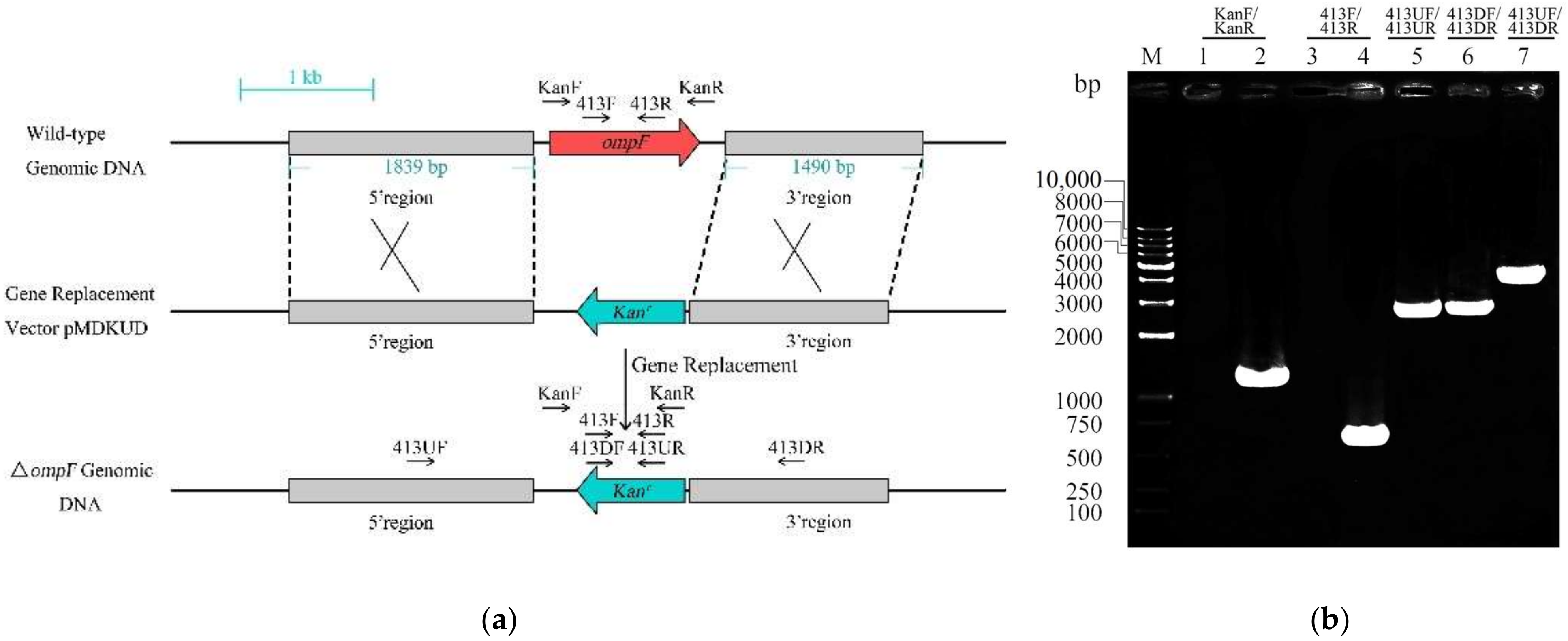
2.2. Construction of ompF Deletion Mutant
2.3. Complementation Study
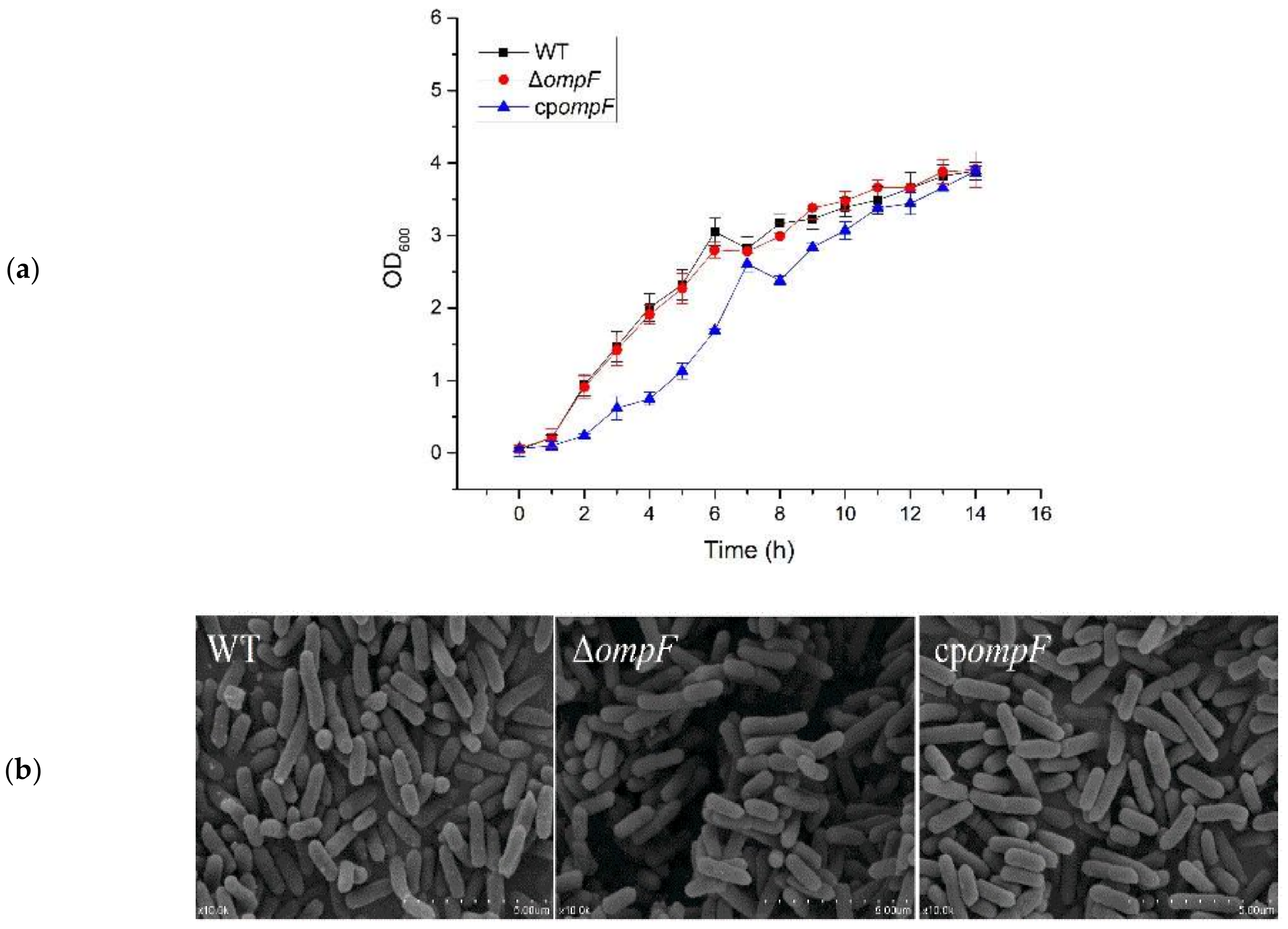
2.4. Growth Curves
2.5. Morphological Differences
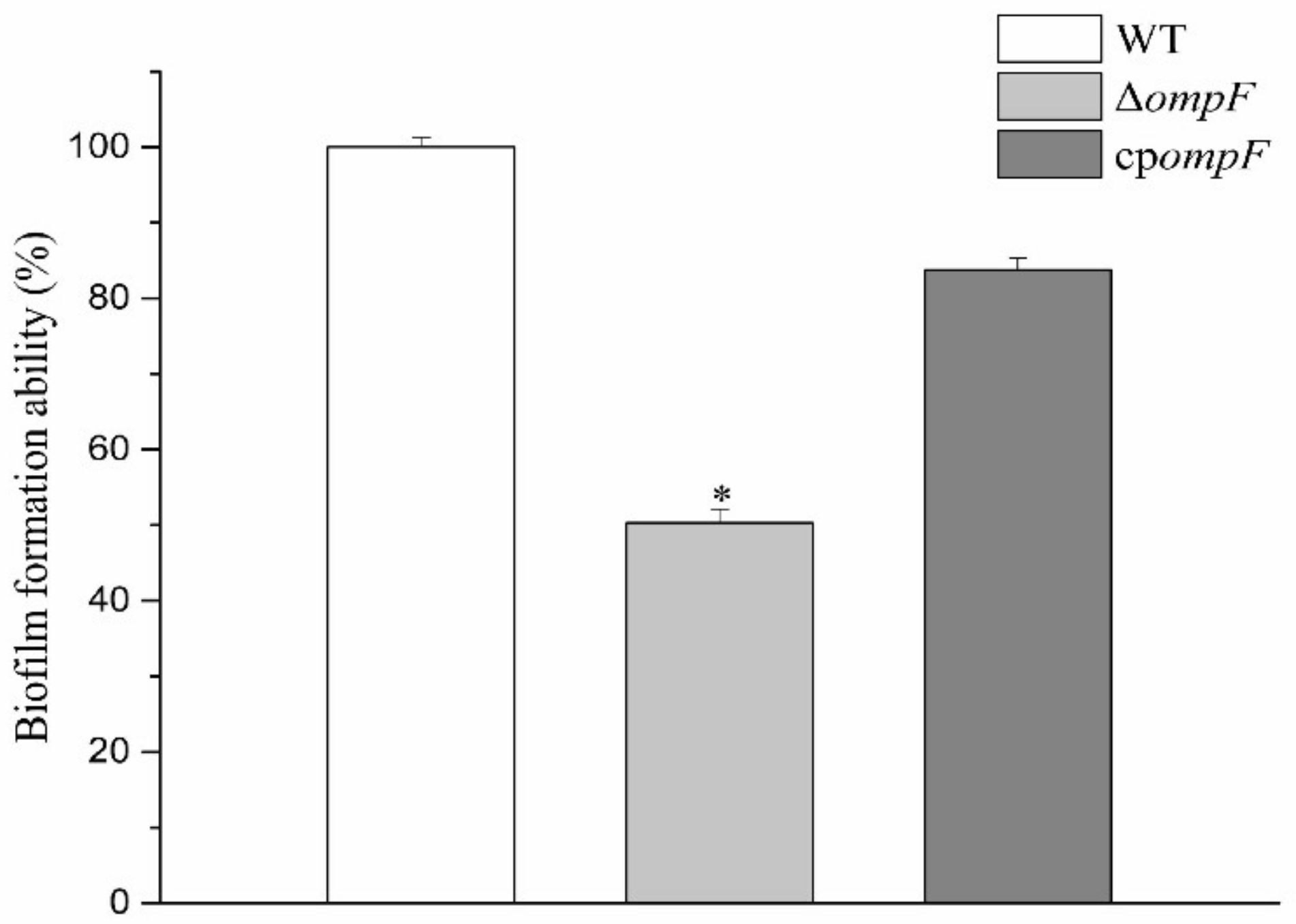
2.6. Analysis of Biofilm Formation Ability
2.7. Determination of Biofilm Biochemical Components
2.8. Analysis of C. sakazakii LPS by SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.9. Adhesion/Invasion Assay
2.10. Cell Permeability Assay
2.11. Antimicrobial Susceptibility Testing
2.12. Statistical Analysis
3. Results
3.1. Verification and Growth Characterization of the ompF Mutant
3.2. Estimation of the Ability to Form Biofilms
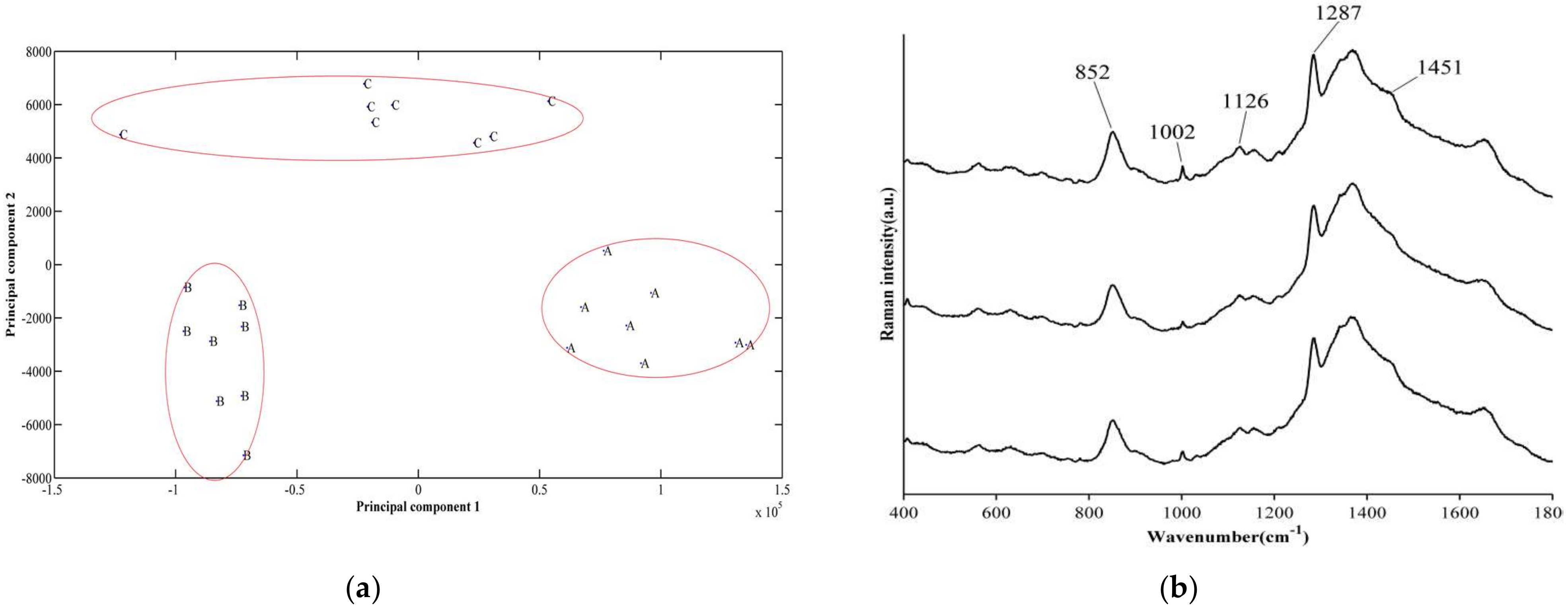
3.3. Differences in Biofilm Composition Examined by Raman Spectroscopy
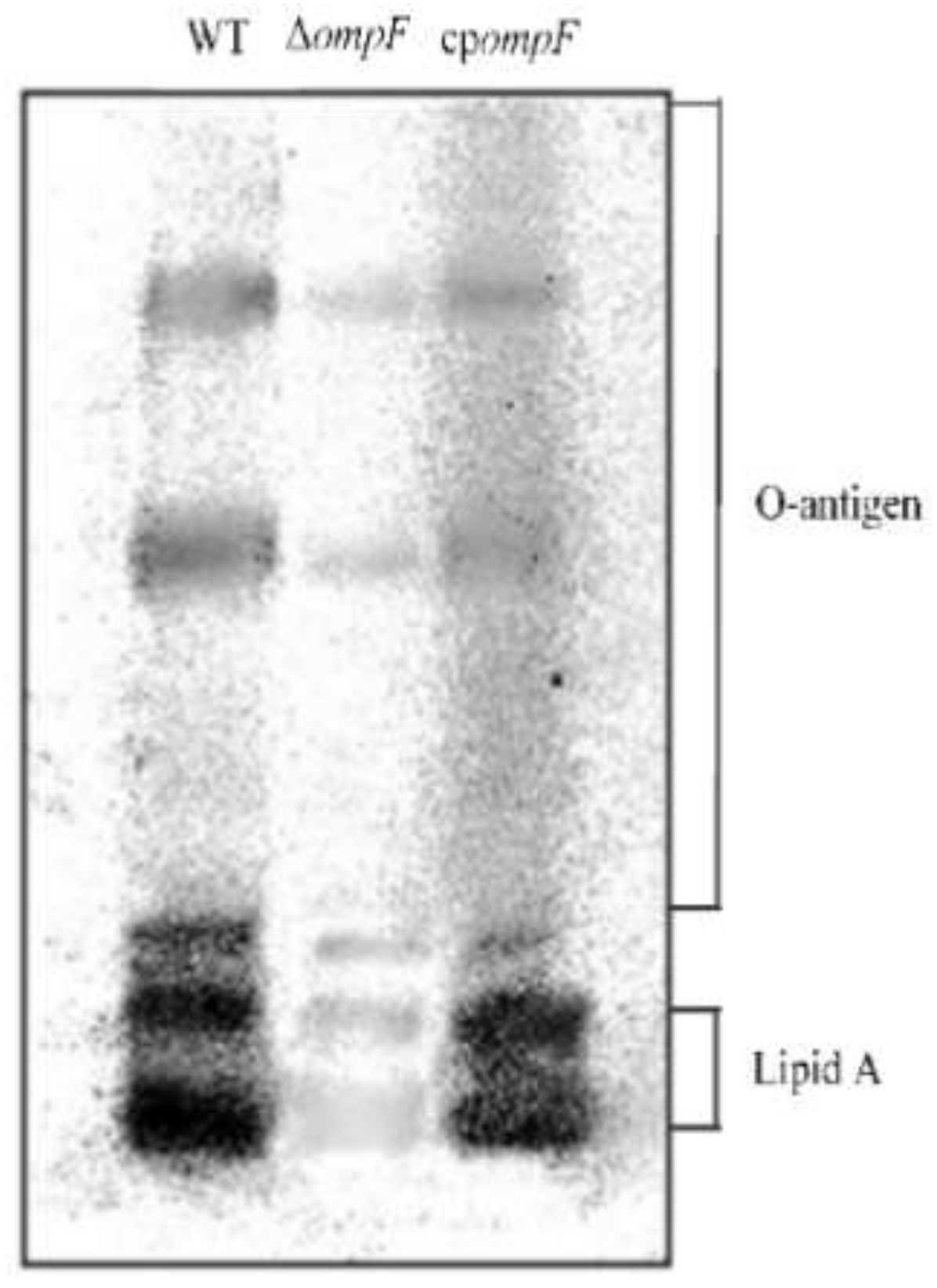
3.4. Analysis of the LPS Content of the ompF Mutant and WT Strains
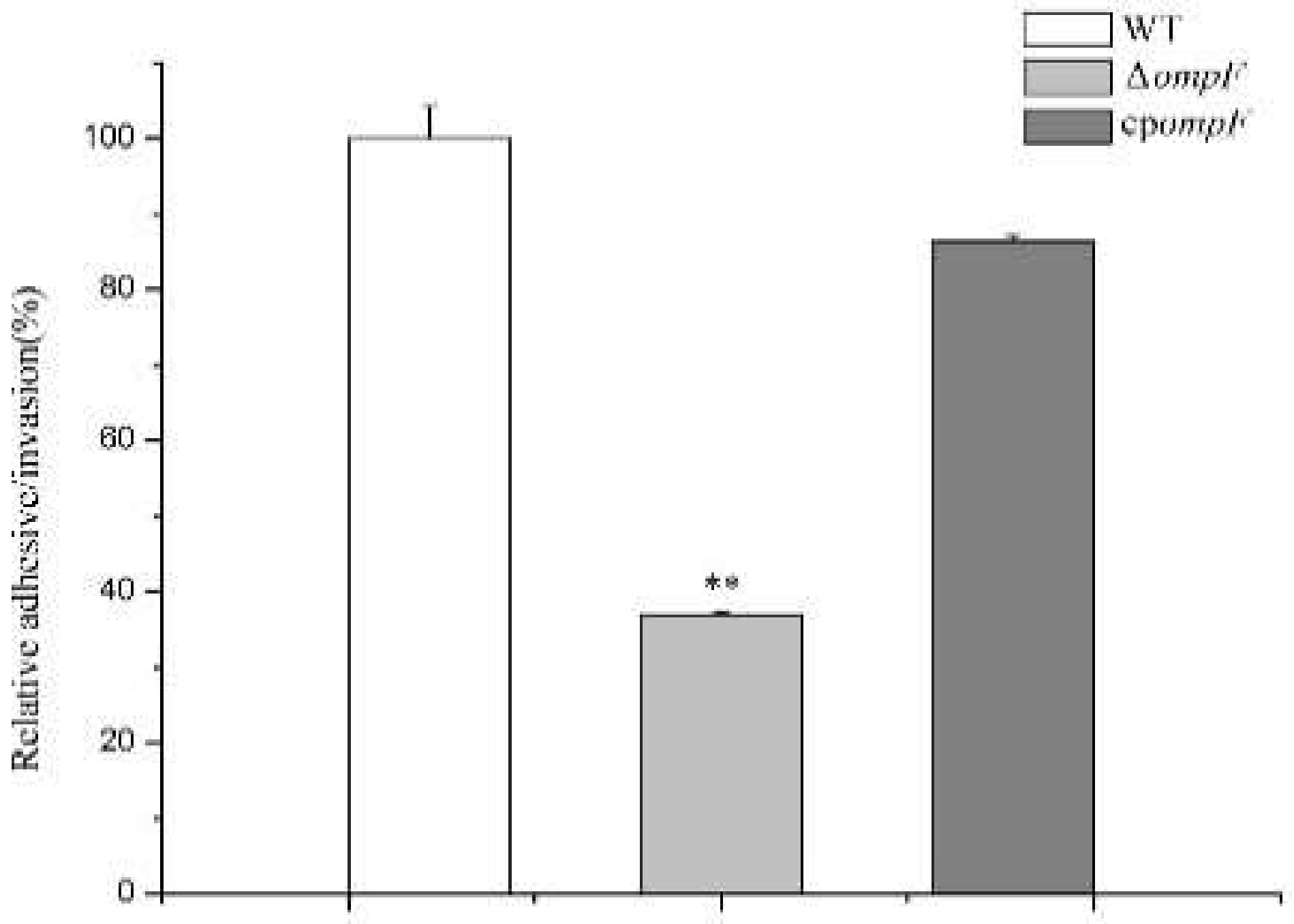
3.5. ompF Affects Adhesion/Invasion
3.6. Evaluation of Cell Permeability
3.7. Estimation of Antibiotic Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Blackwood, B.P.; Hunter, C.J. Cronobacter Spp. Microbiol. Spectr. 2016, 4, 255. [Google Scholar] [CrossRef]
- Jin, T.; Guan, N.; Du, Y.; Zhang, X.; Li, J.; Xia, X. Cronobacter sakazakii ATCC 29544 translocated human brain microvascular endothelial cells via endocytosis, apoptosis Induction, and disruption of tight junction. Front. Microbiol. 2021, 12, 675020. [Google Scholar] [CrossRef]
- Forsythe, S.J.; Dickins, B.; Jolley, K.A. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genom. 2014, 15, 1121. [Google Scholar] [CrossRef]
- Joseph, S.; Desai, P.; Ji, Y.; Cummings, C.A.; Shih, R.; Degoricija, L.; Rico, A.; Brzoska, P.; Hamby, S.E.; Masood, N.; et al. Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS ONE 2012, 7, e49455. [Google Scholar] [CrossRef] [PubMed]
- Morato Rodríguez, M.D.; Velandia Rodríguez, D.; Castañeda, S.; Crosby, M.; Vera, H. Cronobacter spp. in Common Breast Milk Substitutes, Bogotá, Colombia. Emerg. Infect. Dis. 2018, 24, 1907–1909. [Google Scholar] [CrossRef]
- Ling, N.; Jiang, Y.; Zeng, H.; Ding, Y.; Forsythe, S. Advances in our understanding and distribution of the Cronobacter genus in China. Int. J. Food Microbiol. J. Food Sci. 2021, 86, 276–283. [Google Scholar]
- Marotta, S.M.; Giarratana, F.; Calvagna, A.; Ziino, G.; Giuffrida, A.; Panebianco, A. Study on microbial communities in domestic kitchen sponges: Evidence of Cronobacter sakazakii and Extended Spectrum Beta Lactamase (ESBL) producing bacteria. Ital. J. Food Saf. 2018, 7, 7672. [Google Scholar] [CrossRef]
- Almajed, F.S.; Forsythe, S.J. Cronobacter sakazakii clinical isolates overcome host barriers and evade the immune response. Microb. Pathog. 2016, 90, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Kuchma, S.L.; O’Toole, G.A. Keeping their options open: Acute versus persistent infections. J. Bacteriol. 2006, 188, 1211–1217. [Google Scholar] [CrossRef]
- Lehner, A.; Riedel, K.; Eberl, L.; Breeuwer, P.; Diep, B.; Stephan, R. Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: Aspects promoting environmental persistence. J. Food Prot. 2005, 68, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ryu, J.H.; Beuchat, L.R. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 2006, 72, 5846–5856. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, I.; Carranza, P.; Lehner, A.; Stephan, R.; Eberl, L.; Riedel, K. Genes involved in Cronobacter sakazakii biofilm formation. Appl. Environ. Microbiol. 2010, 76, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Achouak, W.; Heulin, T.; Pagès, J.M. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 2001, 199, 1–7. [Google Scholar] [CrossRef]
- Hatfaludi, T.; Al-Hasani, K.; Boyce, J.D.; Adler, B. Outer membrane proteins of Pasteurella multocida. Vet. Microbiol. 2010, 144, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Im, W.; Roux, B. Ion permeation and selectivity of OmpF porin: A theoretical study based on molecular dynamics, brownian dynamics, and continuum electrodiffusion theory. J. Mol. Biol. 2002, 322, 851–869. [Google Scholar] [CrossRef]
- Varma, S.; Chiu, S.W.; Jakobsson, E. The influence of amino acid protonation states on molecular dynamics simulations of the bacterial porin OmpF. Biophys. J. 2006, 90, 112–123. [Google Scholar] [CrossRef]
- Prehna, G.; Zhang, G.; Gong, X.; Duszyk, M.; Okon, M.; McIntosh, L.P.; Weiner, J.H.; Strynadka, N.C. A protein export pathway involving Escherichia coli porins. Structure 2012, 20, 1154–1166. [Google Scholar] [CrossRef]
- Nestorovich, E.M.; Rostovtseva, T.K.; Bezrukov, S.M. Residue ionization and ion transport through OmpF channels. Biophys. J. 2003, 85, 3718–3729. [Google Scholar] [CrossRef]
- Kojima, S.; Nikaido, H. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc. Natl. Acad. Sci. USA 2013, 110, 2629–2634. [Google Scholar] [CrossRef]
- Kim, K.; Kim, K.P.; Choi, J.; Lim, J.A.; Lee, J.; Hwang, S.; Ryu, S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef]
- Chang, A.C.; Cohen, S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978, 134, 1141–1156. [Google Scholar] [CrossRef]
- Hu, L.; Grim, C.J.; Franco, A.A.; Jarvis, K.G.; Sathyamoorthy, V.; Kothary, M.H.; McCardell, B.A.; Tall, B.D. Analysis of the cellulose synthase operon genes, bcsA, bcsB, and bcsC in Cronobacter species: Prevalence among species and their roles in biofilm formation and cellecell aggregation. Food Microbiol. 2015, 52, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.X.; Li, P.; Du, X.J.; Han, Z.H.; Xue, R.; Liang, B.; Wang, S. A Negative Regulator of Cellulose Biosynthesis, bcsR, Affects Biofilm Formation, and Adhesion/Invasion Ability of Cronobacter sakazakii. Front. Microbiol. 2017, 8, 1839. [Google Scholar] [CrossRef]
- Hong, T.P.; Carter, M.Q.; Struffi, P.; Casonato, S.; Hao, Y.; Lam, J.S.; Lory, S.; Jousson, O. Conjugative type IVb pilus recognizes lipopolysaccharide of recipient cells to initiate PAPI-1 pathogenicity island transfer in Pseudomonas aeruginosa. BMC Microbiol. 2017, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Jarvis, K.G.; Chase, H.R.; Hebert, K.; Trach, L.H.; Lee, C.; Sadowski, J.; Lee, B.; Hwang, S.; Sathyamoorthy, V.; et al. A proposed harmonized LPS molecular-subtyping scheme for Cronobacter species. Food Microbiol. 2015, 50, 38–43. [Google Scholar] [CrossRef]
- Rogers, T.J.; Thorpe, C.M.; Paton, A.W.; Paton, J.C. Role of lipid rafts and flagellin in invasion of colonic epithelial cells by Shiga-Toxigenic Escherichia coli O113:H21. Infect. Immun. 2012, 80, 2858–2867. [Google Scholar] [CrossRef]
- Han, Z.H.; Liu, B.; Niu, Z.Y.; Zhang, Y.; Gao, J.X.; Shi, L.; Wang, S.J.; Wang, S. Role of α-Dicarbonyl Compounds in the Inhibition Effect of Reducing Sugars on the Formation of 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. J. Agric. Food Chem. 2017, 65, 10084–10092. [Google Scholar] [CrossRef] [PubMed]
- Mccullagh, P. Discussion of “Analysis of variance-why it is more important than ever” by A. Gelman. Ann. Stat. 2005, 33, 34–40. [Google Scholar]
- Du, X.J.; Zhang, X.; Li, P.; Xue, R.; Wang, S. Screening of genes involved in interactions with intestinal epithelial cells in Cronobacter sakazakii. AMB Express 2016, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Gelder, J.D.; Gussem, K.D.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Gieroba, B.; Krysa, M.; Wojtowicz, K.; Wiater, A.; Pleszczy’nska, M.; Tomczyk, M.; Sroka-Bartnicka, A. The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci. Int. J. Mol. Sci. 2020, 21, 3811. [Google Scholar] [CrossRef]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef] [PubMed]
- Kolter, R.; Greenberg, E.P. Microbial sciences-The superficial life of microbes. Nature 2006, 441, 300–302. [Google Scholar] [CrossRef]
- Puttamreddy, S.; Cornick, N.A.; Minion, F.C. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect. Immun. 2010, 78, 2377–2384. [Google Scholar] [CrossRef]
- De Araujo, C.; Balestrino, D.; Roth, L.; Charbonnel, N.; Forestier, C. Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in Klebsiella pneumoniae. Res. Microbiol. 2010, 161, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.C.; Lindhout, T.; Beveridge, T.J.; Dutcher, J.R.; Lam, J.S. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2009, 191, 6618–6631. [Google Scholar] [CrossRef]
- Hathroubi, S.; Hancock, M.A.; Bosse, J.T.; Langford, P.R.; Tremblay, Y.D.; Labrie, J.; Jacques, M. Surface polysaccharide mutants reveal that absence of O antigen reduces biofilm formation of Actinobacillus pleuropneumoniae. Infect. Immun. 2016, 84, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Quinn, P.J. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog. Lipid Res. 2010, 49, 97–107. [Google Scholar] [CrossRef]
- Brabetz, W.; Muller-Loennies, S.; Holst, O.; Brade, H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in a Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem. 1997, 247, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Fourel, D.; Mizushima, S.; Bernadac, A.; Pagès, J.M. Specific regions of Escherichia coli OmpF protein involved in antigenic and colicin receptor sites and in stable trimerization. J. Bacteriol. 1993, 175, 2754–2757. [Google Scholar] [CrossRef][Green Version]
- Efremov, R.G.; Sazanov, L.A. Structure of Escherichia coli OmpF porin from lipidic mesophase. J. Struct. Biol. 2012, 178, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Aparna, M.S.; Yadav, S. Biofilms: Microbes and disease. Braz. J. Infect. Dis. 2008, 12, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Byrd, M.S.; Pang, B.; Mishra, M.; Swords, W.E.; Wozniak, D.J. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappa B activation in A549 cells. mBio 2010, 1, e00140-10. [Google Scholar] [CrossRef] [PubMed]
- Kunyanee, C.; Kamjumphol, W.; Taweechaisupapong, S.; Kanthawong, S.; Wongwajana, S.; Wongratanacheewin, S.; Hahnvajanawong, C.; Chareonsudjai, S. Burkholderia pseudomallei biofilm promotes adhesion, internalization and stimulates proinflammatory cytokines in human epithelial A549 Cells. PLoS ONE 2016, 11, e0160741. [Google Scholar] [CrossRef] [PubMed]
- Jap, B.K.; Walian, P.J. Structure and functional mechanism of porins. Physiol. Rev. 1996, 76, 1073–1088. [Google Scholar] [CrossRef] [PubMed]
- Bekhit, A.; Fukamachi, T.; Saito, H.; Kobayashi, H. The role of OmpC and OmpF in acidic resistance in Escherichia Coli. Biol. Pharm. Bull. 2011, 34, 330–334. [Google Scholar] [CrossRef]
- Jeanteur, D.; Schirmer, T.; Fourel, D.; Simonet, V.; Rummel, G.; Widmer, C.; Pages, J.M. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia Coli. Proc. Natl. Acad. Sci. USA 1994, 91, 10675–10679. [Google Scholar] [CrossRef]
- Jaktaji, R.P.; Ebadi, R. Study the expression of marA gene in ciprofloxacin and tetracycline resistant mutants of Esherichia Coli. Iran J. Pharm. Res. 2013, 12, 923–928. [Google Scholar]
- Law, C.J.; Penfold, C.N.; Walker, D.C.; Moore, G.R.; James, R.; Kleanthous, C. OmpF enhances the ability of BtuB to protect susceptible Escherichia coli cells from colicin E9 cytotoxicity. FEBS Lett. 2003, 545, 127–132. [Google Scholar] [CrossRef]
- Thanassi, D.G.; Suh, G.S.; Nikaido, H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia Coli. J. Bacterial. 1995, 177, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Kishii, R.; Takei, M. Relationship between the expression of ompF and quinolone resistance in Escherichia Coli. J. Infect. Chemother. 2009, 15, 361–366. [Google Scholar] [CrossRef]
- Tavío, M.M.; Vila, J.; Ruiz, J.; Ruiz, J.; Martín-Sánchez, A.M.; Jiménez de Anta, M.T. Mechanisms involved in the development of resistance to fluoroquinolones in Escherichia coli isolates. J. Antimicrob. Chemother. 1999, 44, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; O’Neill, A.J.; Chopra, I. Escherichia coli mutators present an enhanced risk for emergence of antibiotic resistance during urinary tract infections. Antimicrob. Agents Chemother. 2004, 48, 23–29. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, H.; Kim, K.P.; Yoon, H.; Kang, D.H.; Ryu, S. hfq Plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 2015, 83, 2089–2098. [Google Scholar] [CrossRef] [PubMed]

| Samples | Arginine (µmol·mL−1) | Lysine (µmol·mL−1) |
|---|---|---|
| WT | 0.12 ± 0.01 a | 0.27 ± 0.04 a |
| ΔompF | BDL | 0.11 ± 0.03 b |
| cpompF | 0.09 ± 0.03 a | 0.27 ± 0.02 a |
| Antibiotics | Inhibition Zone (mm) | |||
|---|---|---|---|---|
| WT | ΔompF | cpompF | 25922 | |
| gentamicin | 19.2 ± 0.3 a | 16.2 ± 0.3 b | 18.7 ± 0.3 a | 15.3 |
| amoxicillin | 19.7 ± 0.4 a | 17.6 ± 0.4 b | 19.7 ± 0.4 a | 17.4 |
| kanamycin | 14.7 ± 0.2 a | 0.0 | 14.7 ± 0.3 a | 17.5 |
| chloramphenicol | 19.3 ± 0.9 a | 17.8 ± 0.5 b | 18.3 ± 0.4 a | - |
| tetracycline | 17.2 ± 0.8 a | 15.1 ± 0.3 b | 16.4 ± 0.4 a | - |
| ciprofloxacin | 24.6 ± 0.3 a | 21.5 ± 0.3 b | 22.4 ± 0.9 a | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Han, Z.; Li, P.; Zhang, H.; Du, X.; Wang, S. Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii. Microorganisms 2021, 9, 2338. https://doi.org/10.3390/microorganisms9112338
Gao J, Han Z, Li P, Zhang H, Du X, Wang S. Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii. Microorganisms. 2021; 9(11):2338. https://doi.org/10.3390/microorganisms9112338
Chicago/Turabian StyleGao, Jianxin, Zhonghui Han, Ping Li, Hongyan Zhang, Xinjun Du, and Shuo Wang. 2021. "Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii" Microorganisms 9, no. 11: 2338. https://doi.org/10.3390/microorganisms9112338
APA StyleGao, J., Han, Z., Li, P., Zhang, H., Du, X., & Wang, S. (2021). Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii. Microorganisms, 9(11), 2338. https://doi.org/10.3390/microorganisms9112338






