Isolation and Identification of a Rumen Lactobacillus Bacteria and Its Degradation Potential of Gossypol in Cottonseed Meal during Solid-State Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Gossypol Degradation Bacteria
2.2. Morphological Characteristics of the Strain
2.3. DNA Extraction and Bacteria Identification
2.4. Growth Study of Isolated Bacteria in Different Carbon Source
2.5. Solid-State Fermentation and Chemical Analysis
2.6. Statistical Analysis
3. Results
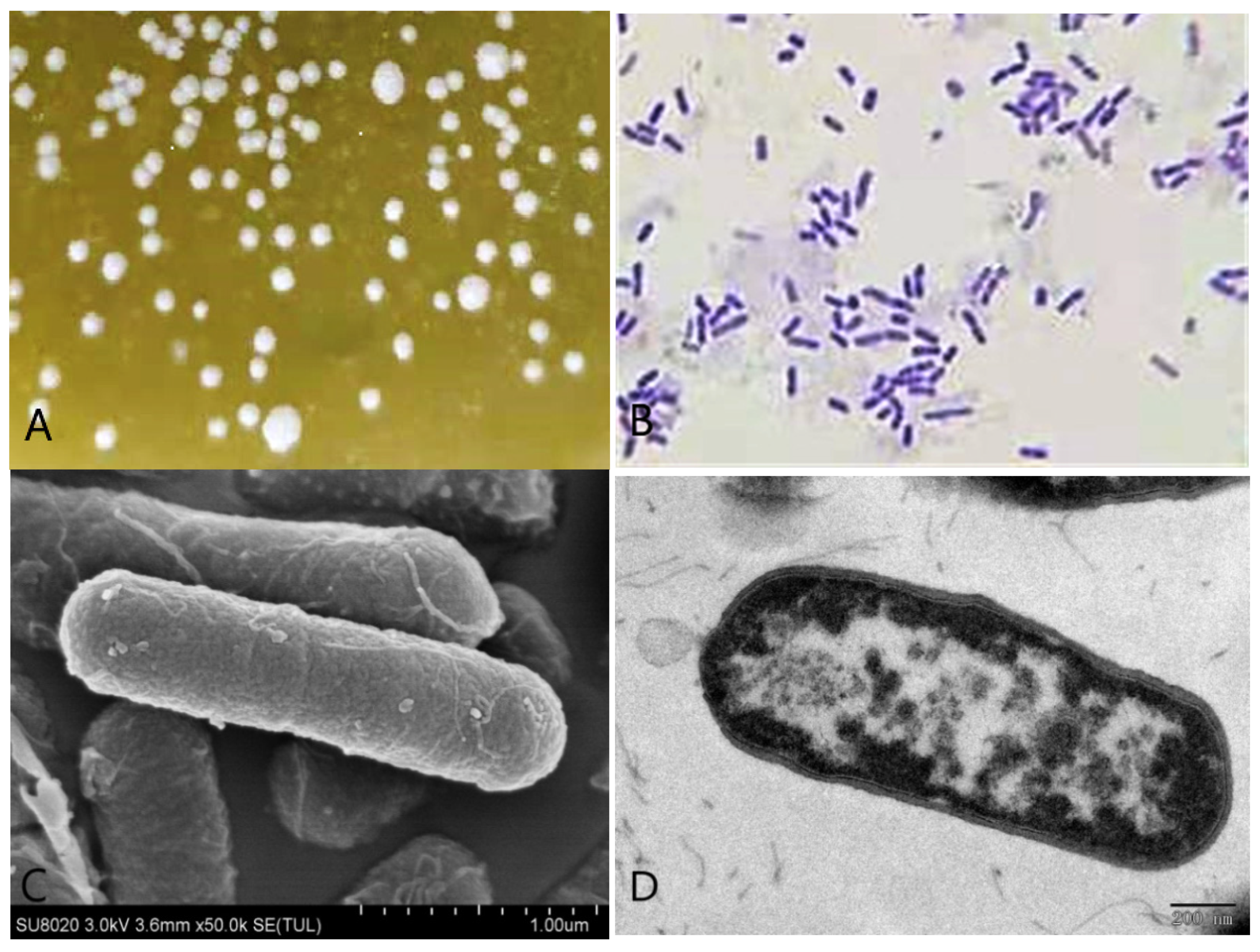
3.1. Morphological Characteristics
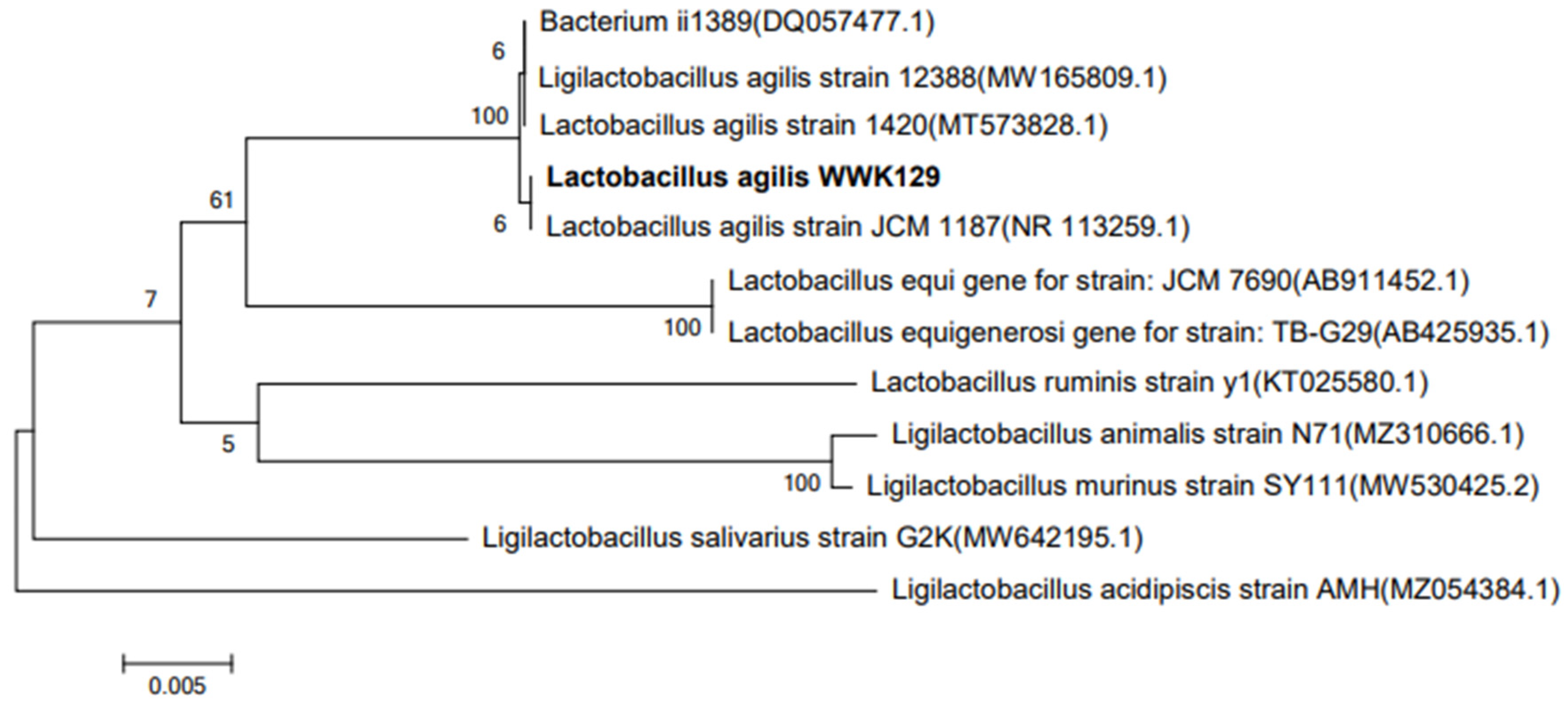
3.2. 16S rDNA Homology
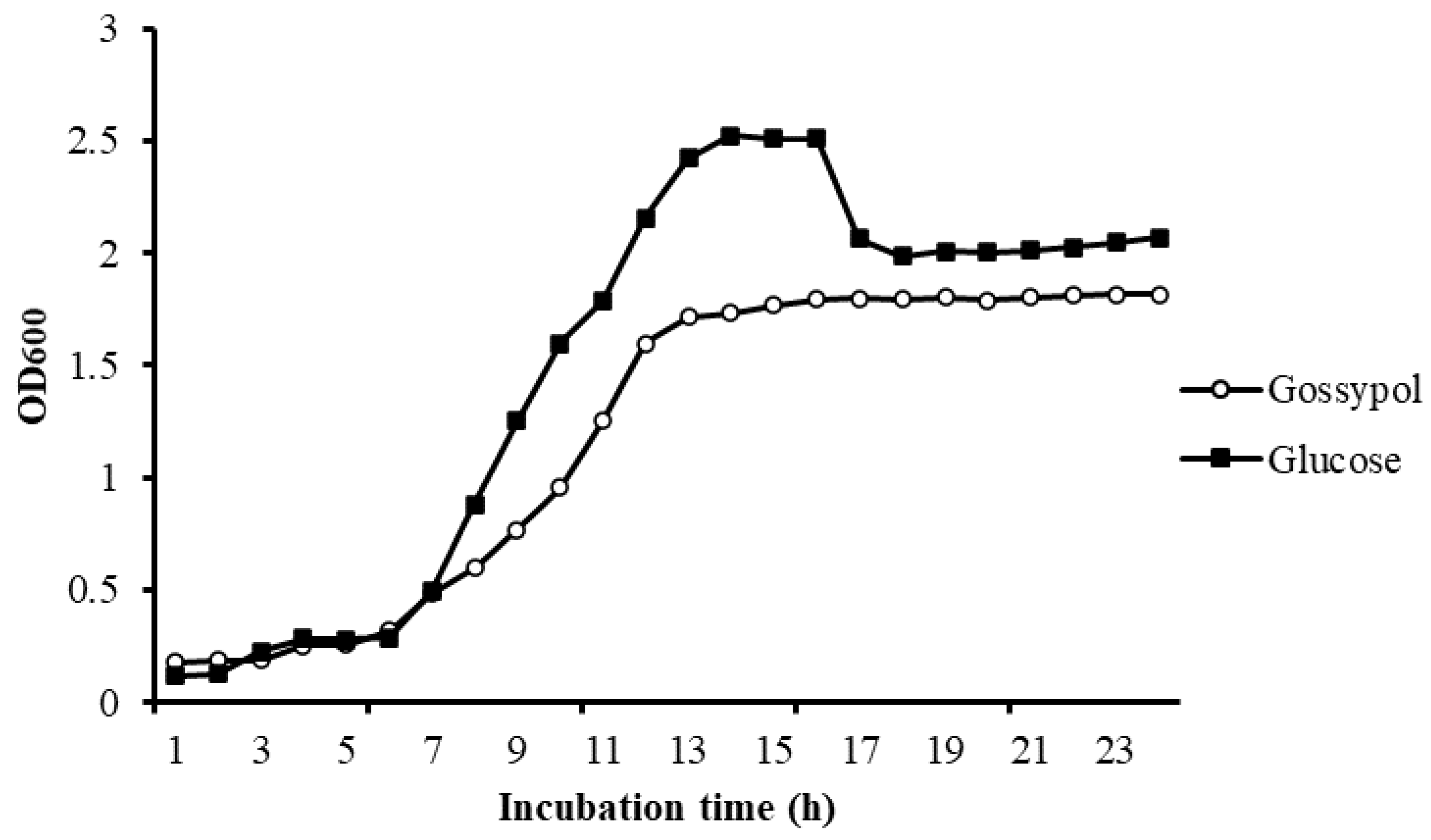
3.3. Growth Study
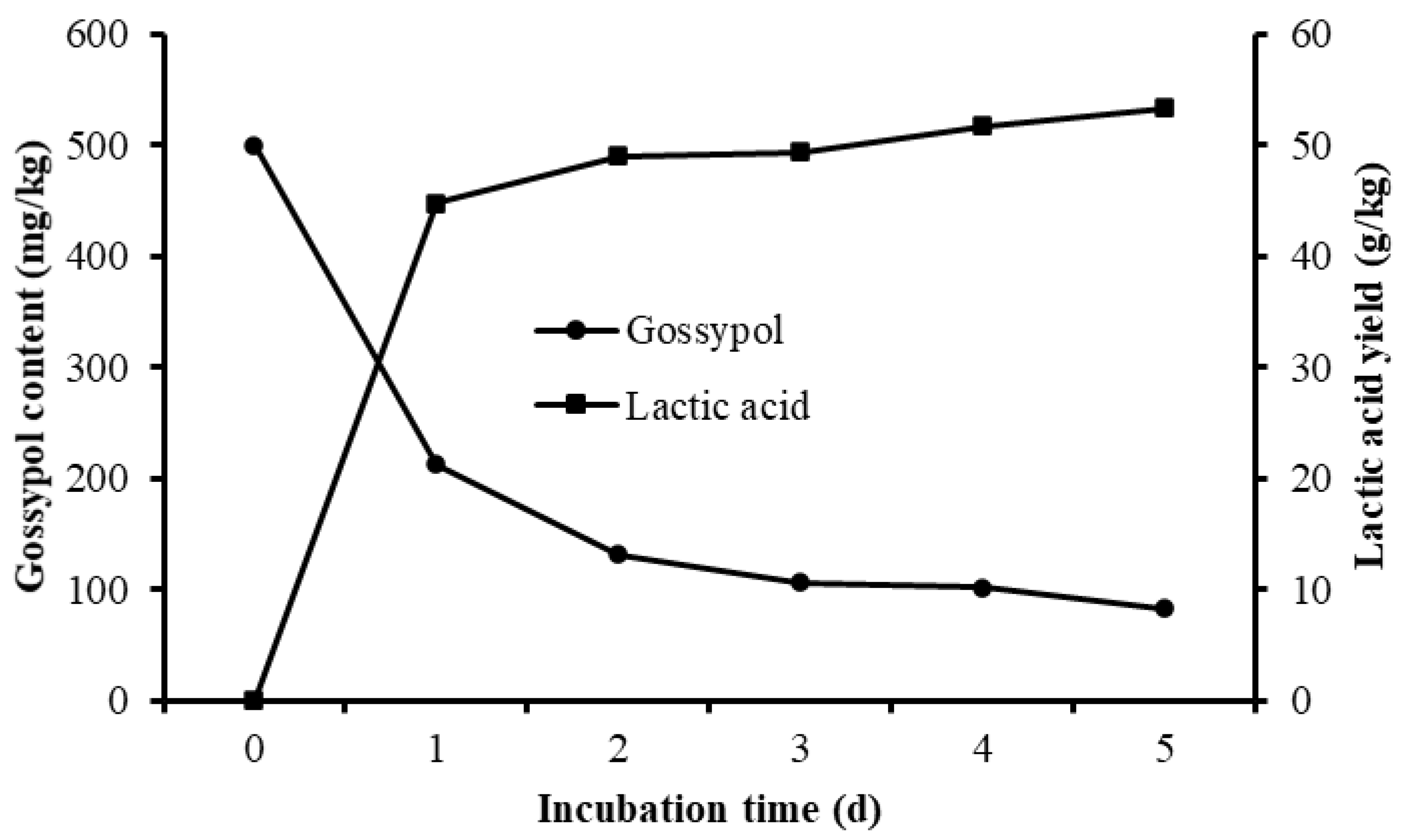
3.4. Gossypol Disappearance and Nutrient Shifts of Fermented CSM
4. Discussion
4.1. Identification of L. agilis WWK129
4.2. Effect of L. agilis WWK129 Fermentation on the Nutritional Value of CSM
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Santos, J.E.P.; Villaseňor, M.; Depeters, E.J.; Robinson, P.H.; Baldwin, B.C., Jr. Type of cottonseed and level of gossypol in diets of lactating dairy cows: Effects on lactation performance and plasma gossypol. J. Dairy Sci. 2002, 85, 1491–1501. [Google Scholar] [CrossRef]
- Krempl, C.; Heidel-Fischer, H.M.; Jiménez-Alemán, G.H.; Reichelt, M.; Menezes, R.C.; Boland, W.; Vogel, H.; Heckel, D.G.; Joußen, N. Gossypol toxicity and detoxification in Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 78, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.H.; Getachew, G.; De Peters, E.J.; Calhoun, M.C. Influence of variety and storage for up to 22 days on nutrient composition and gossypol level of Pima cottonseed (Gossypium spp.). Anim. Feed. Sci. Technol. 2001, 91, 149–156. [Google Scholar] [CrossRef]
- Velasquez-Pereira, J.; Arechiga, C.F.; McDowell, L.R.; Hansen, P.J.; Chenoweth, P.J.; Calhoun, M.C.; Risco, C.A.; Batra, T.R.; Williams, S.N.; Wilkinson, N.S. Effects of gossypol from cottonseed meal and dietary vitamin E on the reproductive characteristics of superovulated beef heifers. J. Anim. Sci. 2002, 80, 2485–2492. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.E.P.; Villaseñor, M.; Robinson, P.H.; Depeters, E.J.; Holmberg, C.A. Type of Cottonseed and Level of Gossypol in Diets of Lactating Dairy Cows: Plasma Gossypol, Health, and Reproductive Performance. J. Dairy Sci. 2003, 86, 892–905. [Google Scholar] [CrossRef]
- Wang, X.; Howell, C.P.; Chen, F.; Yin, J.J.; Jiang, Y.M. Gossypol—A polyphenolic compound from cotton plant. Adv. Food Nutr. Res. 2009, 58, 215–263. [Google Scholar]
- Reiser, R.; Fu, H.C. The Mechanism of Gossypol Detoxification by Ruminant Animals. J. Nutr. 1962, 76, 215–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Wang, Y. The nutrition value and metabolism of cottonseed in ruminants. Feed. Res. 2011, 4, 13–16. [Google Scholar]
- Rahma, E.H.; Rao, M.S.N. Gossypol Removal and Functional Properties of Protein Produced by Extraction of Glanded Cottonseed with Different Solvents. J. Food Sci. 1984, 49, 1057–1060. [Google Scholar] [CrossRef]
- Tabatabai, F.; Golian, A.; Salarmoeini, M. Determination and detoxification methods of cottonseed meal gossypol for broiler chicken rations. Agric. Sci. Technol. 2002, 16, 3–15. [Google Scholar]
- Nagalakshmi, D.; Sastry, V.R.B.; Pawde, A. Rumen fermentation patterns and nutrient digestion in lambs fed cottonseed meal supplemental diets. Anim. Feed. Sci. Technol. 2003, 103, 1–14. [Google Scholar] [CrossRef]
- Wu, X.Y.; Chen, J.X. The utilization of microbes to break down FG in cottonseed meal. Sci. Agric. Sin. 1989, 22, 82–86. [Google Scholar]
- Weng, X.-Y.; Sun, J.-Y. Biodegradation of free gossypol by a new strain of Candida tropicalis under solid state fermentation: Effects of fermentation parameters. Process. Biochem. 2006, 41, 1663–1668. [Google Scholar] [CrossRef]
- Weng, X.-Y.; Sun, J.-Y. Kinetics of biodegradation of free gossypol by Candida tropicalis in solid-state fermentation. Biochem. Eng. J. 2006, 32, 226–232. [Google Scholar] [CrossRef]
- Khalaf, M.A.; Meleigy, S.A. Reduction of free gossypol levels in cottonseed meal by microbial treatment. Int. J. Agric. Biol. 2008, 10, 185–190. [Google Scholar]
- Yang, X.; Sun, J.-Y.; Guo, J.-L.; Weng, X.-Y. Identification and proteomic analysis of a novel gossypol-degrading fungal strain. J. Sci. Food Agric. 2011, 92, 943–951. [Google Scholar] [CrossRef] [PubMed]
- DeRuiter, J.; Jacyno, J.M.; Davis, R.A.; Cutler, H.G. Studies on Aldose Reductase Inhibitors from Fungi. I. Citrinin and Related Benzopyran Derivatives. J. Enzym. Inhib. 1992, 6, 201–210. [Google Scholar] [CrossRef]
- De Mann, J.C.; Rogosa, M.; Sharpe, M.E. A medium for cultivation of Lactobacill. J. Appl. Microb. 1969, 23, 130–135. [Google Scholar]
- Perfumo, A.; Elsaesser, A.; Littmann, S.; Foster, R.A.; Kuypers, M.M.; Cockell, C.S.; Kminek, G. Epifluorescence, SEM, TEM and nanoSIMS image analysis of the cold phenotype of Clostridium psychrophilum at subzero temperatures. FEMS Microbiol. Ecol. 2014, 90, 869–882. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Luan, H.; Wei, Z.; Hao, Z.; Xi, R.; Liao, X. Exploiting of honey-associated Bacillus strains as plant-growth promoting bacteria for enhancing barley growth in rare earth tailings. Ann. Microbiol. 2015, 66, 559–568. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. Mega 4: Molecular evolutionary gnentics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- AOAC. Official Methods of Analysis of AOAC International, 18th ed.; Association of Official Analytical Chemists International: Gaithersburg, MD, USA, 2005. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Cira, L.A.; Huerta, S.; Hall, G.M.; Shirai, K. Pilot scale lactic acid fermentation of shrimp wastes for chitin recovery. Process. Biochem. 2002, 37, 1359–1366. [Google Scholar] [CrossRef]
- Wang, W.-K.; Wang, Y.-L.; Li, W.-J.; Wu, Q.-C.; Li, S.-L.; Yang, H.-J. Gossypol Exhibited Higher Detrimental Effect on Ruminal Fermentation Characteristics of Low-Forage in Comparison with High-Forage Mixed Feeds. Toxics 2021, 9, 51. [Google Scholar] [CrossRef]
- SAS. Statistical Analytical System (SAS) Users Guides: Statistics; Version 8.2; Statistica Analysis Institute: Cary, NC, USA, 1999. [Google Scholar]
- Chen, L.; Zh, Y.; Ch, X.; Ch, M.; Meng, X.; Cai, H. Gossypol Degradation Strain Coming from Ruminant Rumens and Application Thereof. CN 104328063A, 4 February 2015. [Google Scholar]
- Zhang, Y.; Zhang, Z.; Dai, L.; Liu, Y.; Cheng, M.; Chen, L. Isolation and characterization of a novel gossypol-degrading bac-teriabacillus subtilis strain rumen bacillus subtilis. Asian Austral. J. Anim. 2018, 31, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-K.; Wang, Y.-L.; Li, W.-J.; Wu, Q.-C.; Yang, K.-L.; Li, S.-L.; Yang, H.-J. In situ rumen degradation characteristics and bacterial colonization of whole cottonseed, cottonseed hull and cottonseed meal with different gossypol content. AMB Express 2021, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.W.; Wu, Y.F.; Yao, X.H.; Wang, X. Method for Removing Free Gossypol from Cotton Dregs by Biological Fermentation Method. CN102138632B, 24 July 2013. [Google Scholar]
- Chen, W.; Tian, F.W.; Zhai, Q.X.; Sun, Y.Y.; Zhao, J.X.; Wang, G.; Zhang, Q.X.; Liu, X.M.; Zhang, B.X.; Zhang, H. Lactobacillus Plantarum Capable of Degrading Gossypol and Application of Lactobacillus Plantarum. CN105255793A, 20 January 2016. [Google Scholar]
- Heeney, D.D.; Gareau, M.G.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebeer, S.; Bron, P.A.; Marco, M.L.; Pijkeren, J.P.V.; Motherway, M.O.C.; Hill, C.; Pot, B.; Roos, S.; Klaenhammer, T. Identification of probiotic effector molecules: Present state and future perspectives. Curr. Opin. Biotechnol. 2018, 49, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L., Jr. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef]
- Doyle, N.; Mbandlwa, P.; Kelly, W.J.; Attwood, G.; Li, Y.; Ross, R.P.; Stanton, C.; Leahy, S. Use of Lactic Acid Bacteria to Re-duce Methane Production in Ruminants, a Critical Review. Front. Microbiol. 2019, 10, 2207. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.L. Ruminant Nutrition; Science Press: Beijing, China, 2004; pp. 10–11. [Google Scholar]
- Stephenson, D.P.; Moore, R.J.; Allison, G.E. Transformation of, and Heterologous Protein Expression in, Lactobacillus agilis and Lactobacillus vaginalis Isolates from the Chicken Gastrointestinal Tract. Appl. Environ. Microbiol. 2011, 77, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Kajikawa, A.; Midorikawa, E.; Masuda, K.; Kondo, K.; Irisawa, T.; Igimi, S.; Okada, S. Characterization of flagellins isolated from a highly motile strain of Lactobacillus agilis. BMC Microbiol. 2016, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Palop, M.L.; Smiths, J.P.; Brink, B.T. Degradation of sinigrin by Lactobacillus agilis strain R16. Int. J. Food Microbiol. 1995, 26, 219–229. [Google Scholar] [CrossRef]
- Luangin, V. Influence of Human Gut Microbiota on the Metabolic Fate of Glucosinolates; Imperial College London: London, UK, 2013. [Google Scholar]
- Zhu, Y.; Liu, J.; Lopez, J.M.; Mills, D.A. Inulin Fermentation by Lactobacilli and Bifidobacteria from Dairy Calves. Appl. Environ. Microbiol. 2020, 87, e01738-20. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y. A Study on Screening and Breeding of High-Yield Lactic Acid Bacteria and the Nutritional Characteristics of Fermented Cottonseed Meal; Shihezi University: Shihezi, China, 2013. [Google Scholar]
- Zhang, W.-J.; Xu, Z.-R.; Zhao, S.-H.; Sun, J.-Y.; Yang, X. Development of a microbial fermentation process for detoxification of gossypol in cottonseed meal. Anim. Feed. Sci. Technol. 2007, 135, 176–186. [Google Scholar] [CrossRef]
- Hong, Z. Microbial Solid Fermentation to Reduce Free Gossypol Content in the Cottonseed Meal; Wuhan Polytechnic University: Wuhan, China, 2016. [Google Scholar]
- Peran, L.; Sierra, S.; Comalada, M.; Lara-Villoslada, F.; Bailón, E.; Nieto, A.; Concha, Á.; Olivares, M.; Zarzuelo, A.; Xaus, J.; et al. A comparative study of the preventative effects exerted by two probiotics, Lactobacillus reuteri and Lactobacillus fermentum, in the trinitrobenzene sulfonic acid model of rat colitis. Br. J. Nutr. 2007, 97, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Śliżewska, K.; Chlebicz-Wójcik, A.; Nowak, A. Probiotic Properties of New Lactobacillus Strains Intended to Be Used as Feed Additives for Monogastric Animals. Probiotics Antimicrob. Proteins 2021, 13, 146–162. [Google Scholar] [CrossRef]
- Slover, C.M.; Danziger, L. Lactobacillus: A Review. Clin. Microbiol. Newsl. 2008, 30, 23–27. [Google Scholar] [CrossRef]


| Item 1 | Fermented CSM at Different Incubation Time | S.E.M | p Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Con 2 | 1 Day | 2 Day | 3 Day | 4 Day | 5 Day | Time | L | Q | ||
| NDF | 16.3 a | 15.3 ab | 14.9 ab | 14.6 ab | 13.6 bc | 12.7 c | 0.46 | <0.01 | 0.45 | 0.04 |
| ADF | 11.3 a | 6.8 b | 6.5 c | 6.4 cd | 6.3 de | 6.1 e | 0.05 | <0.01 | <0.01 | <0.01 |
| CP | 57.6 b | 60.2 a | 60.7 a | 60.9 a | 61.3 a | 61.7 a | 0.41 | 0.03 | 0.81 | <0.01 |
| Aspartic acid | 4.28 a | 4.24 ab | 4.24 ab | 4.20 bc | 4.17 d | 4.17 d | 0.011 | <0.01 | 0.29 | <0.01 |
| Threonine | 1.29 c | 1.36 c | 1.36 c | 1.44 b | 1.51 a | 1.55 a | 0.020 | <0.01 | <0.01 | 0.01 |
| Serine | 1.77 a | 1.64 b | 1.65 b | 1.65 b | 1.65 b | 1.67 b | 0.028 | 0.10 | 0.42 | 0.20 |
| Glutamic acid | 11.36 a | 10.74 ab | 10.52 b | 10.42 b | 10.32 b | 9.94 b | 0.222 | 0.04 | 0.50 | 0.01 |
| Proline | 1.46 a | 1.28 b | 1.27 b | 1.19 b | 1.07 c | 0.99 c | 0.029 | <0.01 | 0.11 | <0.01 |
| Glycine | 1.87 | 1.90 | 1.91 | 1.92 | 1.93 | 1.99 | 0.034 | 0.36 | 0.31 | 0.13 |
| Alanine | 1.76 c | 1.78 c | 1.80 c | 1.87 b | 1.98 a | 1.98 a | 0.021 | <0.01 | <0.01 | 0.10 |
| Cysteine | 0.74 b | 0.74 b | 0.76 ab | 0.78 ab | 0.79 a | 0.79 a | 0.016 | 0.09 | 0.07 | 0.09 |
| Valine | 2.11 d | 2.18 cd | 2.35 bc | 2.40 ab | 2.48 ab | 2.54 a | 0.050 | <0.01 | 0.01 | <0.01 |
| Methionine | 0.56 c | 0.60 c | 0.61 c | 0.68 b | 0.74 a | 0.75 a | 0.016 | <0.01 | <0.01 | 0.01 |
| Isoleucine | 1.53 d | 1.53 d | 1.58 cd | 1.64 bc | 1.67 ab | 1.73 a | 0.019 | <0.01 | 0.01 | <0.01 |
| Leucine | 2.71 d | 2.79 c | 2.80 c | 2.85 b | 2.87 ab | 2.91 a | 0.033 | <0.01 | <0.01 | <0.01 |
| Tyrosine | 1.52 c | 1.53 bc | 1.56 abc | 1.58 ab | 1.60 a | 1.61 a | 0.015 | 0.02 | 0.06 | 0.05 |
| Phenylalanine | 2.53 d | 2.75 c | 2.81 bc | 2.90 b | 3.21 a | 3.21 a | 0.037 | <0.01 | <0.01 | <0.01 |
| Lysine | 1.90 b | 1.91 b | 1.93 b | 2.18 a | 2.28 a | 2.31 a | 0.044 | <0.01 | <0.01 | 0.04 |
| Histidine | 1.27 | 1.25 | 1.27 | 1.28 | 1.27 | 1.27 | 0.016 | 0.59 | 0.19 | 0.35 |
| Arginine | 5.66 a | 5.53 ab | 5.44 ab | 5.47 ab | 5.21 bc | 5.09 c | 0.089 | 0.03 | 0.40 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-K.; Li, W.-J.; Wu, Q.-C.; Wang, Y.-L.; Li, S.-L.; Yang, H.-J. Isolation and Identification of a Rumen Lactobacillus Bacteria and Its Degradation Potential of Gossypol in Cottonseed Meal during Solid-State Fermentation. Microorganisms 2021, 9, 2200. https://doi.org/10.3390/microorganisms9112200
Wang W-K, Li W-J, Wu Q-C, Wang Y-L, Li S-L, Yang H-J. Isolation and Identification of a Rumen Lactobacillus Bacteria and Its Degradation Potential of Gossypol in Cottonseed Meal during Solid-State Fermentation. Microorganisms. 2021; 9(11):2200. https://doi.org/10.3390/microorganisms9112200
Chicago/Turabian StyleWang, Wei-Kang, Wen-Juan Li, Qi-Chao Wu, Yan-Lu Wang, Sheng-Li Li, and Hong-Jian Yang. 2021. "Isolation and Identification of a Rumen Lactobacillus Bacteria and Its Degradation Potential of Gossypol in Cottonseed Meal during Solid-State Fermentation" Microorganisms 9, no. 11: 2200. https://doi.org/10.3390/microorganisms9112200
APA StyleWang, W.-K., Li, W.-J., Wu, Q.-C., Wang, Y.-L., Li, S.-L., & Yang, H.-J. (2021). Isolation and Identification of a Rumen Lactobacillus Bacteria and Its Degradation Potential of Gossypol in Cottonseed Meal during Solid-State Fermentation. Microorganisms, 9(11), 2200. https://doi.org/10.3390/microorganisms9112200







