Role of Commensal Microbes in the γ-Ray Irradiation-Induced Physiological Changes in Drosophila melanogaster
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Husbandry and Generation of Axenic (Axe) D. melanogaster
2.2. γ-Ray Irradiation
2.3. Quantitative Analysis of Bacteria
2.4. Pyrosequencing of the 16S rRNA Gene
2.5. Lifespan Assay
2.6. γH2AX Staining
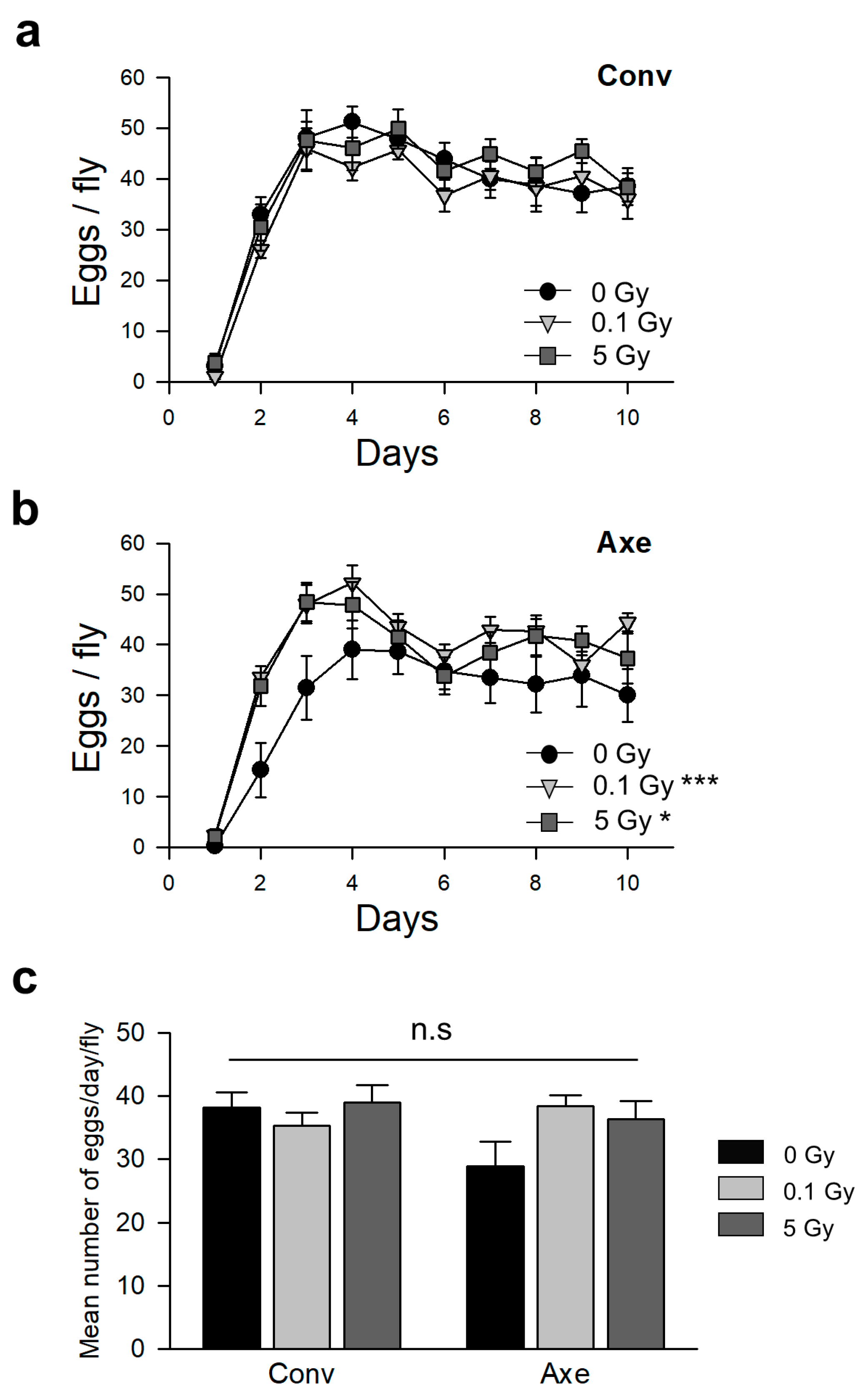
2.7. Measurement of Fecundity
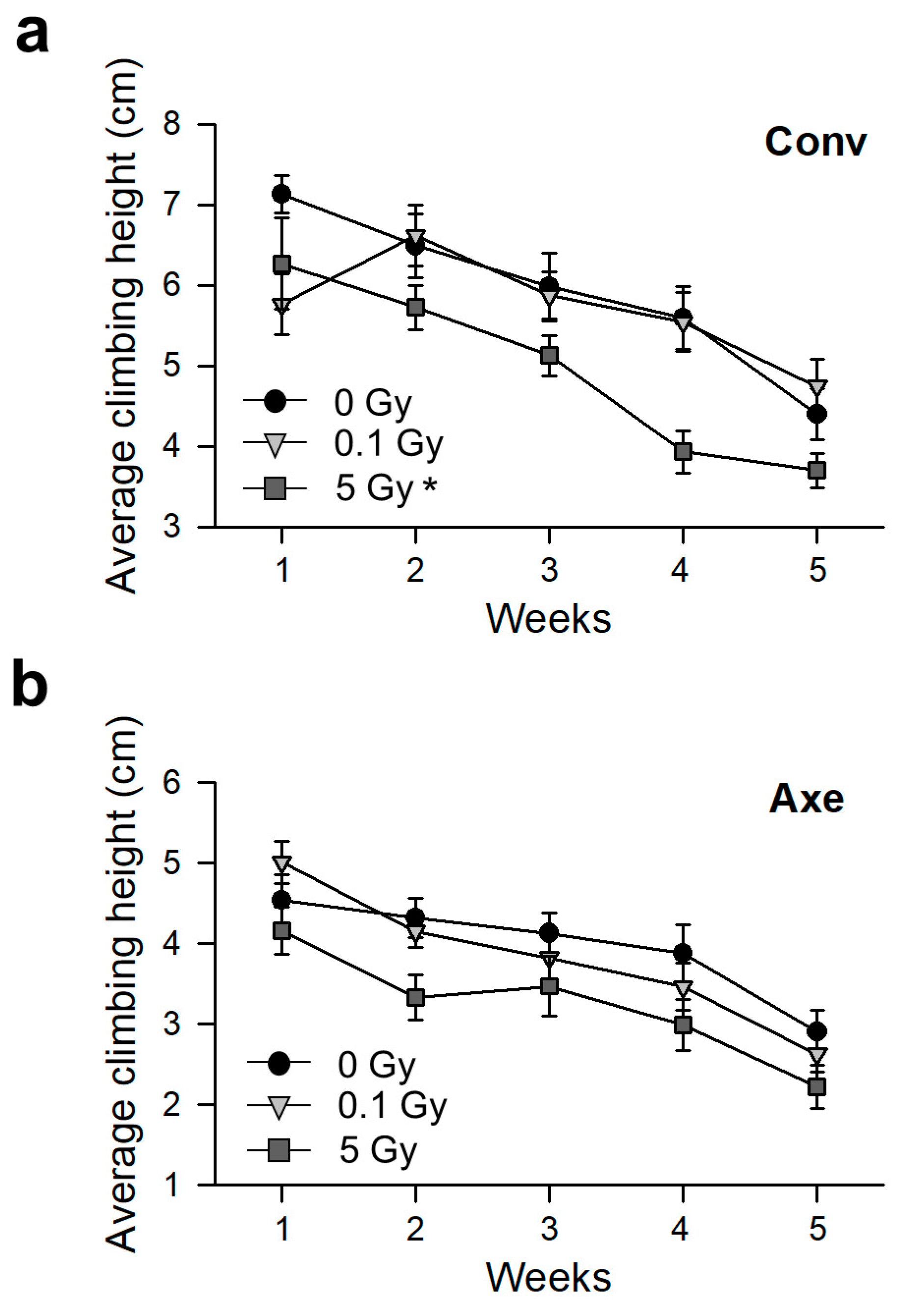
2.8. Measurement of Physical Activity
2.9. ROS Detection
2.10. MitoTracker Red Staining
2.11. Measurement of Mitochondrial DNA
2.12. Statistical Analysis
3. Results
3.1. γ-Ray Irradiation Changes Commensal Microbe Flora in D. melanogaster
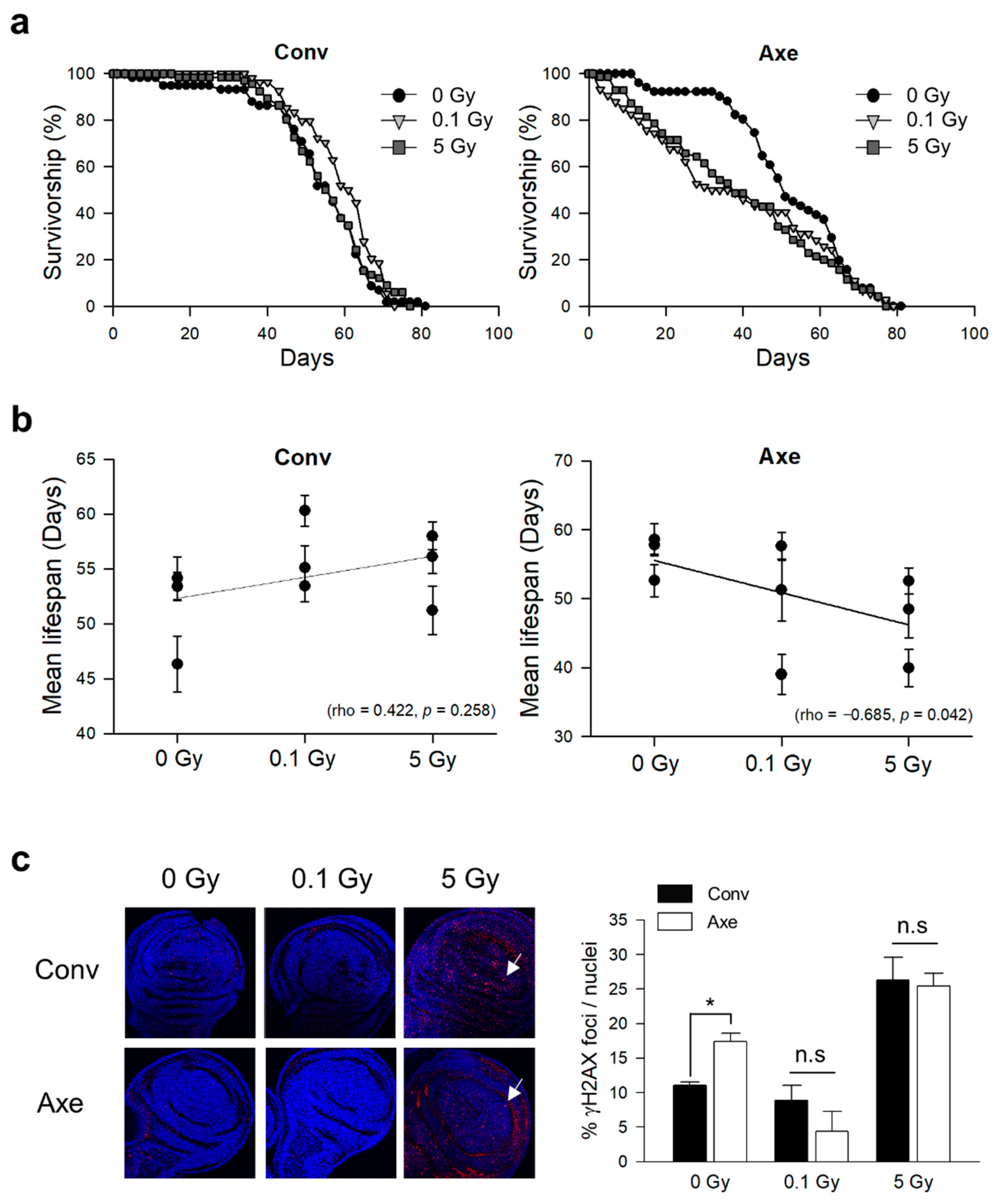
3.2. Effects of γ-Ray Irradiation on Lifespan and DNA Damage Response in Conventional and Axenic D. melanogaster
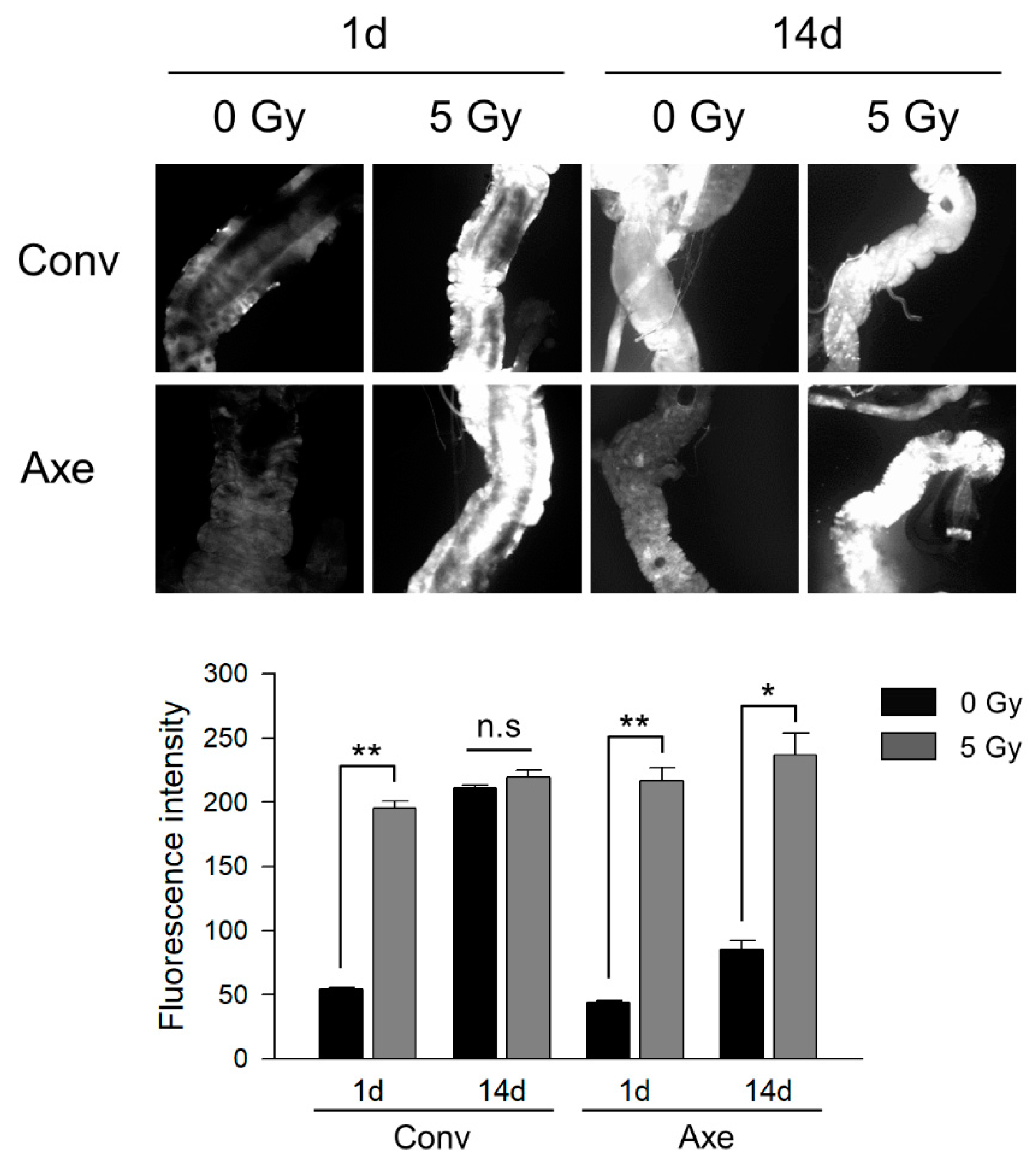
3.3. Effects of Commensal Microbes on ROS Generation by γ-Ray Irradiation
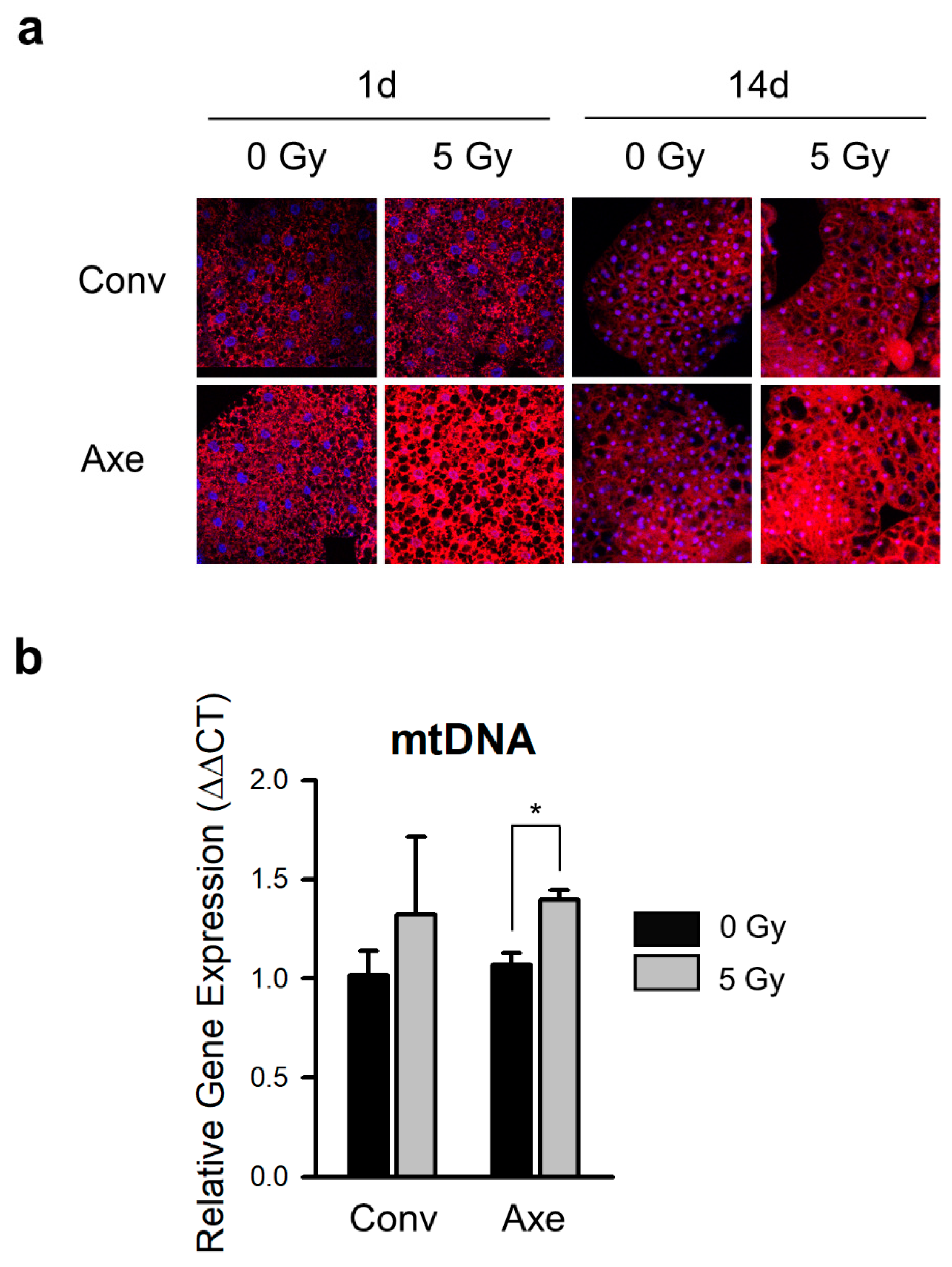
3.4. Effects of Commensal Microbes on Mitochondrial Change by γ-Ray Irradiation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Burnette, B.; Weichselbaum, R.R. Radiation as an immune modulator. Semin. Radiat. Oncol. 2013, 23, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Abuhanoglu, G.; Ozer, A.Y. Radiation sterilization of new drug delivery systems. Interv. Med. Appl. Sci. 2014, 6, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014, 1, 24. [Google Scholar] [CrossRef] [PubMed]
- Bortolin, E.; De Angelis, C.; Quattrini, M.C.; Barlascini, O.; Fattibene, P. Detection of Ionizing Radiation Treatment in Glass Used for Healthcare Products. Radiat. Prot. Dosim. 2019. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.; Nowacka, M.; Rybak, K.; Rzepna, M. The effects of electron beam radiation on material properties and degradation of commercial PBAT/PLA blend. J. Appl. Polym. Sci. 2019. [Google Scholar] [CrossRef]
- Williams, D. Cancer after nuclear fallout: Lessons from the Chernobyl accident. Nat. Rev. Cancer 2002, 2, 543–549. [Google Scholar] [CrossRef]
- Kamiya, K.; Ozasa, K.; Akiba, S.; Niwa, O.; Kodama, K.; Takamura, N.; Zaharieva, E.K.; Kimura, Y.; Wakeford, R. Long-term effects of radiation exposure on health. Lancet 2015, 386, 469–478. [Google Scholar] [CrossRef]
- Seo, S.; Ha, W.H.; Kang, J.K.; Lee, D.; Park, S.; Kwon, T.E.; Jin, Y.W. Health effects of exposure to radon: Implications of the radon bed mattress incident in Korea. Epidemiol. Health 2019, 41, e2019004. [Google Scholar] [CrossRef]
- Lubin, J.H.; Boice, J.D., Jr. Lung cancer risk from residential radon: Meta-analysis of eight epidemiologic studies. J. Natl. Cancer Inst. 1997, 89, 49–57. [Google Scholar] [CrossRef]
- Klatt, A.; Salzmann, E.; Schneider, L.J.; Reifschneider, A.; Korneck, M.; Hermle, P.; Burkle, A.; Stoll, D.; Kadereit, S. Toxicity of ionizing radiation (IR) in a human induced pluripotent stem cell (hiPSC)-derived 3D early neurodevelopmental model. Arch. Toxicol. 2019. [Google Scholar] [CrossRef]
- Bedford, J.S. Sublethal damage, potentially lethal damage, and chromosomal aberrations in mammalian cells exposed to ionizing radiations. Int. J. Radiat. Oncol. Biol. Phys. 1991, 21, 1457–1469. [Google Scholar] [CrossRef]
- Natarajan, A.T.; Berni, A.; Marimuthu, K.M.; Palitti, F. The type and yield of ionising radiation induced chromosomal aberrations depend on the efficiency of different DSB repair pathways in mammalian cells. Mutat. Res. 2008, 642, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Rombouts, C.; Moreels, M.; Aerts, A.; Quintens, R.; Tabury, K.; Michaux, A.; Janssen, A.; Neefs, M.; Ernst, E.; et al. Modulation of gene expression in endothelial cells in response to high LET nickel ion irradiation. Int. J. Mol. Med. 2014, 34, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Shameer, P.M.; Sowmithra, K.; Harini, B.P.; Chaubey, R.C.; Jha, S.K.; Shetty, N.J. Does exposure of male Drosophila melanogaster to acute gamma radiation influence egg to adult development time and longevity of F-1-F-3 offspring? Entomol. Sci. 2015, 18, 368–376. [Google Scholar] [CrossRef]
- Sudmeier, L.J.; Howard, S.P.; Ganetzky, B. A Drosophila model to investigate the neurotoxic side effects of radiation exposure. Dis. Model Mech. 2015, 8, 669–677. [Google Scholar] [CrossRef]
- Pyo, J.H.; Park, J.S.; Na, H.J.; Jeon, H.J.; Lee, S.H.; Kim, J.G.; Park, S.Y.; Jin, Y.W.; Kim, Y.S.; Yoo, M.A. Functional modification of Drosophila intestinal stem cells by ionizing radiation. Radiat. Res. 2014, 181, 376–386. [Google Scholar] [CrossRef]
- Moskalev, A.A.; Plyusnina, E.N.; Shaposhnikov, M.V. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: The role of cellular stress-resistance mechanisms. Biogerontology 2011, 12, 253–263. [Google Scholar] [CrossRef]
- Morciano, P.; Iorio, R.; Iovino, D.; Cipressa, F.; Esposito, G.; Porrazzo, A.; Satta, L.; Alesse, E.; Tabocchini, M.A.; Cenci, G. Effects of reduced natural background radiation on Drosophila melanogaster growth and development as revealed by the FLYINGLOW program. J. Cell Physiol. 2018, 233, 23–29. [Google Scholar] [CrossRef]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef]
- Lomax, M.E.; Folkes, L.K.; O’Neill, P. Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin. Oncol. R Coll. Radiol. 2013, 25, 578–585. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Scherthan, H.; Sarioglu, H.; Barjaktarovic, Z.; Conrad, M.; Vogt, A.; Calzada-Wack, J.; Neff, F.; Aubele, M.; Buske, C.; et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 2011, 11, 3299–3311. [Google Scholar] [CrossRef] [PubMed]
- Graziewicz, M.A.; Day, B.J.; Copeland, W.C. The mitochondrial DNA polymerase as a target of oxidative damage. Nucleic Acids Res. 2002, 30, 2817–2824. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.N.; Gordon, D.M.; De Toledo, S.M.; Pain, D.; Azzam, E.I. Normal human fibroblasts exposed to high- or low-dose ionizing radiation: Differential effects on mitochondrial protein import and membrane potential. Antioxid. Redox. Signal 2006, 8, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.; Marples, B.; Balasubramaniam, M.; Thomas, R.A.; Tucker, J.D. Mitochondrial gene expression changes in normal and mitochondrial mutant cells after exposure to ionizing radiation. Radiat. Res. 2010, 173, 635–644. [Google Scholar] [CrossRef]
- Lin, R.X.; Zhao, H.B.; Li, C.R.; Sun, Y.N.; Qian, X.H.; Wang, S.Q. Proteomic analysis of ionizing radiation-induced proteins at the subcellular level. J. Proteome Res. 2009, 8, 390–399. [Google Scholar] [CrossRef]
- Shimura, T.; Sasatani, M.; Kawai, H.; Kamiya, K.; Kobayashi, J.; Komatsu, K.; Kunugita, N. A comparison of radiation-induced mitochondrial damage between neural progenitor stem cells and differentiated cells. Cell Cycle 2017, 16, 565–573. [Google Scholar] [CrossRef]
- DiMauro, S.; Hirano, M. Pathogenesis and treatment of mitochondrial disorders. Adv. Exp. Med. Biol. 2009, 652, 139–170. [Google Scholar] [CrossRef]
- Lane, R.K.; Hilsabeck, T.; Rea, S.L. The role of mitochondrial dysfunction in age-related diseases. Biochim. Biophys. Acta 2015, 1847, 1387–1400. [Google Scholar] [CrossRef]
- Keebaugh, E.S.; Yamada, R.; Obadia, B.; Ludington, W.B.; Ja, W.W. Microbial Quantity Impacts Drosophila Nutrition, Development, and Lifespan. iScience 2018, 4, 247–259. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Littman, D.R. Modulation of immune homeostasis by commensal bacteria. Curr. Opin. Microbiol. 2011, 14, 106–114. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, S.H.; Lee, J.H.; Lee, W.J.; Min, K.J. The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging Albany N. Y. 2019, 11, 4611–4640. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Lee, H.J.; Jeong, S.E.; Jeon, C.O.; Hyun, S. Comparative Analysis of Drosophila melanogaster Gut Microbiota with Respect to Host Strain, Sex, and Age. Microb. Ecol. 2017, 74, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Webster, P.; Finkel, S.E.; Tower, J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007, 6, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Moschen, A.R.; Wieser, V.; Tilg, H. Dietary Factors: Major Regulators of the Gut’s Microbiota. Gut Liver 2012, 6, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Chaston, J.M.; Dobson, A.J.; Newell, P.D.; Douglas, A.E. Host Genetic Control of the Microbiota Mediates the Drosophila Nutritional Phenotype. Appl. Environ. Microbiol. 2015, 82, 671–679. [Google Scholar] [CrossRef]
- Dalby, M.J.; Ross, A.W.; Walker, A.W.; Morgan, P.J. Dietary Uncoupling of Gut Microbiota and Energy Harvesting from Obesity and Glucose Tolerance in Mice. Cell Rep. 2017, 21, 1521–1533. [Google Scholar] [CrossRef]
- Storelli, G.; Defaye, A.; Erkosar, B.; Hols, P.; Royet, J.; Leulier, F. Lactobacillus plantarum Promotes Drosophila Systemic Growth by Modulating Hormonal Signals through TOR-Dependent Nutrient Sensing. Cell Metab. 2011, 14, 403–414. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.A.; Yoon, J.H.; Ryu, J.H.; Lee, W.J. Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef]
- Schwarzer, M.; Makki, M.; Storelli, G.; Machuca-Gayet, I.; Srutkova, D.; Hermanova, P.; Martino, M.E.; Balmand, S.; Hudcovic, T.; Heddi, A.; et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 2016, 351, 854–857. [Google Scholar] [CrossRef]
- Casero, D.; Gill, K.; Sridharan, V.; Koturbash, I.; Nelson, G.; Hauer-Jensen, M.; Boerma, M.; Braun, J.; Cheema, A.K. Space-type radiation induces multimodal responses in the mouse gut microbiome and metabolome. Microbiome 2017, 5, 105. [Google Scholar] [CrossRef]
- Burns, E.M.; Ahmed, H.; Isedeh, P.N.; Kohli, I.; Van Der Pol, W.; Shaheen, A.; Muzaffar, A.F.; Al-Sadek, C.; Foy, T.M.; Abdelgawwad, M.S.; et al. Ultraviolet radiation, both UVA and UVB, influences the composition of the skin microbiome. Exp. Dermatol. 2019, 28, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.D.; Kim, H.J.; Seo, J.G.; Kang, S.W.; Bae, J.W. Impact of Pelvic Radiotherapy on Gut Microbiota of Gynecological Cancer Patients Revealed by Massive Pyrosequencing. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.M.; Dacquisto, M.P.; Jacobus, D.P.; Horowitz, R.E. Effects of the Germfree State on Responses of Mice to Whole-Body Irradiation. Radiat. Res. 1964, 23, 333–349. [Google Scholar] [CrossRef]
- Crawford, P.A.; Gordon, J.I. Microbial regulation of intestinal radiosensitivity. Proc. Natl. Acad. Sci. USA 2005, 102, 13254–13259. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Xu, Z.W.; Zhang, C.G. The effects of gut commensal bacteria depletion on mice exposed to acute lethal irradiation. J. Radiat. Res. 2007, 48, 347–350. [Google Scholar] [CrossRef]
- Wong, C.N.; Ng, P.; Douglas, A.E. Low-diversity bacterial community in the gut of the fruit fly Drosophila melanogaster. Environ. Microbiol. 2011, 13, 1889–1900. [Google Scholar] [CrossRef]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef]
- Cox, C.R.; Gilmore, M.S. Native Microbial Colonization of Drosophila melanogaster and Its Use as a Model of Enterococcus faecalis Pathogenesis. Infect. Immun. 2007, 75, 1565–1576. [Google Scholar] [CrossRef]
- Ryu, J.H.; Kim, S.H.; Lee, H.Y.; Bai, J.Y.; Nam, Y.D.; Bae, J.W.; Lee, D.G.; Shin, S.C.; Ha, E.M.; Lee, W.J. Innate Immune Homeostasis by the Homeobox Gene Caudal and Commensal-Gut Mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef]
- Chandler, J.A.; Lang, J.M.; Bhatnagar, S.; Eisen, J.A.; Kopp, A. Bacterial Communities of Diverse Drosophila Species: Ecological Context of a Host–Microbe Model System. PLoS Genet. 2011, 7, e1002272. [Google Scholar] [CrossRef]
- Lamb, M.J. The effects of radiation on the longevity of female Drosophila subobscura. J. Ins. Physiol. 1964, 10, 487–497. [Google Scholar] [CrossRef]
- Paithankar, J.G.; Deeksha, K.; Patil, R.K. Gamma radiation tolerance in different life stages of the fruit fly Drosophila melanogaster. Int. J. Radiat. Biol. 2017, 93, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Biosa, A.; Sanchez-Martinez, A.; Filograna, R.; Terriente-Felix, A.; Alam, S.M.; Beltramini, M.; Bubacco, L.; Bisaglia, M.; Whitworth, A.J. Superoxide dismutating molecules rescue the toxic effects of PINK1 and parkin loss. Hum. Mol. Genet 2018, 27, 1618–1629. [Google Scholar] [CrossRef] [PubMed]
- Rera, M.; Bahadorani, S.; Cho, J.; Koehler, C.L.; Ulgherait, M.; Hur, J.H.; Ansari, W.S.; Lo, T., Jr.; Jones, D.L.; Walker, D.W. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011, 14, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.M.; Kim, C.S.; Lee, B.S.; Nam, S.Y.; Yang, K.H.; Kim, J.Y.; Park, J.J.; Min, K.J.; Jin, Y.W. Low-dose radiation induces Drosophila innate immunity through Toll pathway activation. J. Radiat. Res. 2012, 53, 242–249. [Google Scholar] [CrossRef]
- Seong, K.M.; Yu, M.; Lee, K.S.; Park, S.; Jin, Y.W.; Min, K.J. Curcumin mitigates accelerated aging after irradiation in Drosophila by reducing oxidative stress. Biomed. Res. Int. 2015, 2015, 425380. [Google Scholar] [CrossRef]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef]
- Heys, C.; Lize, A.; Blow, F.; White, L.; Darby, A.; Lewis, Z.J. The effect of gut microbiota elimination in Drosophila melanogaster: A how-to guide for host-microbiota studies. Ecol. Evol. 2018, 8, 4150–4161. [Google Scholar] [CrossRef]
- Panganiban, R.A.; Snow, A.L.; Day, R.M. Mechanisms of radiation toxicity in transformed and non-transformed cells. Int. J. Mol. Sci. 2013, 14, 15931–15958. [Google Scholar] [CrossRef]
- Wentworth, C.C.; Alam, A.; Jones, R.M.; Nusrat, A.; Neish, A.S. Enteric commensal bacteria induce extracellular signal-regulated kinase pathway signaling via formyl peptide receptor-dependent redox modulation of dual specific phosphatase 3. J. Biol. Chem. 2011, 286, 38448–38455. [Google Scholar] [CrossRef]
- Jones, R.M.; Luo, L.; Ardita, C.S.; Richardson, A.N.; Kwon, Y.M.; Mercante, J.W.; Alam, A.; Gates, C.L.; Wu, H.; Swanson, P.A.; et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013, 32, 3017–3028. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Suman, S.; Kallakury, B.V.; Fornace, A.J., Jr. Exposure to heavy ion radiation induces persistent oxidative stress in mouse intestine. PLoS ONE 2012, 7, e42224. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.M.; Yang, H.S.; Kang, S.W.; Ho, J.N.; Lee, S.B.; Um, H.D. Amplification of the gamma-irradiation-induced cell death pathway by reactive oxygen species in human U937 cells. Cell Signal. 2008, 20, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Yamamori, T.; Sasagawa, T.; Ichii, O.; Hiyoshi, M.; Bo, T.; Yasui, H.; Kon, Y.; Inanami, O. Analysis of the mechanism of radiation-induced upregulation of mitochondrial abundance in mouse fibroblasts. J. Radiat. Res. 2017, 58, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.R. The Hiroshima/Nagasaki Survivor Studies: Discrepancies between Results and General Perception. Genetics 2016, 203, 1505–1512. [Google Scholar] [CrossRef]
- Dalke, C.; Neff, F.; Bains, S.K.; Bright, S.; Lord, D.; Reitmeir, P.; Rossler, U.; Samaga, D.; Unger, K.; Braselmann, H.; et al. Lifetime study in mice after acute low-dose ionizing radiation: A multifactorial study with special focus on cataract risk. Radiat. Environ. Biophys. 2018, 57, 99–113. [Google Scholar] [CrossRef]
- Prasanth, M.I.; Santoshram, G.S.; Bhaskar, J.P.; Balamurugan, K. Ultraviolet-A triggers photoaging in model nematode Caenorhabditis elegans in a DAF-16 dependent pathway. Age Dordr. 2016, 38, 27. [Google Scholar] [CrossRef]
- Li, H.; Qi, Y.; Jasper, H. Preventing Age-Related Decline of Gut Compartmentalization Limits Microbiota Dysbiosis and Extends Lifespan. Cell Host Microbe. 2016, 19, 240–253. [Google Scholar] [CrossRef]
- Iatsenko, I.; Boquete, J.P.; Lemaitre, B. Microbiota-Derived Lactate Activates Production of Reactive Oxygen Species by the Intestinal NADPH Oxidase Nox and Shortens Drosophila Lifespan. Immunity 2018, 49, 929–942.e5. [Google Scholar] [CrossRef]
- Clark, R.I.; Salazar, A.; Yamada, R.; Fitz-Gibbon, S.; Morselli, M.; Alcaraz, J.; Rana, A.; Rera, M.; Pellegrini, M.; Ja, W.W.; et al. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell Rep. 2015, 12, 1656–1667. [Google Scholar] [CrossRef]
- Guo, L.; Karpac, J.; Tran, S.L.; Jasper, H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 2014, 156, 109–122. [Google Scholar] [CrossRef] [PubMed]
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895. [Google Scholar] [CrossRef] [PubMed]
- Asimakis, E.D.; Khan, M.; Stathopoulou, P.; Caceres, C.; Bourtzis, K.; Tsiamis, G. The effect of diet and radiation on the bacterial symbiome of the melon fly, Zeugodacus cucurbitae (Coquillett). BMC Biotechnol. 2019, 19, 88. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Torija, M.J.; Mateo, E.; Guillamon, J.M.; Mas, A. Identification and quantification of acetic acid bacteria in wine and vinegar by TaqMan-MGB probes. Food Microbiol. 2010, 27, 257–265. [Google Scholar] [CrossRef]
- Gomes, R.J.; Borges, M.F.; Rosa, M.F.; Castro-Gomez, R.J.H.; Spinosa, W.A. Acetic Acid Bacteria in the Food Industry: Systematics, Characteristics and Applications. Food Technol. Biotechnol. 2018, 56, 139–151. [Google Scholar] [CrossRef]
- Hastings, J.W.; Holzapfel, W.H.; Niemand, J.G. Radiation resistance of lactobacilli isolated from radurized meat relative to growth and environment. Appl. Environ. Microbiol. 1986, 52, 898–901. [Google Scholar] [CrossRef]
- Lu, H.; Gao, G.; Xu, G.; Fan, L.; Yin, L.; Shen, B.; Hua, Y. Deinococcus radiodurans PprI switches on DNA damage response and cellular survival networks after radiation damage. Mol. Cell Proteom. 2009, 8, 481–494. [Google Scholar] [CrossRef]
- El-Saghire, H.; Thierens, H.; Monsieurs, P.; Michaux, A.; Vandevoorde, C.; Baatout, S. Gene set enrichment analysis highlights different gene expression profiles in whole blood samples X-irradiated with low and high doses. Int. J. Radiat. Biol. 2013, 89, 628–638. [Google Scholar] [CrossRef]
- Lumniczky, K.; Candeias, S.M.; Gaipl, U.S.; Frey, B. Editorial: Radiation and the Immune System: Current Knowledge and Future Perspectives. Front Immunol. 2017, 8, 1933. [Google Scholar] [CrossRef]
- Stoecklein, V.M.; Osuka, A.; Ishikawa, S.; Lederer, M.R.; Wanke-Jellinek, L.; Lederer, J.A. Radiation exposure induces inflammasome pathway activation in immune cells. J. Immunol. 2015, 194, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Patra, V.; Byrne, S.N.; Wolf, P. The Skin Microbiome: Is It Affected by UV-induced Immune Suppression? Front Microbiol. 2016, 7, 1235. [Google Scholar] [CrossRef] [PubMed]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fang, P.; Mai, J.; Choi, E.T.; Wang, H.; Yang, X.F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013, 6, 19. [Google Scholar] [CrossRef]
- Barjaktarovic, Z.; Schmaltz, D.; Shyla, A.; Azimzadeh, O.; Schulz, S.; Haagen, J.; Dorr, W.; Sarioglu, H.; Schafer, A.; Atkinson, M.J.; et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS ONE 2011, 6, e27811. [Google Scholar] [CrossRef]
- Clark, A.; Mach, N. The Crosstalk between the Gut Microbiota and Mitochondria during Exercise. Front Physiol. 2017, 8, 319. [Google Scholar] [CrossRef]
- Goubern, M.; Andriamihaja, M.; Nubel, T.; Blachier, F.; Bouillaud, F. Sulfide, the first inorganic substrate for human cells. FASEB J. 2007, 21, 1699–1706. [Google Scholar] [CrossRef]
- Leschelle, X.; Goubern, M.; Andriamihaja, M.; Blottiere, H.M.; Couplan, E.; Gonzalez-Barroso, M.D.; Petit, C.; Pagniez, A.; Chaumontet, C.; Mignotte, B.; et al. Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochim. Biophys. Acta 2005, 1725, 201–212. [Google Scholar] [CrossRef]
- Vermeiren, J.; Van de Wiele, T.; Van Nieuwenhuyse, G.; Boeckx, P.; Verstraete, W.; Boon, N. Sulfide- and nitrite-dependent nitric oxide production in the intestinal tract. Microb. Biotechnol. 2012, 5, 379–387. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Yadav, H.; Lee, J.H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J. Biol. Chem. 2013, 288, 25088–25097. [Google Scholar] [CrossRef] [PubMed]
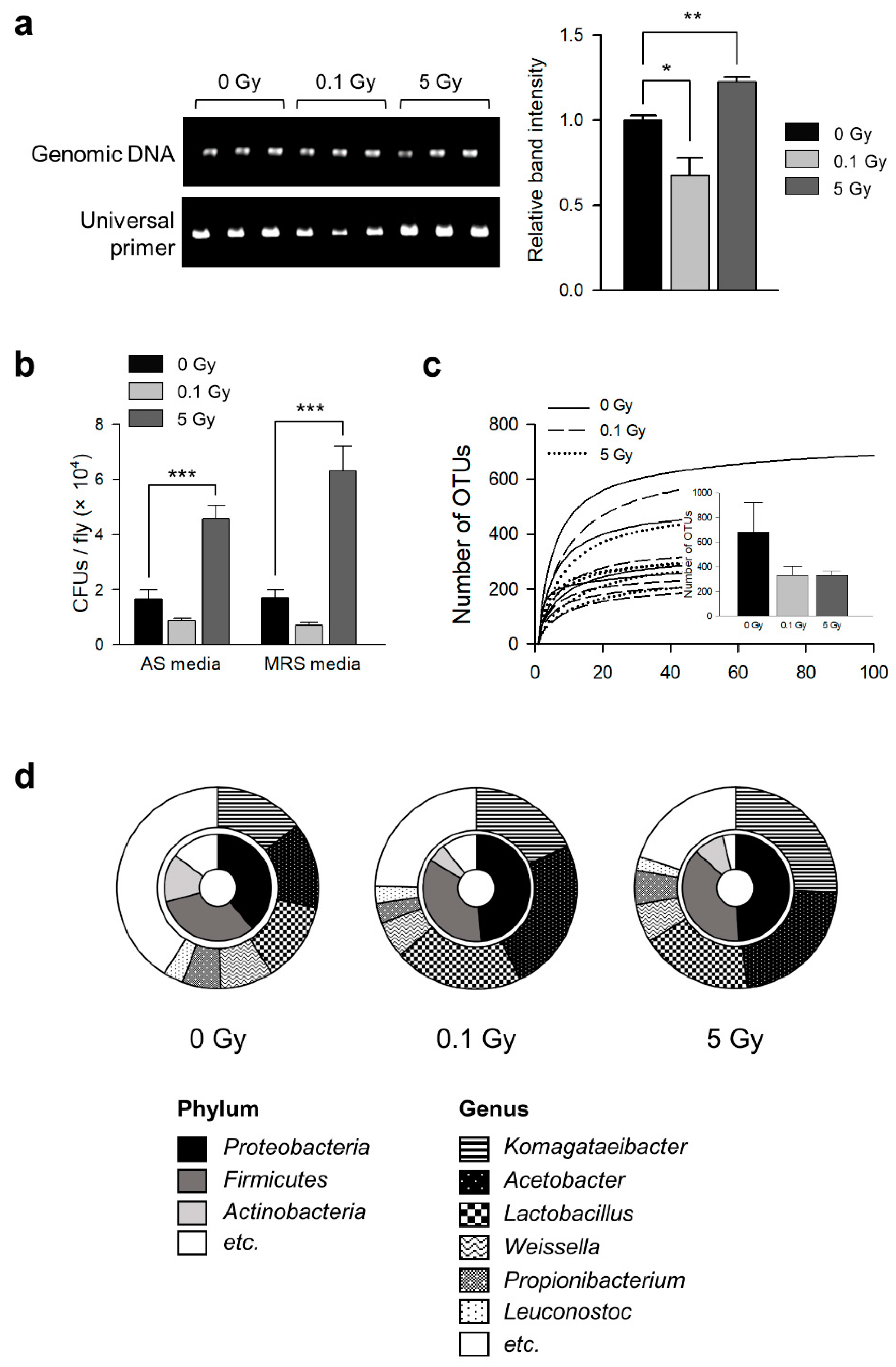
| Group | OTU Number | Shannon | Simpson | Chao |
|---|---|---|---|---|
| 0 Gy | 677.40 ± 241.07 | 4.30 ± 0.54 a | 0.06 ± 0.02 a | 705.34 ± 241.91 a |
| 0.1 Gy | 329.20 ± 76.18 | 2.94 ± 0.30 b | 0.16 ± 0.03 b | 340.07 ± 76.02 a |
| 5 Gy | 329.40 ± 40.26 | 2.81 ± 0.40 b | 0.19 ± 0.06 b | 342.38 ± 42.11 a |
| Trial | Radiation Dose (Gy) | Mean Lifespan (Day) | Median Lifespan (Day) | Maximum Lifespan (Day) | Number of Flies | Log-Rank ‡ | Wilcoxon ‡ | |||
|---|---|---|---|---|---|---|---|---|---|---|
| ꭓ2 | p-Value | ꭓ2 | p-Value | |||||||
| Conv † | 1st | 0 | 53.38 ± 1.29 | 57 | 61 | 140 | ||||
| 0.1 | 53.44 ± 1.45 | 59 | 65 | 154 | 1.5165 | 0.2181 | 0.4455 | 0.5045 | ||
| 5 | 58 ± 1.25 | 61 | 67 | 130 | 8.3963 | 0.0038 * | 8.2578 | 0.0041 * | ||
| 2nd | 0 | 46.32 ± 2.53 | 55 | 62 | 85 | |||||
| 0.1 | 55.13 ± 1.97 | 60 | 66 | 78 | 3.8521 | 0.0497 * | 6.8168 | 0.009 * | ||
| 5 | 51.21 ± 2.21 | 57 | 64 | 71 | 0.2432 | 0.6219 | 0.9642 | 0.3261 | ||
| 3rd | 0 | 54.16 ± 1.95 | 57 | 63 | 69 | |||||
| 0.1 | 60.3 ± 1.42 | 62 | 63 | 68 | 4.9049 | 0.0268 * | 5.3073 | 0.0212 * | ||
| 5 | 56.12 ± 1.53 | 56 | 63 | 66 | 0.1918 | 0.6614 | 0.0317 | 0.8588 | ||
| Axe † | 1st | 0 | 57.79 ± 1.32 | 59 | 67 | 157 | ||||
| 0.1 | 57.6 ± 1.99 | 61 | 69 | 73 | 0.0274 | 0.8686 | 0.0408 | 0.8400 | ||
| 5 | 52.53 ± 1.89 | 57 | 67 | 120 | 1.266 | 0.2605 | 3.204 | 0.0735 | ||
| 2nd | 0 | 58.57 ± 2.31 | 60 | 72 | 70 | |||||
| 0.1 | 51.24 ± 4.48 | 53 | 70 | 25 | 1.8119 | 0.1783 | 2.1248 | 0.1449 | ||
| 5 | 48.48 ± 4.18 | 60 | 68 | 44 | 1.4508 | 0.2284 | 2.1308 | 0.1444 | ||
| 3rd | 0 | 52.61 ± 2.34 | 51 | 66 | 51 | |||||
| 0.1 | 39.01 ± 2.88 | 35 | 63 | 74 | 2.5024 | 0.1137 | 8.1454 | 0.0043 * | ||
| 5 | 39.94 ± 2.66 | 38 | 57 | 70 | 4.2501 | 0.0392 * | 9.4652 | 0.0021 * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-J.; Lee, S.-H.; Lee, J.-H.; Kim, Y.; Seong, K.M.; Jin, Y.W.; Min, K.-J. Role of Commensal Microbes in the γ-Ray Irradiation-Induced Physiological Changes in Drosophila melanogaster. Microorganisms 2021, 9, 31. https://doi.org/10.3390/microorganisms9010031
Lee H-J, Lee S-H, Lee J-H, Kim Y, Seong KM, Jin YW, Min K-J. Role of Commensal Microbes in the γ-Ray Irradiation-Induced Physiological Changes in Drosophila melanogaster. Microorganisms. 2021; 9(1):31. https://doi.org/10.3390/microorganisms9010031
Chicago/Turabian StyleLee, Hwa-Jin, Shin-Hae Lee, Ji-Hyeon Lee, Yongjoong Kim, Ki Moon Seong, Young Woo Jin, and Kyung-Jin Min. 2021. "Role of Commensal Microbes in the γ-Ray Irradiation-Induced Physiological Changes in Drosophila melanogaster" Microorganisms 9, no. 1: 31. https://doi.org/10.3390/microorganisms9010031
APA StyleLee, H.-J., Lee, S.-H., Lee, J.-H., Kim, Y., Seong, K. M., Jin, Y. W., & Min, K.-J. (2021). Role of Commensal Microbes in the γ-Ray Irradiation-Induced Physiological Changes in Drosophila melanogaster. Microorganisms, 9(1), 31. https://doi.org/10.3390/microorganisms9010031






