An Assessment of the Molecular Diversity of Ticks and Tick-Borne Microorganisms of Small Ruminants in Pakistan
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Tick Samples
2.2. Morphological Identification of Ticks and DNA Extraction
2.3. Molecular Characterization of Ticks
2.4. Microfluidic PCR-Based Detection of Tick-Borne Microorganisms
2.5. Sequence and Phylogenetic Analyses
2.6. Statistical Analyses
3. Results
3.1. Morphological Characterization of Ticks
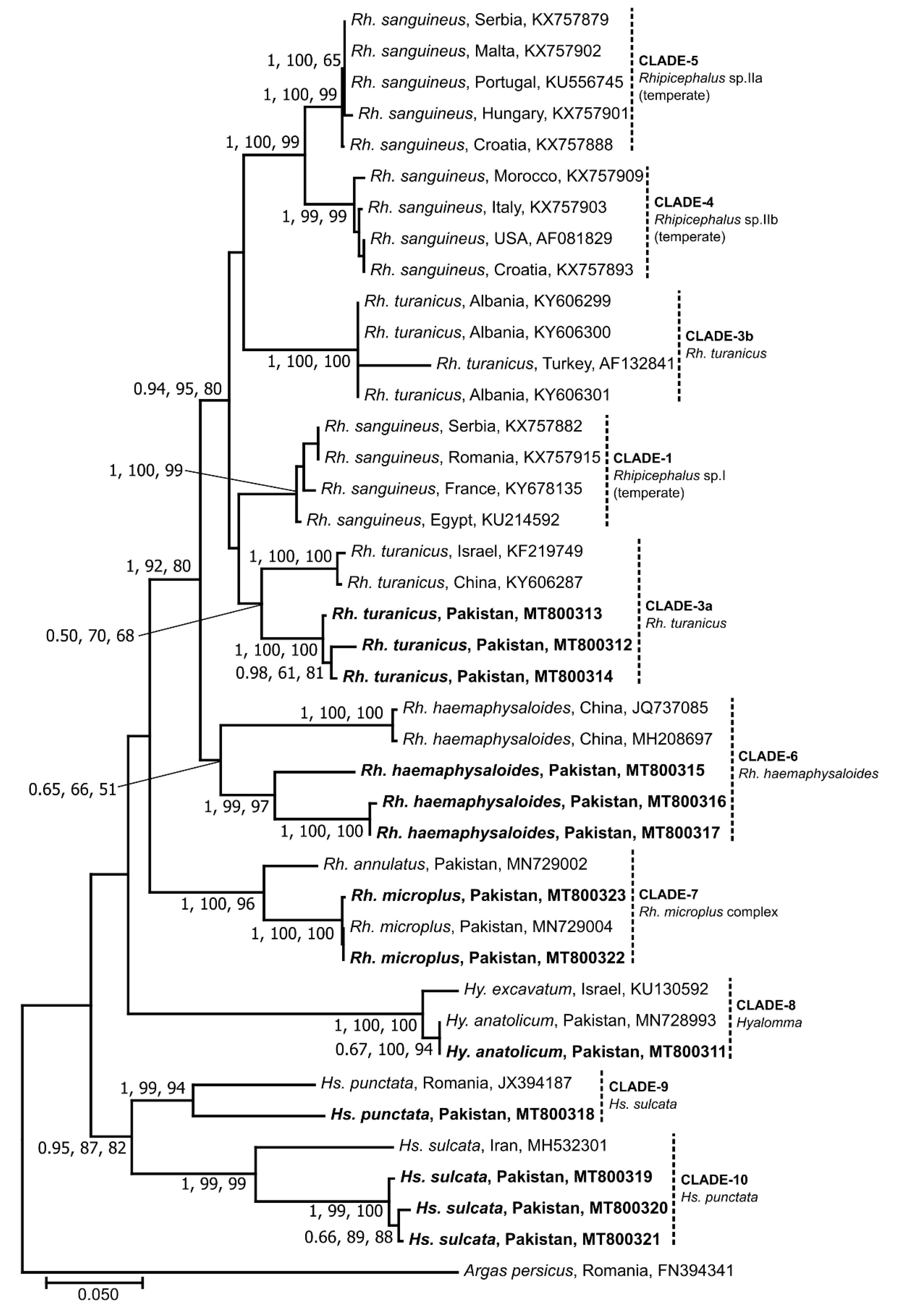
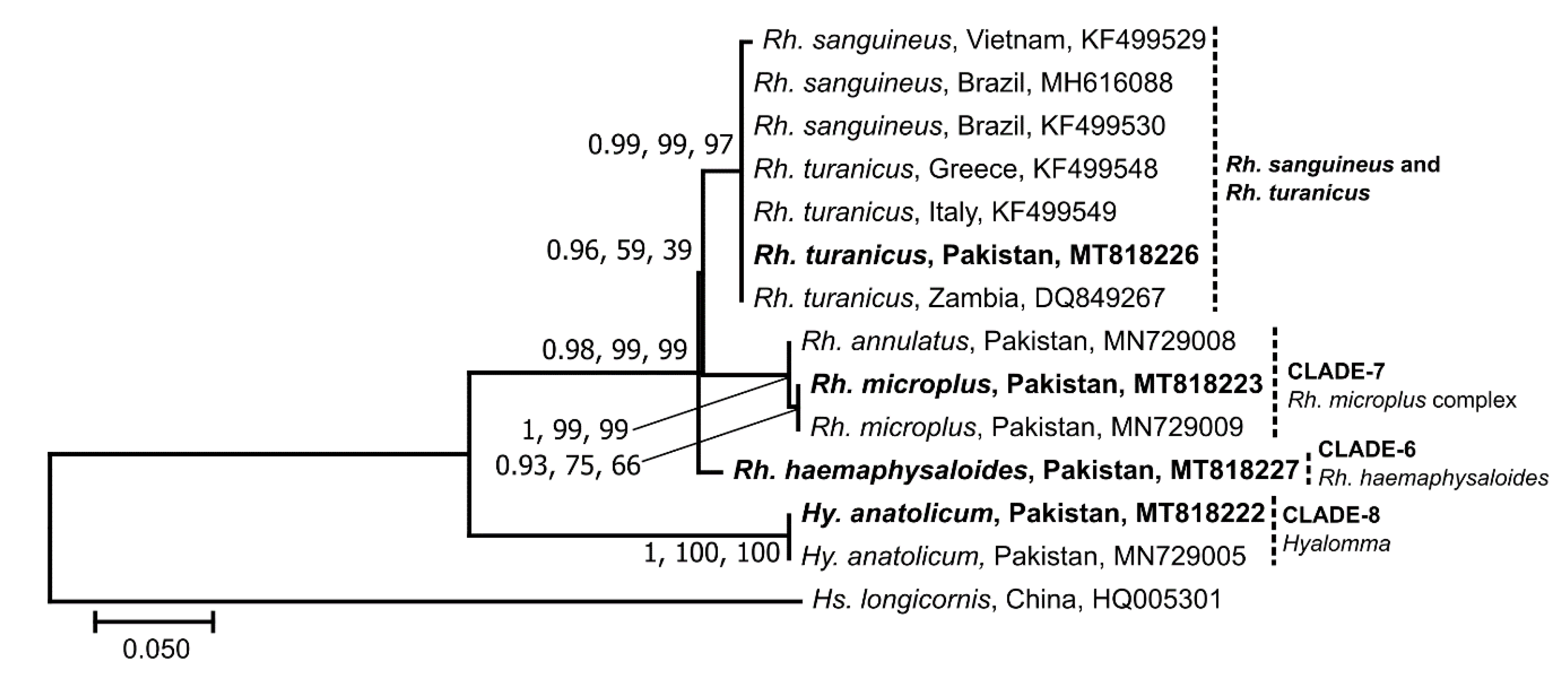
3.2. Sequence and Phylogenetic Analyses of Ticks
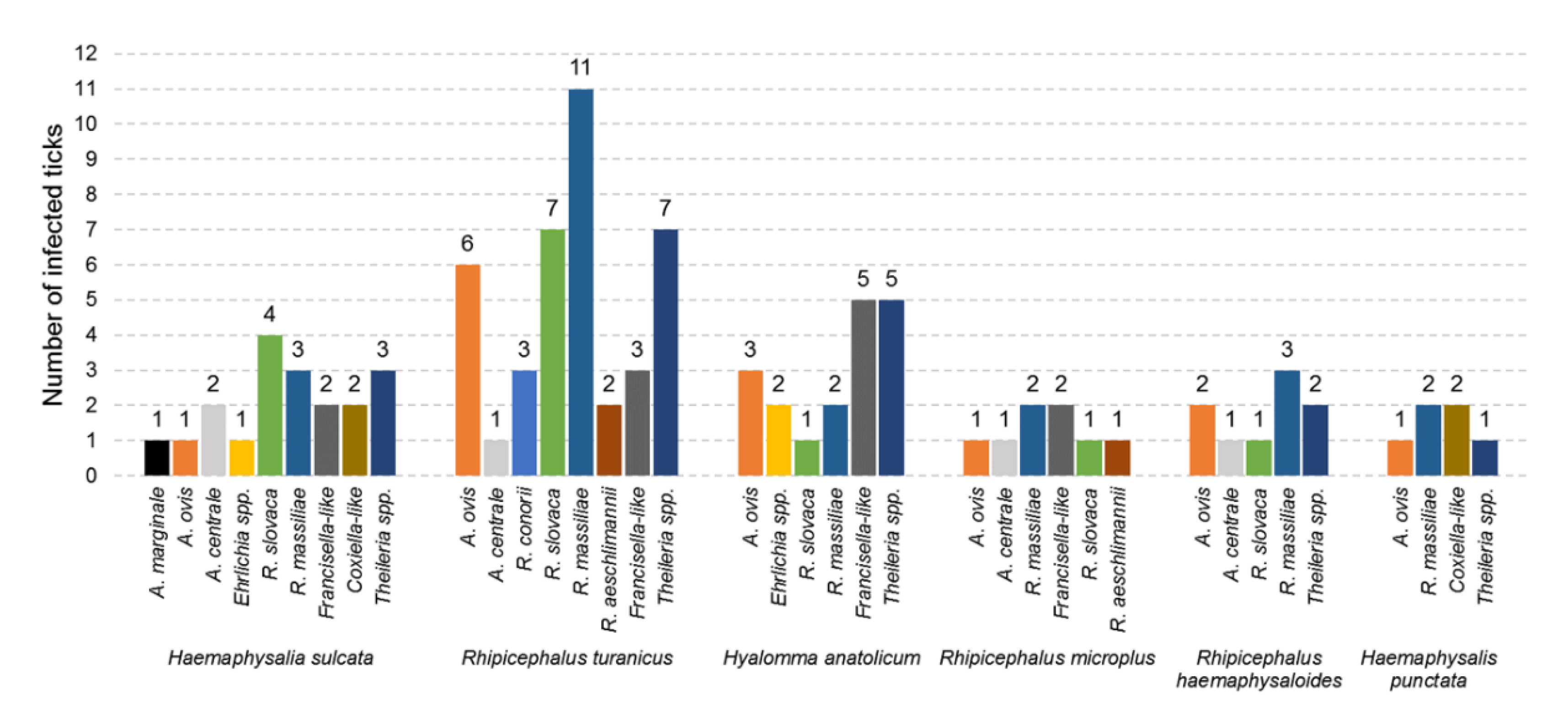
3.3. Diversity of Microorganisms in Ticks
3.4. Co-Occurrence of Microorganisms in Ticks
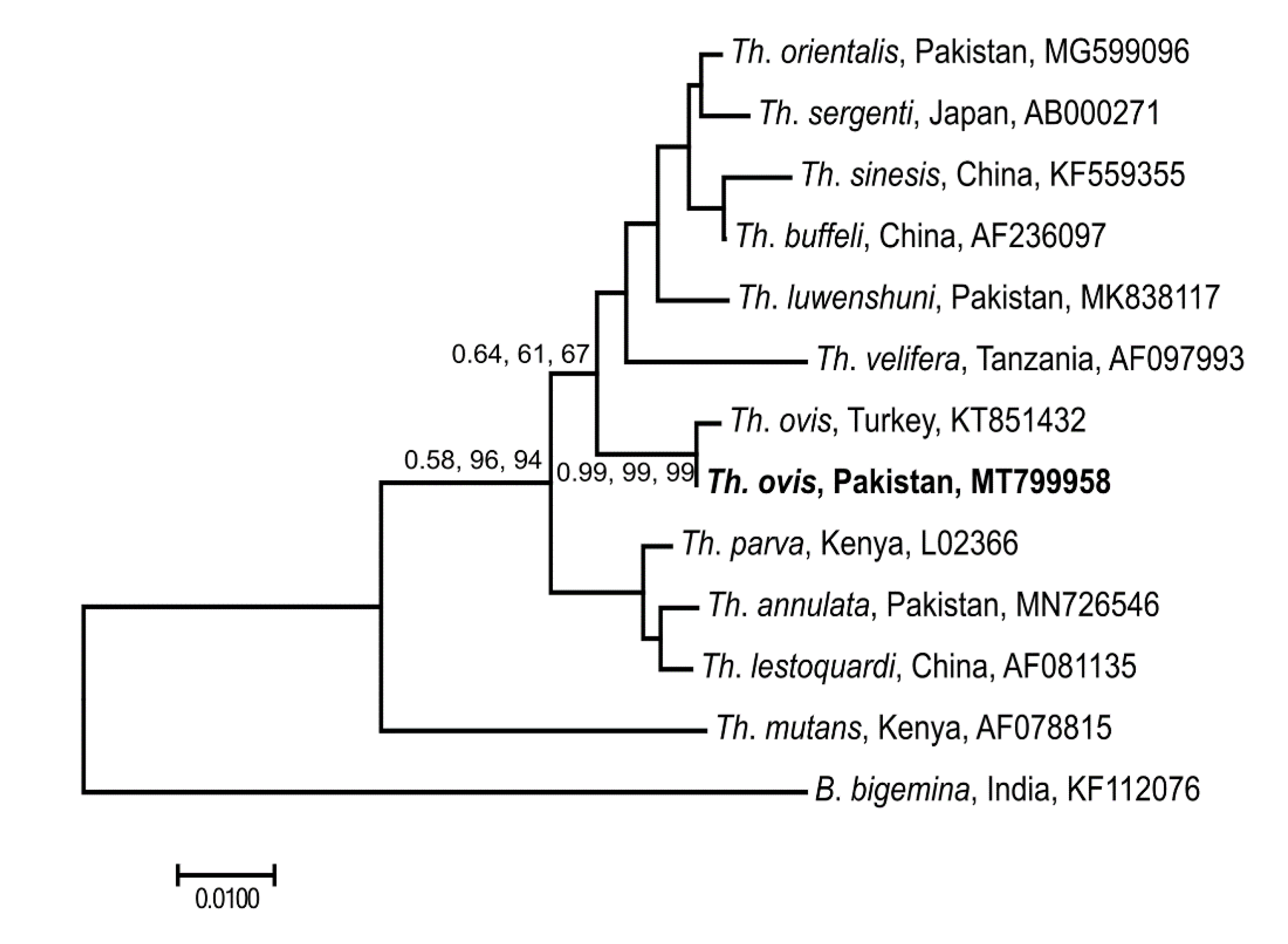
3.5. Genetic Relationship of Selected Microorganisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Material
References
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.F.; Magnarelli, L.A. Biology of Ticks. Infect. Dis. Clin. North Am. 2008, 22, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Sonenshine, D.E.; Roe, R.M. Biology of Ticks, 2nd ed.; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Government of Pakistan, Ministry of Finance, Islamabad. Pakistan Economic Survey 2019–2020; pp. 17–41. Available online: http://www.finance.gov.pk/survey_1920.html (accessed on 22 August 2020).
- Shah, S.A.A.; Parveen, S.; Khalil, J. Governance challenges in mainstreaming of Federally Administered Tribal Areas into Khyber Pakhtunkhwa. FWU J. Soc. Sci. 2019, 13, 131–145. [Google Scholar]
- Noor, S.; Hashmi, A.S.; Bukhari, S.T. Fata Merger with Khyber Pakhtunkhwa: Prospects and Opportunities. ISSRA Papers 2018. Available online: https://prdb.pk/article/fata-merger-with-khyber-pakhtunkhwa-prospects-and-opportuni-4653 (accessed on 22 August 2020).
- Nieto, N.C.; Khan, K.; Uhllah, G.; Teglas, M.B. The emergence and maintenance of vector-borne diseases in the Khyber Pakhtunkhwa province, and the Federally Administered Tribal Areas of Pakistan. Front. Physiol. 2012, 3, 250. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Khan, M.A.; Zahid, H.; Yaseen, P.M.; Khan, M.Q.; Nawab, J.; Rehman, Z.U.; Ateeq, M.; Khan, S.; Ibrahim, M. Seasonal dynamics, record of Ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front. Physiol. 2019, 10, 793. [Google Scholar] [CrossRef]
- Hussain, S.I.; Kumar, G.A. The incidence of ticks (Ixodoidea: Ixodidae) infesting sheep and goat in Sind Province Pakistan. Pak. J. Zool. 1985, 17, 89–97. [Google Scholar]
- Iqbal, A.; Siddique, F.; Mahmood, M.S.; Shamim, A.; Zafar, T.; Rasheed, I.; Saleem, I.; Ahmad, W. Prevalence and impacts of ectoparasitic fauna infesting goats (Capra hircus) of district Toba Tek Singh, Punjab, Pakistan. Glob. Vet. 2014, 12, 158–164. [Google Scholar]
- Iqbal, F.; Khattak, R.; Ozubek, S.; Khattak, M.; Rasul, A.; Aktas, M. Application of the reverse line blot assay for the molecular detection of Theileria and Babesia sp. in sheep and goat blood samples from Pakistan. Iran. J. Parasitol. 2013, 8, 289–295. [Google Scholar]
- Irshad, N.; Qayyum, M.; Hussain, M.; Khan, M. Prevalence of tick infestation and theileriosis in sheep and goats. Pak. Vet. J. 2010, 30, 178–180. [Google Scholar]
- Karim, S.; Budachetri, K.; Mukherjee, N.; Williams, J.; Kausar, A.; Hassan, M.J.; Adamson, S.; Dowd, S.E.; Apanskevich, D.; Arijo, A.; et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl. Trop. Dis. 2017, 11, e5681. [Google Scholar] [CrossRef]
- Khan, M.A.; Khan, M.; Ahmad, I.; Khan, M.; Anjum, A.; Durrani, A.; Hameed, K.; Kakar, I.; Wajid, A.; Ramazan, M. Risk factors assessment and molecular characterization of Theileria in small ruminants of Balochistan. J. Anim. Plant Sci. 2017, 27, 1190–1196. [Google Scholar]
- Rehman, A.; Nijhof, A.M.; Sauter-Louis, C.; Schauer, B.; Staubach, C.; Conraths, F.J. Distribution of ticks infesting ruminants and risk factors associated with high tick prevalence in livestock farms in the semi-arid and arid agro-ecological zones of Pakistan. Parasit. Vectors 2017, 10, 190. [Google Scholar] [CrossRef]
- Riaz, M.; Nazir, M.M.; Tasawar, Z.; Ahmed, A.N.; Ayaz, M.; Akram, Q.; Lindsay, D.S. Molecular epidemiology and prevalence of Theileria lestoquardi and Theileria ovis infection in goats infested with tick vectors from Multan, Pakistan. J. Med. Entomol. 2019, 56, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.S.; Iqbal, Z.; Khan, M.N.; Muhammad, G.; Needham, G.; Khan, M.K. Prevalence, associated determinants, and in vivo chemotherapeutic control of hard ticks (Acari: Ixodidae) infesting domestic goats (Capra hircus) of lower Punjab, Pakistan. Parasitol. Res. 2011, 108, 601–609. [Google Scholar] [CrossRef]
- Shah, A.; Shah, S.R.; Rafi, M.A.; Noorrahim, M.S.; Mitra, A. Identification of the prevalent ticks (Ixodid) in goats and sheep in Peshawar, Pakistan. J. Entomol. Zool. Stud. 2015, 3, 11–14. [Google Scholar]
- Shahzad, W.; Haider, N.; Mansur-ud-Din, A.; Munir, R.; Saghar, M.S.; Mushtaq, M.H.; Ahmad, N.; Akbar, G.; Mehmood, F. Prevalence and molecular diagnosis of Babesia ovis and Theileria ovis in Lohi sheep at Livestock Experiment Station (LES), Bahadurnagar, Okara, Pakistan. Iran. J. Parasitol. 2013, 8, 570–578. [Google Scholar]
- Khan, A.; Nasreen, N.; Niaz, S.; Shah, S.S.A.; Mitchell, R.D., III; Ayaz, S.; Naeem, H.; Khan, L.; De León, A.P. Tick burden and tick species prevalence in small ruminants of different agencies of the Federally Administered Tribal Areas (FATA), Pakistan. Int. J. Acarol. 2019, 45, 374–380. [Google Scholar] [CrossRef]
- Diarra, A.Z.; Almeras, L.; Laroche, M.; Bérenger, J.M.; Koné, A.K.; Bocoum, Z.; Dabo, A.; Doumbo, O.; Raoult, D.; Parola, P. Molecular and MALDI-TOF identification of ticks and tick-associated bacteria in Mali. PLoS Negl. Trop. Dis. 2017, 11, e5762. [Google Scholar] [CrossRef]
- Caporale, D.A.; Rich, S.M.; Spielman, A.; Telford, S.R.; Kocher, T. Discriminating between Ixodes ticks by means of mitochondrial DNA sequences. Mol. Phylogenet. Evol. 1995, 4, 361–365. [Google Scholar] [CrossRef]
- Walker, A.; Bouattour, A.; Camicas, J.; Estrada-Peña, A.; Horak, I.; Latif, A.; Pegram, R.G.; Preston, P.M.A. Ticks of Domestic Animals in Africa. A Guide to Identification of Species; Bioscience Reports: London, UK, 2003. [Google Scholar]
- Cuickshank, R.H. Molecular markers for the phylogenetics of mites and ticks. Syst. Appl. Acarol. 2002, 7, 3–14. [Google Scholar]
- Black, W.C.; Piesman, J. Phylogeny of hard- and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc. Natl. Acad. Sci. USA 1994, 91, 10034–10038. [Google Scholar] [CrossRef] [PubMed]
- Zahler, M.; Filippova, N.A.; Morel, P.C.; Gothe, R.; Rinder, H. Relationships between species of the Rhipicephalus sanguineus group: A molecular approach. J. Parasitol. 1997, 83, 302. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Noël, V.; McCoy, K.D.; Bonazzi, M.; Sidi-Boumedine, K.; Morel, O.; Vavre, F.; Zenner, L.; Jourdain, E.; Durand, P.; et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 2015, 11, e1004892. [Google Scholar] [CrossRef]
- Ghafar, A.; Cabezas-Cruz, A.; Galon, C.; Obregon, D.; Gasser, R.B.; Moutailler, S.; Riaz, N. Bovine ticks harbor a diverse array of microorganisms in Pakistan. Parasit. Vectors 2020, 13, 1. [Google Scholar] [CrossRef]
- Kasi, K.K.; Sas, M.A.; Sauter-Louis, C.; Von Arnim, F.; Gethmann, J.M.; Schulz, A.; Wernike, K.; Groschup, M.H.; Conraths, F.J. Epidemiological investigations of Crimean-Congo hemorrhagic fever virus infection in sheep and goats in Balochistan, Pakistan. Ticks Tick Borne Dis. 2020, 11, 101324. [Google Scholar] [CrossRef]
- Salih, D.; El Hussein, A.; Singla, L. Diagnostic approaches for tick-borne haemoparasitic diseases in livestock. J. Veter Med. Anim. Health 2015, 7, 45–56. [Google Scholar] [CrossRef]
- Michelet, L.; Delannoy, S.; Devillers, E.; Umhang, G.; Aspan, A.; Juremalm, M.; Chirico, J.; van der Wal, F.J.; Sprong, H.; Pihl, T.P.B.; et al. High-throughput screening of tick-borne pathogens in Europe. Front. Cell Infect. Microbiol. 2014, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Allain, E.; Ahmad, A.S.; Saeed, M.A.; Rashid, M.I.; Ashraf, K.; Yousfi, L.; Shehzad, W.; Indjein, L.; Rodriguez-Valle, M.; et al. Low genetic diversity of Ehrlichia canis associated with high co-infection rates in Rhipicephalus sanguineus (sl). Parasit. Vectors 2019, 12, 12. [Google Scholar] [CrossRef]
- Gondard, M.; Delannoy, S.; Pinarello, V.; Aprelon, R.; Devillers, E.; Galon, C.; Pradel, J.; Vayssier-Taussat, M.; Albina, E.; Moutailler, S. Upscaling the surveillance of tick-borne pathogens in the French Caribbean islands. Pathogens 2020, 9, 176. [Google Scholar] [CrossRef]
- Grech-Angelini, S.; Stachurski, F.; Vayssier-Taussat, M.; Devillers, E.; Casabianca, F.; Lancelot, R.; Uilenberg, G.; Moutailler, S. Tick-borne pathogens in ticks (Acari: Ixodidae) collected from various domestic and wild hosts in Corsica (France), a Mediterranean island environment. Transbound. Emerg. Dis. 2020, 67, 745–757. [Google Scholar] [CrossRef]
- Lejal, E.; Moutailler, S.; Šimo, L.; Vayssier-Taussat, M.; Pollet, T. Tick-borne pathogen detection in midgut and salivary glands of adult Ixodes ricinus. Parasit. Vectors 2019, 12, 152. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Joncour, G.; Devillers, E.; Torina, A.; Vayssier-Taussat, M.; Bonnet, S.I.; Moutailler, S. Tick species, tick-borne pathogens and symbionts in an insular environment off the coast of Western France. Ticks Tick Borne Dis. 2016, 7, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- FATA Secretariat, Government of Pakistan; Food Agriculture Organization of the United Nations. Agriculture Policy for FATA: Policy period (2016–2025). Available online: http://extwprlegs1.fao.org/docs/pdf/pak173416.pdf (accessed on 22 August 2020).
- Barker, S.C.; Walker, A.R. Ticks of Australia. The species that infest domestic animals and humans. Zootaxa 2014, 3816, 1–144. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.B.; Keirans, J.E.; Horak, I.G. The Genus Rhipicephalus (Acari, Ixodidae): A Guide to the Brown Ticks of the World; Cambridge University Press: New York, NY, USA, 2000. [Google Scholar]
- Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Rees, D.; Dioli, M.; Kirkendall, L.R. Molecules and morphology: Evidence for cryptic hybridization in African Hyalomma (Acari: Ixodidae). Mol. Phylogenet. Evol. 2003, 27, 131–142. [Google Scholar] [CrossRef]
- Gubbels, M.J.; De Vos, A.P.; Van der Weide, M.; Viseras, J.; Schouls, L.M.; De Vries, E.; Jongejan, F. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 1999, 37, 1782–1789. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ghafar, A.; Gasser, R.B.; Rashid, I.; Ghafoor, A.; Riaz, N. Exploring the prevalence and diversity of bovine ticks in five agro-ecological zones of Pakistan using phenetic and genetic tools. Ticks Tick Borne Dis. 2020, 11, 101472. [Google Scholar] [CrossRef] [PubMed]
- Bakkes, D.K.; Chitimia-Dobler, L.; Matloa, D.; Oosthuysen, M.; Mumcuoglu, K.Y.; Mans, B.J.; Matthee, C.A. Integrative taxonomy and species delimitation of Rhipicephalus turanicus (Acari: Ixodida: Ixodidae). Int. J. Parasitol. 2020, 50, 577–594. [Google Scholar] [CrossRef]
- Bakkes, D.K.; De Klerk, D.; Latif, A.A.; Mans, B.J. Integrative taxonomy of Afrotropical Ornithodoros (Ornithodoros) (Acari: Ixodida: Argasidae). Ticks Tick Borne Dis. 2018, 9, 1006–1037. [Google Scholar] [CrossRef]
- Chitimia-Dobler, L.; Langguth, J.; Pfeffer, M.; Kattner, S.; Küpper, T.; Friese, D.; Dobler, G.; Guglielmone, A.A.; Nava, S. Genetic analysis of Rhipicephalus sanguineus sensu lato ticks parasites of dogs in Africa north of the Sahara based on mitochondrial DNA sequences. Vet. Parasitol. 2017, 239, 1–6. [Google Scholar] [CrossRef]
- Lado, P.; Nava, S.; Mendoza-Uribe, L.; Cáceres, A.G.; De La Mora, J.D.; Licona-Enriquez, J.D.; La Mora, D.D.-D.; Labruna, M.B.; Durden, L.A.; Allerdice, M.E.; et al. The Amblyomma maculatum Koch, 1844 (Acari: Ixodidae) group of ticks: Phenotypic plasticity or incipient speciation? Parasit. Vectors 2018, 11, 610. [Google Scholar] [CrossRef]
- Li, L.H.; Zhang, Y.; Wang, J.Z.; Li, X.S.; Yin, S.Q.; Zhu, D.; Xue, J.B.; Li, S.G. High genetic diversity in hard ticks from a China-Myanmar border county. Parasit. Vectors 2018, 11, 469. [Google Scholar] [CrossRef]
- Mans, B.J.; Featherston, J.; Kvas, M.; Pillay, K.A.; De Klerk, D.G.; Pienaar, R.; De Castro, M.H.; Schwan, T.G.; Lopez, J.E.; Teel, P.; et al. Argasid and ixodid systematics: Implications for soft tick evolution and systematics, with a new argasid species list. Ticks Tick Borne Dis. 2019, 10, 219–240. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, S.; Pei, T.; Yu, Z.; Liu, J. Tick mitochondrial genomes: Structural characteristics and phylogenetic implications. Parasit. Vectors 2019, 12, 451. [Google Scholar] [CrossRef] [PubMed]
- Gorski, P.; Niznikowski, R.; Strzelec, E.; Popielarczyk, D.; Gajewska, A.; Wedrychowicz, H. Prevalence of protozoan and helminth internal parasite infections in goat and sheep flocks in Poland. Arch. Tierzucht 2004, 47, 43–49. [Google Scholar]
- Azmi, K.; Al-Jawabreh, A.; Abdeen, Z. Molecular Detection of Theileria ovis and Theleiria equi in Livestock from Palestine. Sci. Rep. 2019, 9, 11557. [Google Scholar] [CrossRef] [PubMed]
- Chochlakis, D.; Ioannou, I.; Sharif, L.; Kokkini, S.; Hristophi, N.; Dimitriou, T.; Tselentis, Y.; Psaroulaki, A. Prevalence of Anaplasma sp. in goats and sheep in Cyprus. Vector Borne Zoonotic Dis. 2009, 9, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Rjeibi, M.R.; Gharbi, M.; Mhadhbi, M.; Mabrouk, W.; Ayari, B.; Nasfi, I.; Jedidi, M.; Sassi, L.; Rekik, M.; Darghouth, M.A. Prevalence of piroplasms in small ruminants in North-West Tunisia and the first genetic characterization of Babesia ovis in Africa. Parasite 2014, 21, 23. [Google Scholar] [CrossRef]
- Ben Saïd, M.; Belkahia, H.; Alberti, A.; Zobba, R.; Bousrih, M.; Yahiaoui, M.; Daaloul-Jedidi, M.; Mamlouk, A.; Gharbi, M.; Messadi, L. Molecular survey of Anaplasma species in small ruminants reveals the presence of novel strains closely related to A. phagocytophilum in Tunisia. Vector Borne Zoonotic Dis. 2015, 15, 580–590. [Google Scholar] [CrossRef]
- Guizzo, M.G.; Parizi, L.F.; Nunes, R.D.; Schama, R.; Albano, R.M.; Tirloni, L.; Oldiges, D.P.; Vieira, R.P.; Oliveira, W.H.C.; Leite, M.D.S.; et al. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci. Rep. 2017, 7, 17554. [Google Scholar] [CrossRef]
- Macaluso, K.R.; Sonenshine, D.E.; Ceraul, S.M.; Azad, A.F. Rickettsial Infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J. Med. Entomol. 2002, 39, 809–813. [Google Scholar] [CrossRef]
- Satjanadumrong, J.; Robinson, M.T.; Hughes, T.; Blacksell, S.D. Distribution and ecological drivers of spotted fever group Rickettsia in Asia. EcoHealth 2019, 16, 611–626. [Google Scholar] [CrossRef]
- Kuttler, K.L. Anaplasma infections in wild and domestic ruminants: A review. J. Wildl. Dis. 1984, 20, 12–20. [Google Scholar] [CrossRef]
- Chochlakis, D.; Ioannou, I.; Tselentis, Y.; Psaroulaki, A. Human anaplasmosis and Anaplasma ovis variant. Emerg. Infect. Dis. 2010, 16, 1031–1032. [Google Scholar] [CrossRef] [PubMed]
- Duan, D.Y.; Liu, G.H.; Cheng, T.Y. Microbiome analysis of the saliva and midgut from partially or fully engorged female adult Dermacentor silvarum ticks in China. Exp. Appl. Acarol. 2020, 80, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Yang, Z.N.; Lu, B.; Ma, X.F.; Zhang, C.X.; Xu, H.J. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick Borne Dis. 2014, 5, 864–870. [Google Scholar] [CrossRef]
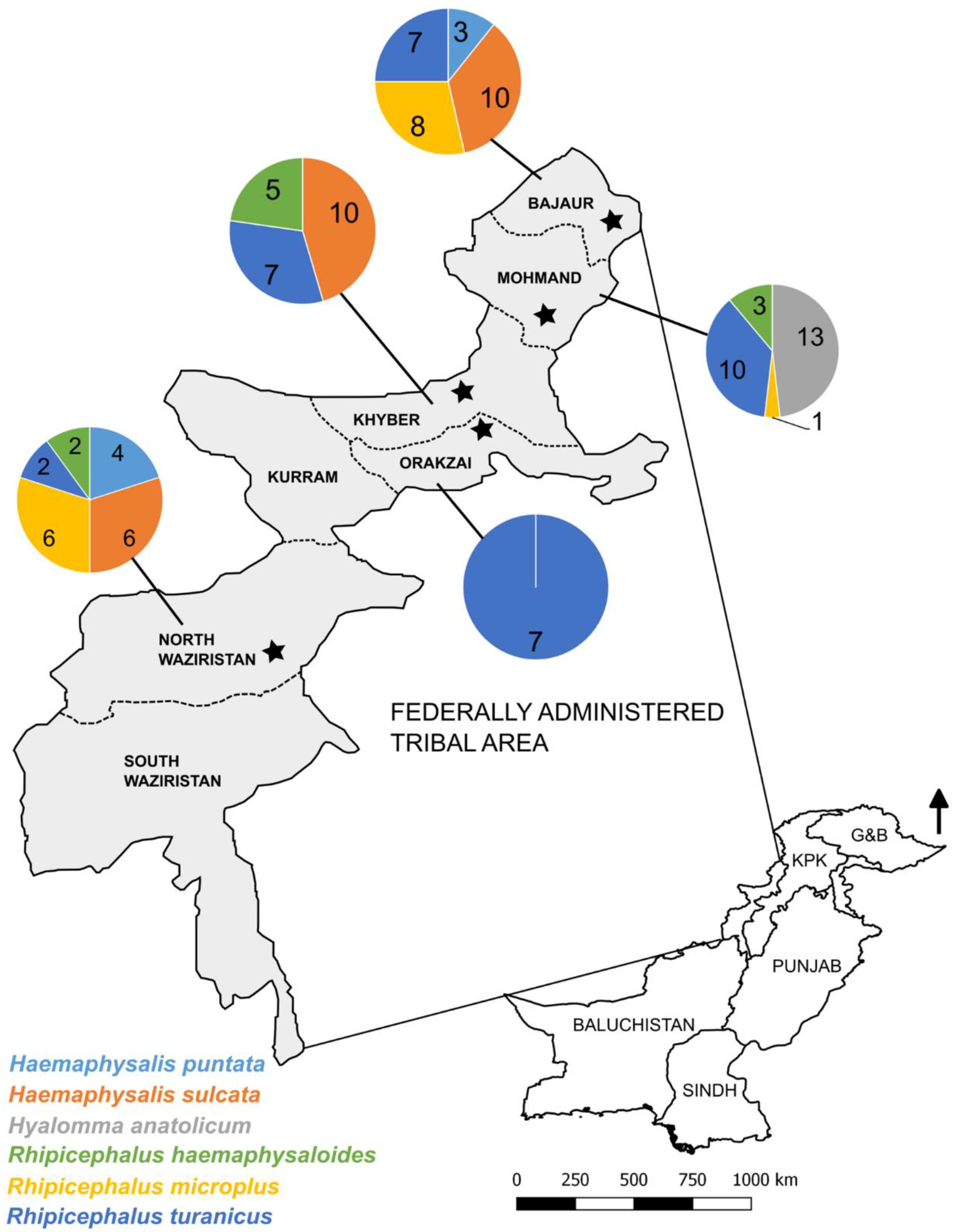

| Tribal Districts (Coordinates) | Host (Number) | Tick Species (Based on Microscopy) | No. of Ticks | GenBank Accession Numbers (Only Unique Sequences) | ||
|---|---|---|---|---|---|---|
| 16S | cox1 | ITS-2 | ||||
| Bajaur (34.856902° N, 71.429936° E) | Sheep (n = 5) | Haemaphysalis sulcata | 4 | MT799946 | MT800319 | - |
| Rhipicephalus microplus | 6 | MT799951 | MT800322 | - | ||
| Rhipicephalus turanicus | 3 | - | - | - | ||
| Goat (n = 7) | Haemaphysalis punctata | 3 | MT799944 | MT800318 | - | |
| Hs. sulcata | 6 | MT799946 | MT800319 | - | ||
| MT799947 | MT800320 | - | ||||
| Rh. microplus | 2 | - | - | - | ||
| Rh. turanicus | 4 | - | - | - | ||
| Khyber (33.940548° N, 71.049777° E) | Sheep (n = 5) | Hs. sulcata | 5 | - | - | - |
| Rh. turanicus | 3 | - | - | - | ||
| Goat (n = 7) | Hs. sulcata | 5 | MT799948 | MT800320 | - | |
| Rh. turanicus | 4 | MT799955 | MT800313 | - | ||
| Rhipicephalus haemaphysaloides | 5 | MT799956 | MT800316 | MT818227 | ||
| MT799956 | MT800315 | MT818227 | ||||
| Mohmand (34.535595° N, 71.287421° E) | Sheep (n = 6) | Hyalomma anatolicum | 6 | MT799950 | MT800311 | MT818222 |
| Rh. turanicus | 5 | MT799954 | MT800312 | MT818226 | ||
| MT799954 | MT800314 | MT818226 | ||||
| Goat (n = 8) | Hy. anatolicum | 7 | MT799950 | MT800311 | MT818222 | |
| Rh. microplus | 1 | - | - | - | ||
| Rh. turanicus | 5 | - | - | - | ||
| Rh. haemaphysaloides | 3 | MT799956 | - | - | ||
| Orakzai (33.667137° N, 70.95468° E) | Sheep (n = 2) | Rh. turanicus | 4 | - | - | - |
| Goat (n = 2) | Rh. turanicus | 3 | - | - | - | |
| North Waziristan (32.320237° N, 69.859741° E) | Sheep (n = 5) | Hs. sulcata | 6 | MT799949 | MT800321 | - |
| Rh. microplus | 2 | MT799952 | MT800323 | MT818223 | ||
| Goat (n = 7) | Hs. punctata | 4 | - | - | - | |
| Rh. microplus | 4 | MT799953 | MT800322 | MT818223 | ||
| Rh. turanicus | 2 | - | - | - | ||
| Rh. haemaphysaloides | 2 | MT799956 | MT800317 | MT818227 | ||
| Total | 54 | 104 | ||||
| Tick Species | District | Host | No. of Ticks Tested | No. of Ticks Infected | Microorganisms Detected |
|---|---|---|---|---|---|
| Haemaphysalis punctata | Bajaur, North Waziristan | Goat | 4 | 3 | Anaplasma ovis, Rickettsia massiliae, Coxiella-like, Theileria spp. |
| Hs. sulcata | Bajaur, Khyber, North Waziristan | Sheep | 8 | 4 | A. marginale, A. ovis, A. centrale, R. slovaca, R. massiliae, Francisella-like, Coxiella-like, Theileria spp. |
| Goat | 6 | 5 | A. centrale, Ehrlichia spp., R. slovaca, R. massiliae, Francisella-like, Coxiella-like, Theileria spp. | ||
| Hyalomma anatolicum | Mohmand | Sheep | 3 | 3 | A. ovis, Ehrlichia spp., R. slovaca, R. massiliae, Francisella-like, Theileria spp. |
| Goat | 3 | 3 | A. ovis, R. massiliae, Francisella-like, Theileria spp. | ||
| Rhipicephalus microplus | Bajaur, Mohmand, North Waziristan | Sheep | 4 | 2 | A. ovis, Francisella-like, R. aeschlimannii, R. massiliae, R. slovaca |
| Goat | 4 | 3 | A. centrale, R. massiliae, Francisella-like | ||
| Rh. turanicus | Bajaur, Khyber, Orakzai, Mohmand, North Waziristan | Sheep | 8 | 4 | A. ovis, R. conorii, R. slovaca, R. massiliae, R. aeschlimannii, Francisella-like, Theileria spp. |
| Goat | 10 | 8 | A. ovis, A. centrale, R. conorii, R. slovaca, R. massiliae, R. aeschlimannii, Francisella-like, Theileria spp. | ||
| Rh. haemaphysaloides | Khyber, Mohmand, North Waziristan | Goat | 4 | 4 | A. ovis, A. centrale, R. slovaca, R. massiliae, Theileria spp. |
| Total | 54 | 39 | |||
| Microorganisms | Bajaur | Khyber | Mohmand | Orakzai | North Waziristan | % Prevalence (Proportion) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sheep (n = 5) | Goat (n = 7) | Sheep (n = 5) | Goat (n = 7) | Sheep (n = 6) | Goat (n = 8) | Sheep (n = 2) | Goat (n = 2) | Sheep (n = 5) | Goat (n = 7) | ||
| Anaplasma marginale | - | - | 1 | - | - | - | - | - | - | - | 1.9 (1/54) |
| A. ovis | 1 | - | 1 | 1 | 3 | 4 | 1 | - | 1 | 2 | 25.9 (14/54) |
| A. centrale | - | 1 | 1 | - | - | 1 | - | - | - | 2 | 9.3 (5/54) |
| Ehrlichia spp. | - | - | - | 1 | 2 | - | - | - | - | - | 5.6 (3/54) |
| Rickettsia conorii | - | - | - | 1 | 1 | 1 | - | - | - | - | 5.6 (3/54) |
| R. slovaca | 1 | 2 | - | 3 | 3 | 1 | 1 | 2 | 1 | - | 25.9 (14/54) |
| R. massiliae | 1 | 1 | 2 | 4 | 3 | 4 | 1 | 2 | 1 | 4 | 42.6 (23/54) |
| R. aeschlimannii | 1 | - | - | - | 1 | 1 | - | - | - | - | 5.6 (3/54) |
| Francisella-like | 2 | - | - | 1 | 4 | 3 | 1 | 1 | - | - | 22.2 (12/54) |
| Coxiella-like | - | - | - | 1 | - | - | - | - | 1 | 2 | 7.4 (4/54) |
| Theileria spp. | 1 | 1 | 2 | 1 | 3 | 5 | 1 | 2 | - | 2 | 33.3 (18/54) |
| Total | 7 | 5 | 7 | 13 | 20 | 20 | 5 | 7 | 4 | 12 | |
| Microorganisms | Number of Ticks Positive for Single and Mixed Infections | |||||
|---|---|---|---|---|---|---|
| Bajaur (n = 12) | Khyber (n = 12) | Mohmand (n = 14) | Orakzai (n = 4) | North Waziristan (n = 12) | Total | |
| Single | ||||||
| Anaplasma ovis | - | - | - | - | 1 | 1 |
| A. centrale | - | - | - | - | 1 | 1 |
| Rickettsia slovaca | 1 | 1 | - | - | 1 | 3 |
| R. massiliae | - | 1 | - | - | 2 | 3 |
| Francisella-like | - | - | 1 | - | - | 1 |
| Theileria spp. | 1 | 1 | - | - | - | 2 |
| Double | ||||||
| A. centrale, R. massiliae | - | - | - | - | 1 | 1 |
| A. ovis, Theileria spp. | - | - | 1 | - | 1 | 2 |
| A. ovis, R. massiliae. | - | 1 | - | - | 1 | |
| A. ovis, Coxiella-like | - | - | - | - | 1 | 1 |
| Francisella-like, Theileria spp. | - | - | 1 | - | - | 1 |
| R. massiliae, R. slovaca | - | 1 | - | - | - | 1 |
| R. massiliae, Coxiella-like | - | - | - | - | 1 | 1 |
| Triple | ||||||
| A. ovis, R. massiliae, Theileria spp. | - | 1 | 1 | - | - | 2 |
| A. ovis, R. massiliae, R. slovaca | - | - | 1 | - | - | 1 |
| A. ovis, Francisella-like, Theileria spp. | 1 | - | - | - | - | 1 |
| A. centrale, R. massiliae, R. slovaca | 1 | - | - | - | - | 1 |
| Coxiella-like, Ehrlichia spp., Francisella-like, | - | 1 | - | - | - | 1 |
| Coxiella-like, R. massiliae, Theileria spp. | - | - | - | - | 1 | 1 |
| Ehrlichia spp., Francisella-like, Theileria spp. | - | - | 1 | - | - | 1 |
| Francisella-like, R. massiliae, R. slovaca | - | - | 1 | - | - | 1 |
| R. conorii, R. massiliae, R. slovaca | - | 1 | - | - | - | 1 |
| R. massiliae, R. slovaca, Theileria spp. | - | - | - | 1 | - | 1 |
| Quadruple | ||||||
| A. centrale, A. marginale, R. massiliae, Theileria spp. | - | 1 | - | - | - | 1 |
| A. centrale, A. ovis, R. massiliae, Theileria spp. | - | 1 | - | - | 1 | |
| A. ovis, Ehrlichia spp., Francisella-like, Theileria spp. | - | - | 1 | - | - | 1 |
| A. ovis, R. massiliae, Francisella-like, Theileria spp. | - | - | 1 | - | - | 1 |
| Francisella-like, R. aeschlimannii, R. massiliae, R. slovaca | 1 | - | - | - | - | 1 |
| Francisella-like, R. massiliae, R. slovaca, Theileria spp. | - | - | - | 1 | - | 1 |
| R. aeschlimannii, R. conorii, R. massiliae, R. slovaca | - | 1 | - | - | 1 | |
| Quintuple | ||||||
| A. ovis, Francisella-like, R. massiliae, R. slovaca, Theileria spp. | - | - | - | 1 | - | 1 |
| Septuple | ||||||
| A. ovis, Francisella-like, R. aeschlimannii, R. conorii, R. massiliae, R. slovaca, Theileria spp. | - | - | 1 | - | - | 1 |
| Total | 5 | 9 | 12 | 3 | 10 | 39 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghafar, A.; Khan, A.; Cabezas-Cruz, A.; Gauci, C.G.; Niaz, S.; Ayaz, S.; Mateos-Hernández, L.; Galon, C.; Nasreen, N.; Moutailler, S.; et al. An Assessment of the Molecular Diversity of Ticks and Tick-Borne Microorganisms of Small Ruminants in Pakistan. Microorganisms 2020, 8, 1428. https://doi.org/10.3390/microorganisms8091428
Ghafar A, Khan A, Cabezas-Cruz A, Gauci CG, Niaz S, Ayaz S, Mateos-Hernández L, Galon C, Nasreen N, Moutailler S, et al. An Assessment of the Molecular Diversity of Ticks and Tick-Borne Microorganisms of Small Ruminants in Pakistan. Microorganisms. 2020; 8(9):1428. https://doi.org/10.3390/microorganisms8091428
Chicago/Turabian StyleGhafar, Abdul, Adil Khan, Alejandro Cabezas-Cruz, Charles G. Gauci, Sadaf Niaz, Sultan Ayaz, Lourdes Mateos-Hernández, Clemence Galon, Nasreen Nasreen, Sara Moutailler, and et al. 2020. "An Assessment of the Molecular Diversity of Ticks and Tick-Borne Microorganisms of Small Ruminants in Pakistan" Microorganisms 8, no. 9: 1428. https://doi.org/10.3390/microorganisms8091428
APA StyleGhafar, A., Khan, A., Cabezas-Cruz, A., Gauci, C. G., Niaz, S., Ayaz, S., Mateos-Hernández, L., Galon, C., Nasreen, N., Moutailler, S., Gasser, R. B., & Jabbar, A. (2020). An Assessment of the Molecular Diversity of Ticks and Tick-Borne Microorganisms of Small Ruminants in Pakistan. Microorganisms, 8(9), 1428. https://doi.org/10.3390/microorganisms8091428










