Mining the Microbiome of Key Species from African Savanna Woodlands: Potential for Soil Health Improvement and Plant Growth Promotion
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sampling
2.3. DNA Extraction and Illumina Sequencing of the 16S rRNA Gene
2.4. Sequence Data Analysis
2.5. Isolation and Characterization of Bacteria from Soils
2.6. Isolation and Characterization of Root Nodule Bacteria Using a Host Legume (Vigna Unguiculata)
2.7. Identification of Isolated Bacteria by 16S rRNA Gene Sequence Analysis
3. Results and Discussion
3.1. Bacterial Diversity in Mopane Soils
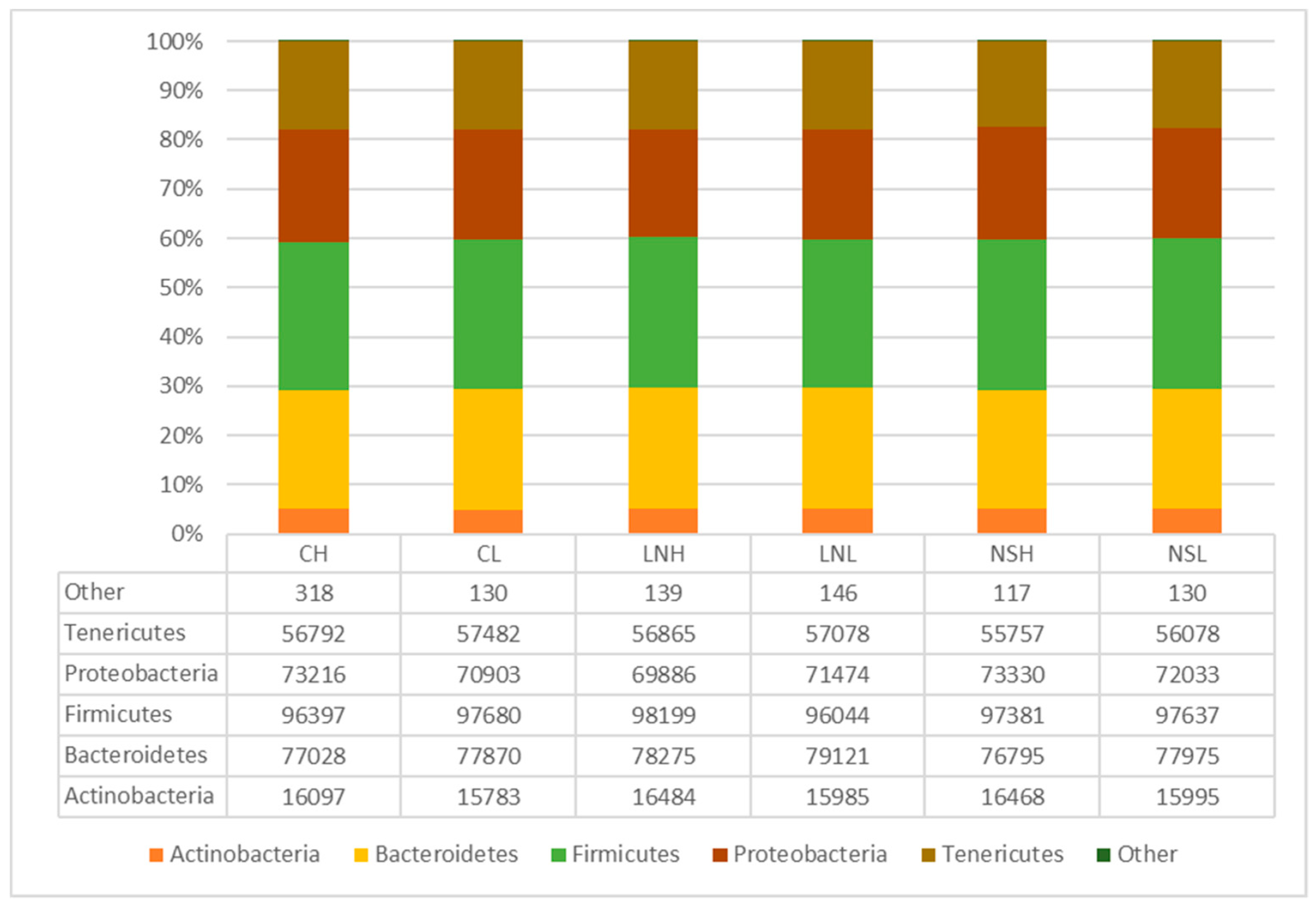
3.2. Taxonomic Composition of the Microbial Communities
3.3. Taxonomic Analysis of Bacteria Isolated from Soils
3.4. In Vitro Activities Related to PGPB
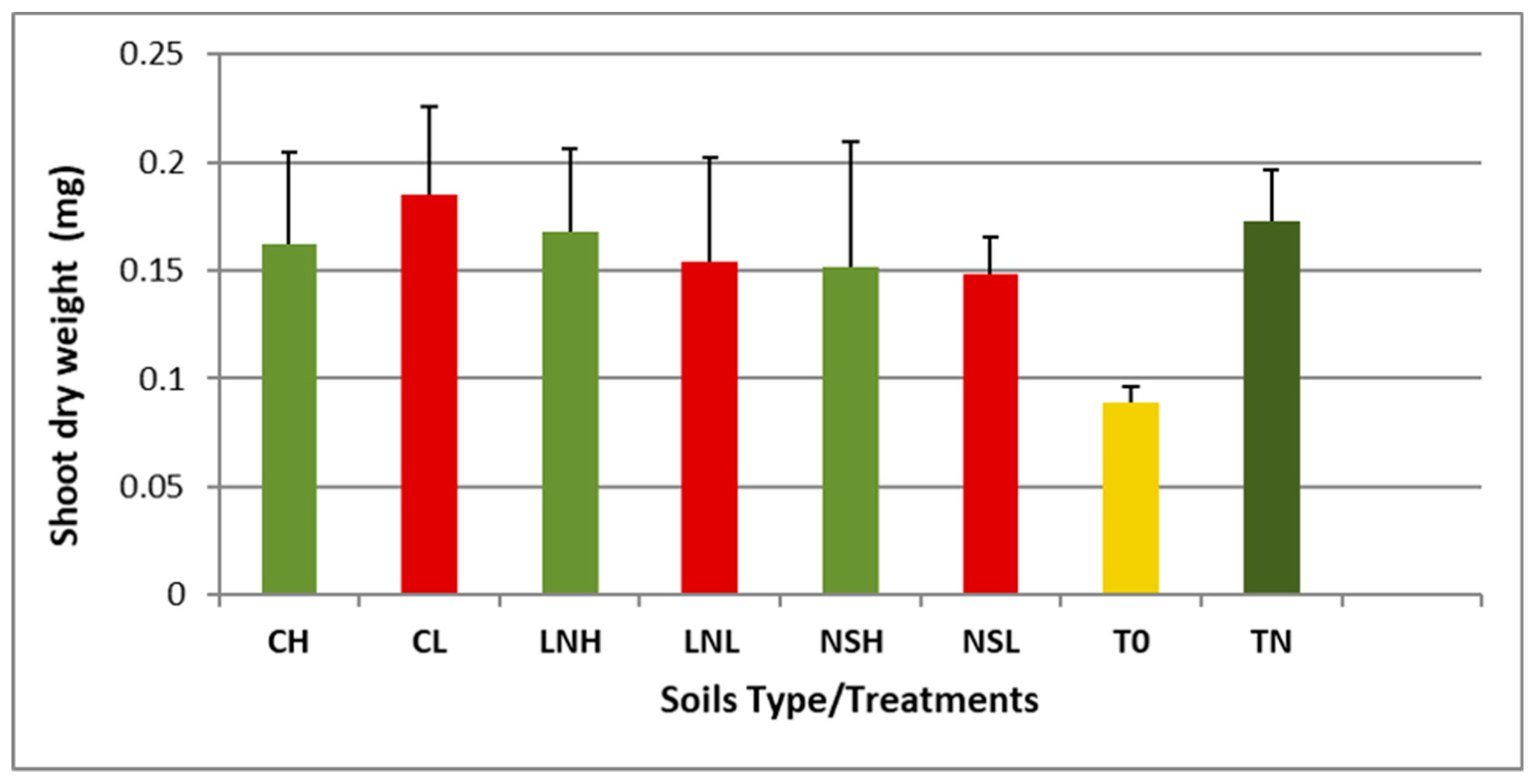
3.5. Vigna unguiculata as a Trap for Rhizobia Bacteria
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fierer, N.; Leff, J.W.; Adams, B.J. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, E.; Kranabetter, J.M.; Hope, G.; Maas, K.R.; Hallam, S.; Mohn, W.W. Forest harvesting reduces the soil metagenomic potential for biomass decomposition. ISME J. 2015, 9, 2465–2476. [Google Scholar] [CrossRef] [PubMed]
- Neary, D.G.; Klopatek, C.C.; deBano, L.F.; Ffolliott, P.F. Fire effects on belowground sustainability: A review and synthesis. For. Ecol. Manag. 1999, 122, 51–71. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Gopal, M.; Gupta, A. Building plant microbiome vault: A future biotechnological resource. Symbiosis 2019, 77, 1–8. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.; Rocha-Granados, M.; Glick, B.R.; Santoyo, G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 2018, 208, 25–31. [Google Scholar] [CrossRef]
- Andreote, F.D.; Pereira, S.M.C. Microbial communities associated with plants: Learning from nature to apply it in agriculture. Curr. Opin. Microbiol. 2017, 37, 29–34. [Google Scholar] [CrossRef]
- Finkel, O.M.; Castrill, G.; Paredes, S.H.; González, I.S.; Dangl, J.L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant. Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef]
- Youssef, N.H.; Elshahed, M.S. Diversity rankings among bacterial lineages in soil. ISME J. 2009, 3, 305–313. [Google Scholar] [CrossRef]
- Cunningham, A.B. African Medicinal Plants: Setting Priorities at the Interface between Conservation and Primary Healthcare; People and Plants Working Paper 1; UNESCO: Paris, France, 1993; Available online: http.//unesdoc.unesco.org/images/0009/000967/096707E.pdf (accessed on 5 June 2020).
- Mittermeier, R.A.; Mittermeier, C.G.; Brooks, T.M.; Pilgrim, J.D.; Konstant, W.R.; da Fonseca, G.A.B.; Kormos, C. Wilderness and biodiversity conservation. Proc. Natl. Acad. Sci. USA 2003, 100, 10309–10313. [Google Scholar] [CrossRef]
- Moura, I.; Maquia, I.; Rija, A.; Ribeiro, N.; Ribeiro-Barros, A.I. Biodiversity studies in key species from the African mopane and miombo woodlands. In Genetic Diversity; Bitz, L., Ed.; IntechOpen: London, UK, 2017; pp. 91–109. [Google Scholar]
- Burgess, N.; Hales, J.D.; Underwood, E.; Dinerstein, E. Terrestrial Ecoregions of Africa and Madagascar: A Conservation Assessment; Island Press: Washington, DC, USA, 2004. [Google Scholar]
- Makhado, R.A.; Mapaure, I.; Potgieter, M.J.; Luus-Powell, W.J.; Saidi, A.T. Factors influencing the adaptation and distribution of Colophospermum mopane in southern Africa’s mopane savannas a review. Bothalia 2014, 44. [Google Scholar] [CrossRef]
- Ribeiro, N.; Ruecker, G.; Govender, N. The influence of fire frequency on the structure and botanical composition of savanna ecosystems. Ecol. Evol. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Frost, P.G.H. The responses and survival of organisms in fire-prone environments. In Ecological Effects of Fire in South African Ecosystems; Booysen, P.V., Tainton, N.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 273–309. [Google Scholar]
- Higgins, S.I.; Bond, W.J.; Trollope, W.S.W. Fire, resprouting and variability: A recipe for grass-tree coexistence in savanna. J. Ecol. 2000, 88, 213–229. [Google Scholar] [CrossRef]
- Higgins, S.I.; Bond, W.J.; February, E.C.; Bronn, A.; Brown, D.I.W.E.; Enslin, B.; Govender, N.; Rademan, L.; O’Regan, S.; Potgieter, A.L.F.; et al. Effects of four decades of fire manipulation on woody vegetation structure in savanna. Ecology 2007, 88, 1119–1125. [Google Scholar] [CrossRef]
- van Wilgen, B.W.; Biggs, H.C. A critical assessment of adaptive ecosystem management in a large savanna protected area in South Africa. Biol. Conserv. 2011, 144, 1179–1187. [Google Scholar] [CrossRef]
- Burbano, C.S.; Grönemeyer, J.L.; Hurek, T.; Reinhold-Hurek, B. Microbial community structure and functional diversity of nitrogen-fixing bacteria associated with Colophospermum mopane. FEMS Microbiol. Ecol. 2015, 9, 1–13. [Google Scholar] [CrossRef]
- Wolmer, W. Transboundary conservation: The politics of ecological integrity in the Great Limpopo Transfrontier Park. J. S. Afr. Stud. 2003, 29, 261–278. [Google Scholar] [CrossRef]
- Hanks, J. Transfrontier Conservation Areas (TFCAs) in Southern Africa: Their role in conserving biodiversity.; socioeconomic development and promoting a culture of peace. J. Sustain. For. 2003, 17, 127–148. [Google Scholar] [CrossRef]
- Goodman, P.; Breen, C. The kruger experience: Ecology and management of savanna heterogeneity. Afr. J. Aquat. Sci. 2004, 29, 121–122. [Google Scholar] [CrossRef]
- Stalmans, M.; Gertenbach, W.P.D.; Carvalho-Serfontein, F. Plant communities and landscapes of the Parque Nacional do Limpopo, Moçambique. Koedoe 2004, 47, 61–81. [Google Scholar] [CrossRef]
- ANAC. Limpopo National Park Management and Development Plan; Mozambique Ministry of Tourism: Maputo, Mozambique, 2003. [Google Scholar]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. Vsearch: A versatile open source tool for metagenomics. Peer J. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Justine, R.H.; Martine, H.; Emily, B.H.; Ryan, A.L.; Brian, B.O.; Donovan, H.P.; Courtney, J.R.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Montagna, M.; Berruti, A.; Bianciotto, V.; Cremones, P.; Giannico, R.; Gusmeroli, F.; Lumini, E.; Pierce, S.; Pizzi, F.; Turri, F. Differential biodiversity responses between kingdoms (plants, fungi, bacteria and metazoa) along an Alpine succession gradient. Mol. Ecol. 2018, 27, 3671–3685. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16s rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, B.D.; Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Carlsen, T.; Kjøller, R.; Kõljalg, U.; Pennanen, T.; Rosendahl, S.; Stenlid, J.; et al. Fungal community analysis by high-throughput sequencing of amplified markers-a user’s guide. New Phytol. 2013, 199, 288–299. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes; a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Lopez-García, A.L.; Quiroga, C.P.; Atxaerandio, R.; Pérez, A.; Hernández, I.; Rodríguez, A.G.; Recio, O.G. Comparison of mothur and QIIME for the analysis of rumen microbiota composition based on 16S rRNA amplicon sequences. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Vegan, Version 2.2-1. 2015. Available online: http://www.worldagroforestry.org/publication/vegan-community-ecology-package-r-package-vegan-vers-22-1 (accessed on 5 June 2020).
- de Cáceres, M.; Jansen, F. Package ‘Indicspecies’ (Version 1.7.6). 2016. Available online: https://mran.microsoft.com/snapshot/2016-09-1/web/packages/indicspecies/indicspecies.pdf (accessed on 5 June 2020).
- Team, R.D.C. R: A language and Environment for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 5 June 2020).
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.M. A manual for practical study of root nodule bacteria. In IBP Handbook; Blackwell Scientific Publishers: Oxford, UK, 1970. [Google Scholar]
- Bertani, G. Studies on lysogenesis. Nippon Saikingaku Zasshi 1951, 11, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, N.L.; Dourado, A.C.; Alves, P.I.; Alícia, M.C.P.; Ana, I.D.R.; Ângela, P.; Nuno, B.; Claudia, S.; Maria, T.B.C.; Paula, F. Annual ryegrass-associated bacteria with potential for plant growth promotion. Microbiol. Res. 2014, 169, 768–779. [Google Scholar] [CrossRef]
- Döbereiner, J.; Marriel, I.; Nery, M. Ecological distribution of Spirillum lipoferum Beijerinck. Can. J. Microbiol. 1976, 22, 1464–1473. [Google Scholar] [CrossRef]
- Martinez-Toledo, M.V.; Gonzalez-Lopez, J.; de la Rubia, T.; Ramos-Cormenzana, A. Isolation and characterization of Azotobacter chroococcum from the roots of Zea mays. FEMS Microbiol. Lett. 1985, 31, 197–203. [Google Scholar] [CrossRef]
- Asghar, H.N.; Zahir, Z.A.; Arshad, M.; Khaliq, A. Relationship between in vitro production of auxins by rhizobacteria and their growth-promoting activities in Brassica juncea, L. Biol. Fert. Soils 2002, 35, 231–237. [Google Scholar] [CrossRef]
- Peix, A.; Rivas-Boyero, A.A.; Mateos, P.F.; Rodriguez-Barrueco, C.; Martínez-Molina, E.; Velazquez, E. Growth promotion of chickpea and barley by a phosphate solubilizing strain of Mesorhizobium mediterraneum under growth chamber conditions. Soil Biol. Biochem. 2001, 33, 103–110. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Meth. 2007, 70, 127–131. [Google Scholar] [CrossRef]
- Somasegaran, P.; Hoben, H.J. Handbook for rhizobia: Methods in legume-rhizobium technology. Q. Rev. Biol. 1995, 70, 224–225. [Google Scholar] [CrossRef]
- Jensen, H.L. Nitrogen fixation in leguminous plants. I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc. Linn. Soc. NSW 1941, 67, 98–108. [Google Scholar]
- Ferreira, E.M.; Marques, J.F. Selection of portuguese Rhizobium leguminosarum bv. trifolii strains for production of legume inoculants. Plant Soil 1992, 147, 151–158. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Havas, S.S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–441. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef]
- Hart, S.C.; DeLuca, T.H.; Newman, G.S.; MacKenzie, M.D.; Boyle, S.I. Post-fire vegetative dynamics as drivers of microbial community structure and function in forest soils. For. Ecol. Manag. 2005, 220, 166–184. [Google Scholar] [CrossRef]
- Cobo-Díaz, J.F.; Fernández-González, A.J.; Villadas, P.J.; Toro, N.; Tringe, S.G.; Fernández-López, M. Taxonomic and functional diversity of a Quercus pyrenaica willd rhizospheric microbiome in the Mediterranean mountains. Forests 2017, 8, 390. [Google Scholar] [CrossRef]
- Rodríguez, J.; González-Pérez, J.A.; Turmero, A.; Hernández, M.; Ball, A.S.; González-Vila, F.J.; Arias, M.E. Physico-chemical and microbial perturbations of Andalusian pine forest soils following a wildfire. Sci. Total Environ. 2018, 634, 650–660. [Google Scholar] [CrossRef]
- Esquilín, A.E.J.; Stromberger, M.E.; Massman, W.J.; Frank, J.M.; Shepperd, W.D. Microbial community structure and activity in a Colorado Rocky Mountain forest soil scarred by slash pile burning. Soil. Biol. Biochem. 2007, 39, 1111–1120. [Google Scholar] [CrossRef]
- Kara, Ö.; Bolat, I.; Bolat, I. Short-term effects of wildfire on microbial biomass and abundance in black pine plantation soils in Turkey. Ecol. Indic. 2009, 9, 1151–1155. [Google Scholar] [CrossRef]
- Goberna, M.; García, C.; Insam, H.; Hernández, M.T.; Verdú, M. Burning fire-prone mediterranean shrublands: Immediate changes in soil microbial community structure and ecosystem functions. Microb. Ecol. 2012, 64, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Hamman, S.T.; Burke, I.C.; Stromberger, M.E. Relationships between microbial community structure and soil environmental conditions in a recently burned system. Soil Biol. Biochem. 2007, 39, 1703–1711. [Google Scholar] [CrossRef]
- Mataix-Solera, J.; Guerrero, C.; García-Orenes, F.; Bárcenas, G.M.; Torres, M.P. Forest fire effects on soil microbiology. In Fire Effects on Soils and Restoration Strategies; Cerda, A., Robichaud, P., Eds.; Science Publisher: Oxford, UK, 2009; pp. 133–175. [Google Scholar]
- Dworkin, M. Prokaryotic life cycles. In The Prokaryotes: A Handbook on Theb of Bacteria, Ecophysiological and Biochemical Aspects; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 140–160. [Google Scholar]
- Prendergast-Miller, M.T.; de Menezes, A.B.; Macdonald, L.M.; Peter, T.; Andrew, B.; Geoff, B.; Mark, F.; Alan, E.R.; Tim, W.; Peter, H.T. Wildfire impact: Natural experiment reveals differential short-term changes in soil microbial communities. Soil Biol. Biochem. 2017, 109, 1–13. [Google Scholar] [CrossRef]
- Ferrenberg, S.; O’Neill, S.P.; Knelman, J.E.; Bryan, T.; Sam, D.; Daniel, B.; Taylor, R.; Steven, K.S.; Alan, R.T.; Mark, W.; et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013, 7, 1102–1111. [Google Scholar] [CrossRef]
- Isobe, K.; Otsuka, S.; Sudiana, I.; Nurkanto, A.; Senoo, K. Community composition of soil bacteria nearly a decade after a fire in a tropical rainforest in East Kalimantan, Indonesia. J. Gen. Appl. Microbiol. 2009, 55, 329–337. [Google Scholar] [CrossRef]
- Pajarillo, E.A.B.; Chae, J.P.; Balolong, M.P.; Kim, H.B.; Kang, D.K. Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J. Gen. Appl. Microbiol. 2011, 60, 140–146. [Google Scholar] [CrossRef]
- Springer, A.; Fichtel, C.; Al-Ghalith, G.A.; Koch, F.; Amato, K.R.; Clayton, J.B.; Knights, D.; Kappeler, P.M. Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild verreaux’s sifakas (Propithecus verreauxi). Ecol. Evol. 2017, 1–14. [Google Scholar] [CrossRef]
- Szekely, B.A.; Singh, J.; Marsh, T.L.; Hagedorn, C.; Were, S.R.; Kaur, T. Fecal bacterial diversity of human-habituated wild chimpanzees (pan troglodytes schweinfurthii) at Mahale Mountains National Park, western Tanzania. Am. J. Primatol. 2010, 72, 566–574. [Google Scholar] [CrossRef]
- Delgado, M.L.; Singh, P.; Funk, J.A.; Moore, J.A.; Cannell, E.M.; Kanesfsky, J.; Manning, S.D.; Scribner, K.T. Intestinal microbial community dynamics of white-tailed deer (Odocoileus virginianus) in an agroecosystem. Microb. Ecol. 2017, 74, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Situmorang, E.C.; Nugroho, Y.A.; Prameswara, A.; Andarini, E.; Hartono, S.R.H.; Toruan-Mathius, N.; Liwang, T. The bacterial diversity investigation in oil palm plantation using terminal restriction length polymorphism. AIP Conf. Proc. 2016, 1744, 020017. [Google Scholar] [CrossRef]
- Pearce, D.; Newsham, K.; Thorne, M.; Calvo-Bado, L.; Krsek, M.; Laskaris, P.; Hodson, A.; Wellington, E. Metagenomic analysis of a Southern maritime antarctic soil. Front. Microbiol. 2012, 3, 403. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Fischer-Le Saux, M.; Zakhia, F. Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassic. Int. J. Syst. Evol. Microbiol. 2006, 56, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.P.; Linardopoulou, E.V.; Samadpour, M. Transfer of 13 species of the genus Burkholderia to the genus Caballeronia and reclassification of Burkholderia jirisanensis as Paraburkholderia jirisanensis comb. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 3846–3853. [Google Scholar] [CrossRef]
- Zuleta, L.F.G.; Cunha, C.D.O.; de Carvalho, F.M. The complete genome of Burkholderia phenoliruptrix strain BR3459a, a symbiont of Mimosa flocculosa: Highlighting the coexistence of symbiotic and pathogenic genes. BMC Genom. 2014, 15, 1–19. [Google Scholar] [CrossRef]
- Castanheira, N.L.; Dourado, A.C.; Pais, I.; José, S.; Paula, S.-C.; Nuno, B.; Gilda, C.; Maria, T.B.C.; Paula, F. Colonization and beneficial effects on annual ryegrass by mixed inoculation with plant growth promoting bacteria. Microbiol. Res. 2017, 198, 47–55. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and genetic diversity within the Pseudomonas fluoresces complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef]
- IIzuku, H.; Komagata, K. New species of pseudomonas belonged to fluorescent group (studies on the microorganisms of cereal grains. Part V). Nippon Nōgeikagaku Kaishi 1963, 37, 137–141. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans.; animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Hashem, A.; Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M.; Kobayashi, H.; Davies, B.W.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The sinorhizobium–medicago model. Nat. Rev. Genet. 2007, 5, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, N.; Dourado, A.C.; Kruz, S.; Alves, P.I.L.; Delgado-Rodríguez, A.I.; Pais, I.; Semedo, J.; Scotti-Campos, P.; Sánchez, C.; Borges, N.; et al. Plant growth-promoting Burkholderia species isolated from annual ryegrass in Portuguese soils. J. Appl. Microbiol. 2016, 120, 724–739. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant. Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Duijff, B.J.; Recorbet, G.; Bakker, P.A.H.M.; Loper, J.E.; Lemanceau, P. Microbial antagonism at the root level is involved in the suppression of Fusarium wilt by the combination of nonpathogenic Fusarium oxysporum Fo47 and Pseudomonas putida WCS358. Phytopathology 1999, 89, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Pule-Meulenberg, F.; Belane, A.K.; Krasova-Wade, T.; Dakora, F.D. Symbiotic functioning and bradyrhizobial biodiversity of cowpea (Vigna unguiculata L. Walp.) in Africa. BMC Microbiol. 2010, 10, 89. [Google Scholar] [CrossRef]
- Leite, J.; Fischer, D.; Rouws, L.F.M.; Júnior, P.I.F.; Hofmann, A.; Kublik, S.; Schloter, M.; Xavier, G.R.; Radl, V. Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front. Plant Sci. 2017, 7. [Google Scholar] [CrossRef]
- Ndungu, S.M.; Messmer, M.M.; Ziegler, D.; Gamper, H.A.; Mészáros, É.; Thuita, M.; Vanlauwe, B.; Frossard, E.; Thonar, C. Cowpea (Vigna unguiculata L. Walp) hosts several widespread bradyrhizobial root nodule symbionts across contrasting agro-ecological production areas in Kenya. Agric. Ecosys. Environ. 2018, 261, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Jordaan, A.; Du Plessis, H.; Wessels, D. Roots of Colophospermum mopane. Are they infected by rhizobia? S. Afr. J. Bot. 2000, 66, 128–130. [Google Scholar] [CrossRef]
- Teixeira, H.; Rodriguez-Echeverria, S. Identification of symbiotic nitrogen-fixing bacteria from three African leguminous trees in Gorongosa National Park. Syst. App. Microbiol. 2016, 39, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.H.; Ten, L.N.; Im, W.T. Cohnella panacarvi sp. nov, a xylanolytic bacterium isolated from ginseng cultivating soil. J. Microbiol. Biotech. 2007, 17, 913–918. [Google Scholar]
- Hameed, A.; Hung, M.H.; Lin, S.Y. Cohnella formosensis sp. nov., a xylanolytic bacterium isolated from the rhizosphere of Medicago sativa L. Int. J Syst. Evol. Microbiol. 2013, 6, 2806–2812. [Google Scholar] [CrossRef]
- García-Fraile, P.; Velázquez, E.; Mateos, P.F.; Martínez-Molina, E.; Rivas, R. Cohnella phaseoli sp. nov., isolated from root nodules of Phaseolus coccineus in Spain, and emended description of the genus Cohnella. Int. J. Syst. Evol. Microbiol. 2008, 58, 1855–1859. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.S.; Zhao, L.F.; Kong, Z.Y.; Yang, W.Q.; Lindström, K.; Wang, E.T.; Wei, G.H. Diversity of endophytic bacteria within nodules of the Sphaerophysa salsula in different regions of Loess Plateau in China. FEMS Microbiol. Ecol. 2011, 76, 463–475. [Google Scholar] [CrossRef]
- Kobayashi, D.Y.; Palumbo, J.D. Bacterial endophytes and their effects on plants and uses in agriculture. In Microbial Endophytes; Bacon, C.W., White, J.F., Eds.; Marcel Dekker: New York, NY, USA, 2000; pp. 199–233. [Google Scholar]
- Teng, Y.; Feng, S.; Ren, W.; Zhu, L.; Ma, W.; Christie, P.; Luo, L. Phytoremediation of diphenylarsinic-acid-contaminated soil by Pteris vittata associated with Phyllobacterium myrsinacearum RC6b. Int. J. Phytoremed. 2017, 19, 463–469. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Luo, Y.; Freitas, H. Phytoextraction of heavy metal polluted soils using sedum plumbizincicola inoculated with metal mobilizing Phyllobacterium myrsinacearum RC6b. Chemosphere 2013, 93, 1386–1392. [Google Scholar] [CrossRef]
- Coenye, T.; Henry, D.; Speert, D.P.; Vandamme, P. Burkholderia phenoliruptrix sp. nov. to accommodate the 2,4,5-trichlorophenoxyacetic acid and halophenol-degrading strain AC1100. Syst. Appl. Microbiol. 2004, 27, 623–627. [Google Scholar] [CrossRef]
- Meng, L.; Li, W.; Bao, M.; Sun, P. Great correlation: Biodegradation and chemotactic adsorption of Pseudomonas synxantha LSH-7’ for oil contaminated seawater bioremediation. Water Res. 2019, 153, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Shreya, D.; Jinal, H.N.; Kartik, V.P.; Amaresan, N. Amelioration effect of chromium-tolerant bacteria on growth, physiological properties and chromium mobilization in chickpea (Cicer arietinum) under chromium stress. Arch. Microbiol. 2020, 202, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wang, Y.; Lin, S.; Deng, Z.; Liang, R. Characterization of an efficient estrogen-degrading bacterium Stenotrophomonas maltophilia SJTH1 in saline-, alkaline-, heavy metal-contained environments or solid soil and identification of four 17β-estradiol-oxidizing dehydrogenases. J. Hazard. Mater. 2020, 385, 121616. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Chimni, S.S.; Saini, H.S.; Chadha, B.S. Pseudomonas gessardii growing cells as a new biocatalyst for asymmetric synthesis of α-bromohydrins. Biocatal. Agric. Biotechnol. 2015, 4, 49–54. [Google Scholar] [CrossRef]
- Ramani, K.; Chockalingam, E.; Sekaran, G. Production of a novel extracellular acidic lipase from Pseudomonas gessardii using slaughterhouse waste as a substrate. J. Ind. Microbiol. Biotechnol. 2010, 37, 531–535. [Google Scholar] [CrossRef]
- Ourique, L.J.; Rocha, C.C.; Gomes, R.C.D.; Rossi, D.M.; Ayub, M.A.Z. Bioreactor production of 2,3-butanediol by Pantoea agglomerans using soybean hull acid hydrolysate as substrate. Bioprocess Biosyst. Eng. 2020. [Google Scholar] [CrossRef]
- Phipps, K.R.; Sulaiman, C.; Simon, R.; Holalagoudar, S.; Kohchi, C.; Nakata, Y. Subchronic (90-day) toxicity assessment of Somacy-FP100, a lipopolysaccharide-containing fermented wheat flour extract from Pantoea agglomerans. J. Appl. Toxicol. 2020, 1–11. [Google Scholar] [CrossRef]
- Williams, A.N.; Stavrinides, J. Pantoea natural product 3 is encoded by an eight-gene biosynthetic gene cluster and exhibits antimicrobial activity against multi-drug resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Microbiol. Res. 2020, 234, 126412. [Google Scholar] [CrossRef]
- Ge, S.; Ma, J.; Liu, L.; Yuan, Z. The impact of exogenous aerobic bacteria on sustainable methane production associated with municipal solid waste biodegradation: Revealed by high-throughput sequencing. Sustainability 2020, 12, 1815. [Google Scholar] [CrossRef]
- Abdelrazek, N.A.; Elkhatib, W.F.; Raafat, M.M.; Aboulwafa, M.M. Production, characterization and bioinformatics analysis of L-asparaginase from a new Stenotrophomonas maltophilia EMCC2297 soil isolate. AMB Express 2020, 10, 71. [Google Scholar] [CrossRef]
- Rustamova, N.; Wubulikasimu, A.; Mukhamedov, N.; Gao, Y.; Egamberdieva, D.; Yili, A. Endophytic bacteria associated with medicinal plant Vernonia anthelmintica: Diversity and characterization. Curr. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]


| Landscape | Soil Type | Fire Frequency | Last Fire | Code | Latitude | Longitude |
|---|---|---|---|---|---|---|
| C | Calcareous | High | 1 year | CH | −23.6139 | 32.0242 |
| 1 year | −23.3311 | 31.7095 | ||||
| Low | 7 years | CL | −23.3823 | 31.7697 | ||
| 10 years | −23.6592 | 32.0485 | ||||
| LN | Rocky | High | 1 year | LNH | −23.8177 | 31.7558 |
| 1 year | −23.7192 | 31.7514 | ||||
| Low | 10 years | LNL | −23.6795 | 31.7329 | ||
| 10 years | −23.7592 | 31.7548 | ||||
| NS | Sandy | High | 1 year | NSH | −23.6321 | 32.1484 |
| 1 year | −23.5752 | 32.0895 | ||||
| Low | 10 years | NSL | −23.5173 | 32.2516 | ||
| 10 years | −23.5285 | 32.2213 |
| Landscape | Soil Type | Fire Frequency | OTU Number | Dominance D | Shannon H | Equitability J | Berger-Parker |
|---|---|---|---|---|---|---|---|
| C | Calcareous | Low | 767 | 0.04023 | 4.085 | 0.6150 | 0.1215 |
| High | 831 | 0.04116 | 4.078 | 0.6066 | 0.1275 | ||
| LN | Rocky | Low | 744 | 0.04109 | 4.067 | 0.6150 | 0.1230 |
| High | 737 | 0.04004 | 4.087 | 0.6190 | 0.1192 | ||
| NS | Sandy | Low | 730 | 0.04157 | 4.065 | 0.6165 | 0.1256 |
| High | 696 | 0.04282 | 4.040 | 0.6172 | 0.1292 | ||
| Between Landscapes | H = 4.571 P = 0.102 | H = 3.714 P = 0.156 | H = 3.429 P = 0.180 | H = 3.271 P = 0.257 | H = 2.571 P = 0.276 | ||
| Between fire regimes | U = 4.005 P = 0.827 | U = 4.000 P = 0.810 | U = 4.550 P = 0.850 | U = 3.005 P = 0.617 | U = 3.000 P = 0.613 |
| Landscape | Soil Type | Fire Frequency | Isolate | Most Related Source Organism | Phylum | GenBank Accession Number | Identity |
|---|---|---|---|---|---|---|---|
| C | Calcareous | High | CH2 | Bacillus sp. strain QW2 | Firmicutes | MT065751.1 | 100.0% |
| CH3 | Pantoea agglomerans strain DSM 3493 | Proteobacteria | KY013009.1 | 98.7% | |||
| CH4 | Phyllobacterium myrsinacearum strain NBRC 100019 | Proteobacteria | NR_113874.1 | 97.3% | |||
| Low | CL1 | Bacillus sp. strain PJA1.5 | Firmicutes | MT275726.1 | 100.0% | ||
| CL2 | Caballeronia concitans strain LMG 29315 | Proteobacteria | NR_145603.1 | 98.7% | |||
| CL3 | Bacillus sp. strain R43 | Firmicutes | MT254891.1 | 100.0% | |||
| LN | Rocky | High | LNH1 | Pseudomonas azotoformans strain LMG 21611 | Proteobacteria | LT629702.1 | 99.9% |
| LNH3 | Pantoea agglomerans strain DSM 3493 | Proteobacteria | MF289172.1 | 99.8% | |||
| Low | LNL1 | Pantoea agglomerans strain KB38 | Proteobacteria | JF327464.1 | 97.9% | ||
| LNL2 | Pseudomonas azotoformans strain BG4 | Proteobacteria | MK875666.1 | 99.9% | |||
| LNL3 | Pseudomonas synxantha strain HNR22 | Proteobacteria | EU373390.1 | 98.9% | |||
| LNL4 | Pseudomonas azotoformans strain BG4 | Proteobacteria | MK875666.1 | 99.6% | |||
| LNL5 | Pseudomonas gessardii strain 4G497 | Proteobacteria | MG972901.1 | 99.8% | |||
| NS | Sandy | High | NSH1 | Stenotrophomonas maltophilia strain PEG-305 | Proteobacteria | CP040437.1 | 100% |
| NSH2 | Stenotrophomonas maltophilia strain PEG-305 | Proteobacteria | CP040437.1 | 100% | |||
| Low | NSL2 | Paraburkholderia phenoliruptrix strain AC1100 | Proteobacteria | NR_042901.1 | 100.0% |
| Landscape | Soil Type | Fire Frequency | Isolate | Most Related Source Organism | Growth in N-Free Media a | IAA Production b | Phosphate Solubilization c | Siderophore Production d | |
|---|---|---|---|---|---|---|---|---|---|
| Aerobic | Microaerophilic | (µg mL−1) | |||||||
| C | Calcareous | High | CH2 | Bacillus sp. strain QW2 | + | − | 10.0 ± 0.1 | + | − |
| CH3 | Pantoea agglomerans strain DSM 3493 | + | − | <5.0 | − | − | |||
| CH4 | Phyllobacterium myrsinacearum strain NBRC 100019 | − | − | <5.0 | − | − | |||
| Low | CL1 | Bacillus sp. strain PJA1.5 | − | − | 6.6 ± 0.9 | − | + | ||
| CL2 | Caballeronia concitans strain LMG 29315 | + | − | <5.0 | − | − | |||
| CL3 | Bacillus sp. strain R43 | + | − | 9.2 ± 0.1 | + | − | |||
| LN | Rocky | High | LNH1 | Pseudomonas azotoformans strain LMG 21611 | − | − | 5.0 ± 0.3 | + | + |
| LNH3 | Pantoea agglomerans strain DSM 3493 | + | − | 29.6 ± 0.5 | + | + | |||
| Low | LNL1 | Pantoea agglomerans strain KB38 | + | − | 5.0 ± 0.1 | − | + | ||
| LNL2 | Pseudomonas azotoformans strain BG4 | + | − | <5.0 | − | + | |||
| LNL3 | Pseudomonas synxantha strain HNR22 | + | − | <5.0 | + | + | |||
| LNL4 | Pseudomonas azotoformans strain BG4 | + | + | <5.0 | − | + | |||
| LNL5 | Pseudomonas gessardii strain 4G497 | + | − | <5.0 | − | + | |||
| NS | Sandy | High | NSH1 | Stenotrophomonas maltophilia strain PEG-305 | − | + | <5.0 | − | − |
| NSH2 | Stenotrophomonas maltophilia strain PEG-305 | − | + | <5.0 | − | − | |||
| Low | NSL2 | Paraburkholderia phenoliruptrix strain AC1100 | − | − | <5.0 | − | + | ||
| Landscape | Soil Type | Fire Frequency | Isolate | Most Related 16S rRNA Gene Sequence (s) | Phylum | GenBank Accession Number | % Identity |
|---|---|---|---|---|---|---|---|
| C | Calcareous | High | CH1.1 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 100.0% |
| CH1.2 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 99.85% | |||
| CH1.3 | Pantoea agglomerans strain CFSAN 047153 | Proteobacteria | CP034469.1 | 99.86% | |||
| CH1.4 | Pantoea agglomerans strain UAEU 18 | Proteobacteria | CP048033.1 | 98.94% | |||
| CH1.5 | Pantoea agglomerans strain CFSAN 047154 | Proteobacteria | CP034474.1 | 99.79% | |||
| Low | CL2.2 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 100.0% | ||
| CL2.3 | Bradyrhizobium sp. strain C-145 | Proteobacteria | CP022221.1 | 99.85% | |||
| CL2.4 | Pantoea agglomerans strain UAEU 18 | Proteobacteria | CP048033.1 | 100.0% | |||
| CL2.5 | Pantoea agglomerans strain C410P1 | Proteobacteria | CP016889.1 | 99.44% | |||
| CL2.6 | Bradyrhizobium sp. ORS 3257 isolate ORS3257 | Proteobacteria | LS398110.1 | 100.0% | |||
| LB | Rocky | High | LNH5.1 | Pantoea agglomerans strain C410P1 | Proteobacteria | CP016889.1 | 99.02% |
| LNH5.2 | Azospirillum zeae strain N7 | Proteobacteria | NR_043934.1 | 98.28% | |||
| LNH5.3 | Bradyrhizobium sp. strain C-145 | Proteobacteria | CP022221.1 | 100% | |||
| LNH5.5 | Rhizobium sp. strain 11515TR | Proteobacteria | MK791683.1 | 99.49% | |||
| LNH5.6 | Pseudomonas nitroreducens strain HJ-3 | Proteobacteria | MH324395.1 | 99.94% | |||
| Low | LNL3.2 | Cohnella rhizosphaerae strain 18JY42-3 | Firmicutes | MH497628.1 | 97.38% | ||
| LNL3.3 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 99.93% | |||
| LNL3.4 | Pantoea agglomerans strain CFSAN 047,153 | Proteobacteria | CP034469.1 | 99.33% | |||
| LNL3.5 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 97.37% | |||
| LNL3.6 | Pantoea agglomerans CFSAN 047153 | Proteobacteria | CP034469.1 | 99.59% | |||
| NS | Sandy | High | NSH9.1 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 99.91% |
| NSH9.3 | Bradyrhizobium sp. strain TUTMGGH52 | Proteobacteria | CP030053.1 | 99.58% | |||
| NSH9.4 | Bradyrhizobium sp. strain C-145 | Proteobacteria | MT229310.1 | 99.72% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maquia, I.S.; Fareleira, P.; Videira e Castro, I.; Brito, D.R.A.; Soares, R.; Chaúque, A.; Ferreira-Pinto, M.M.; Lumini, E.; Berruti, A.; Ribeiro, N.S.; et al. Mining the Microbiome of Key Species from African Savanna Woodlands: Potential for Soil Health Improvement and Plant Growth Promotion. Microorganisms 2020, 8, 1291. https://doi.org/10.3390/microorganisms8091291
Maquia IS, Fareleira P, Videira e Castro I, Brito DRA, Soares R, Chaúque A, Ferreira-Pinto MM, Lumini E, Berruti A, Ribeiro NS, et al. Mining the Microbiome of Key Species from African Savanna Woodlands: Potential for Soil Health Improvement and Plant Growth Promotion. Microorganisms. 2020; 8(9):1291. https://doi.org/10.3390/microorganisms8091291
Chicago/Turabian StyleMaquia, Ivete Sandra, Paula Fareleira, Isabel Videira e Castro, Denise R. A. Brito, Ricardo Soares, Aniceto Chaúque, M. Manuela Ferreira-Pinto, Erica Lumini, Andrea Berruti, Natasha S. Ribeiro, and et al. 2020. "Mining the Microbiome of Key Species from African Savanna Woodlands: Potential for Soil Health Improvement and Plant Growth Promotion" Microorganisms 8, no. 9: 1291. https://doi.org/10.3390/microorganisms8091291
APA StyleMaquia, I. S., Fareleira, P., Videira e Castro, I., Brito, D. R. A., Soares, R., Chaúque, A., Ferreira-Pinto, M. M., Lumini, E., Berruti, A., Ribeiro, N. S., Marques, I., & Ribeiro-Barros, A. I. (2020). Mining the Microbiome of Key Species from African Savanna Woodlands: Potential for Soil Health Improvement and Plant Growth Promotion. Microorganisms, 8(9), 1291. https://doi.org/10.3390/microorganisms8091291









