Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Isolates and Culture Conditions
2.2. Plant Material and Experiment Design
2.3. Aphanomyces euteiches Root Infection
2.4. Assessment of Plant Growth
2.5. Molecular Identification of Trichoderma Species by Barcoding
2.6. Bioinformatic Analysis
2.7. In Vitro Biocontrol Assay
2.8. Statistical Analysis
3. Results
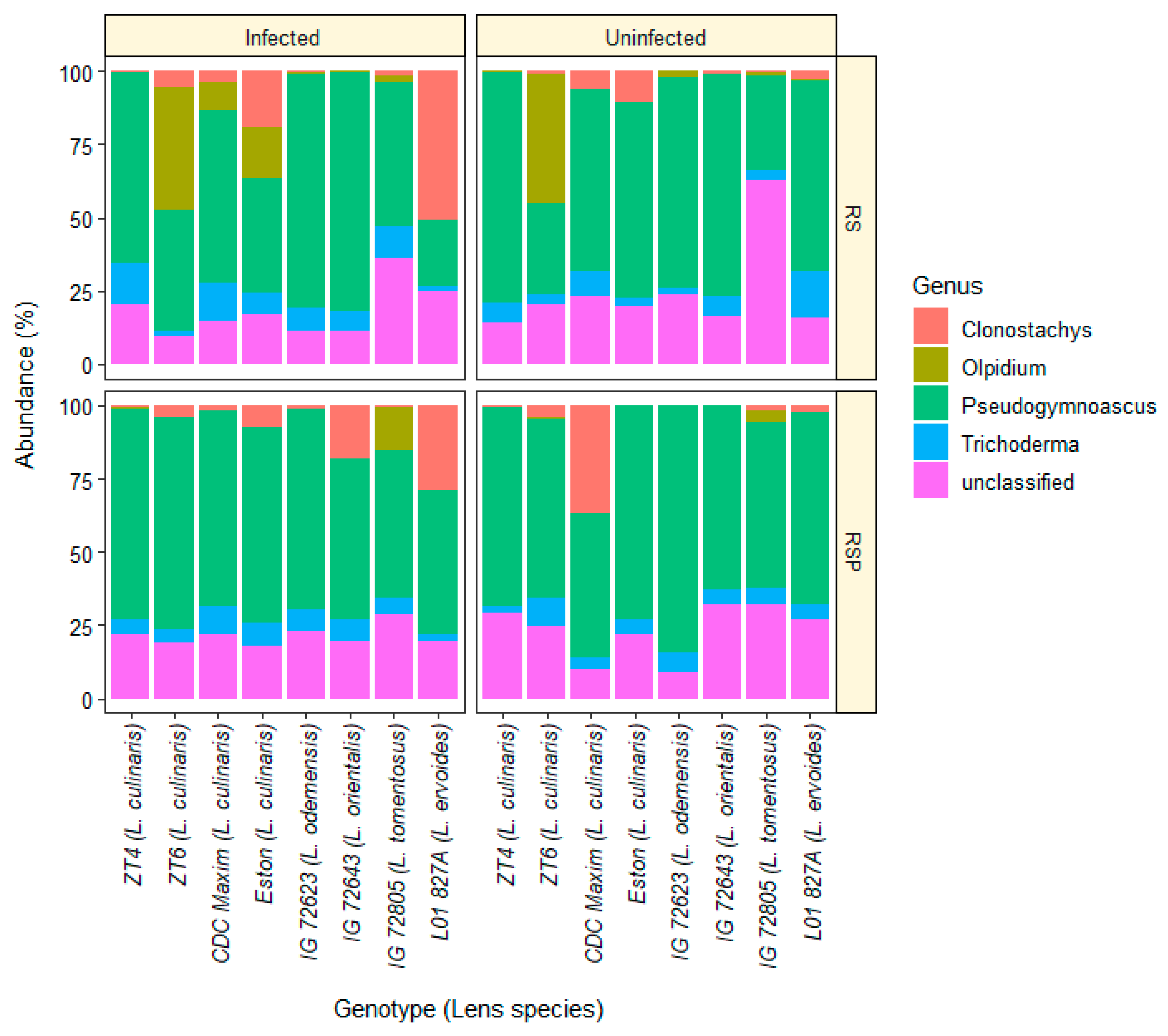
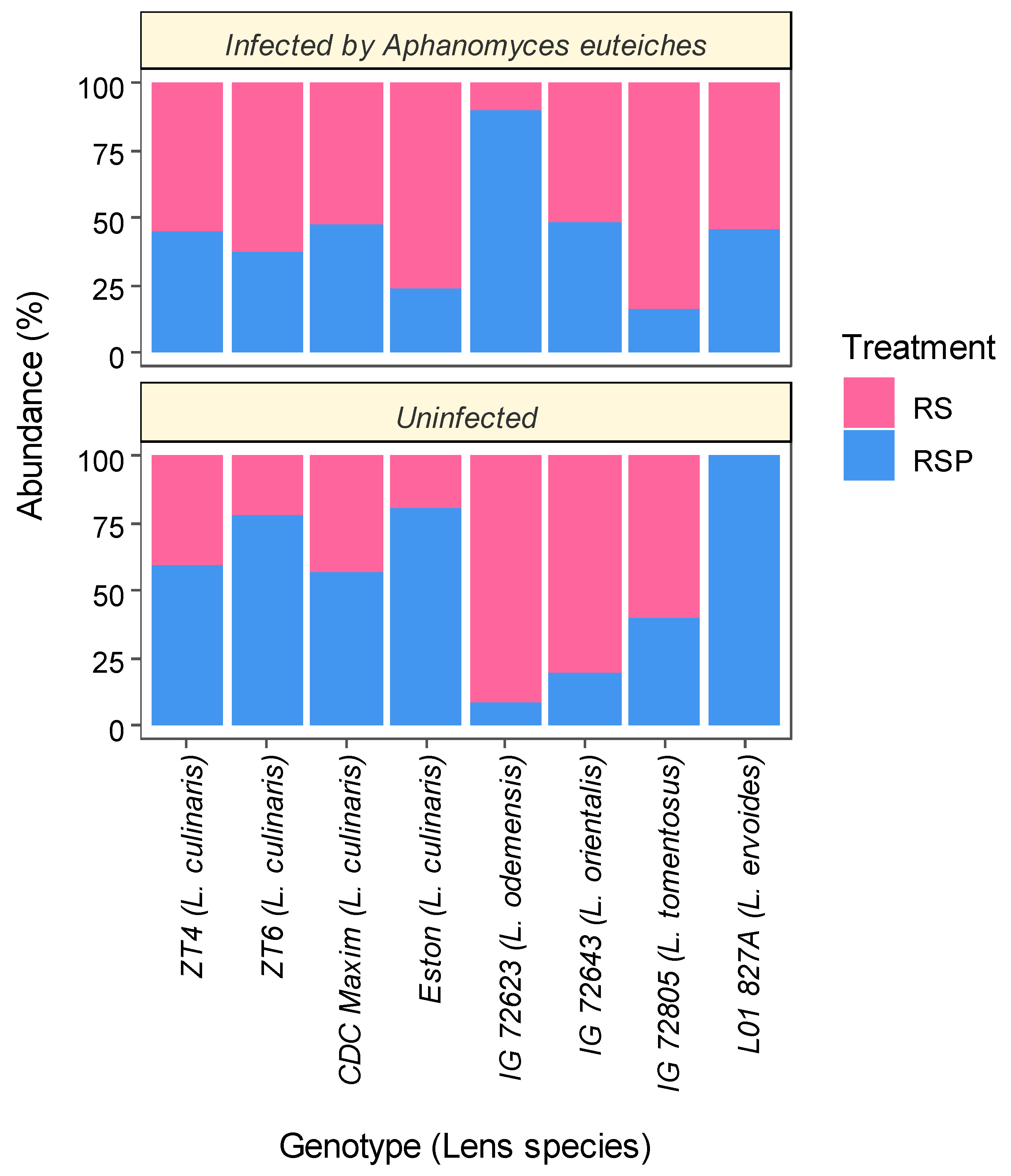
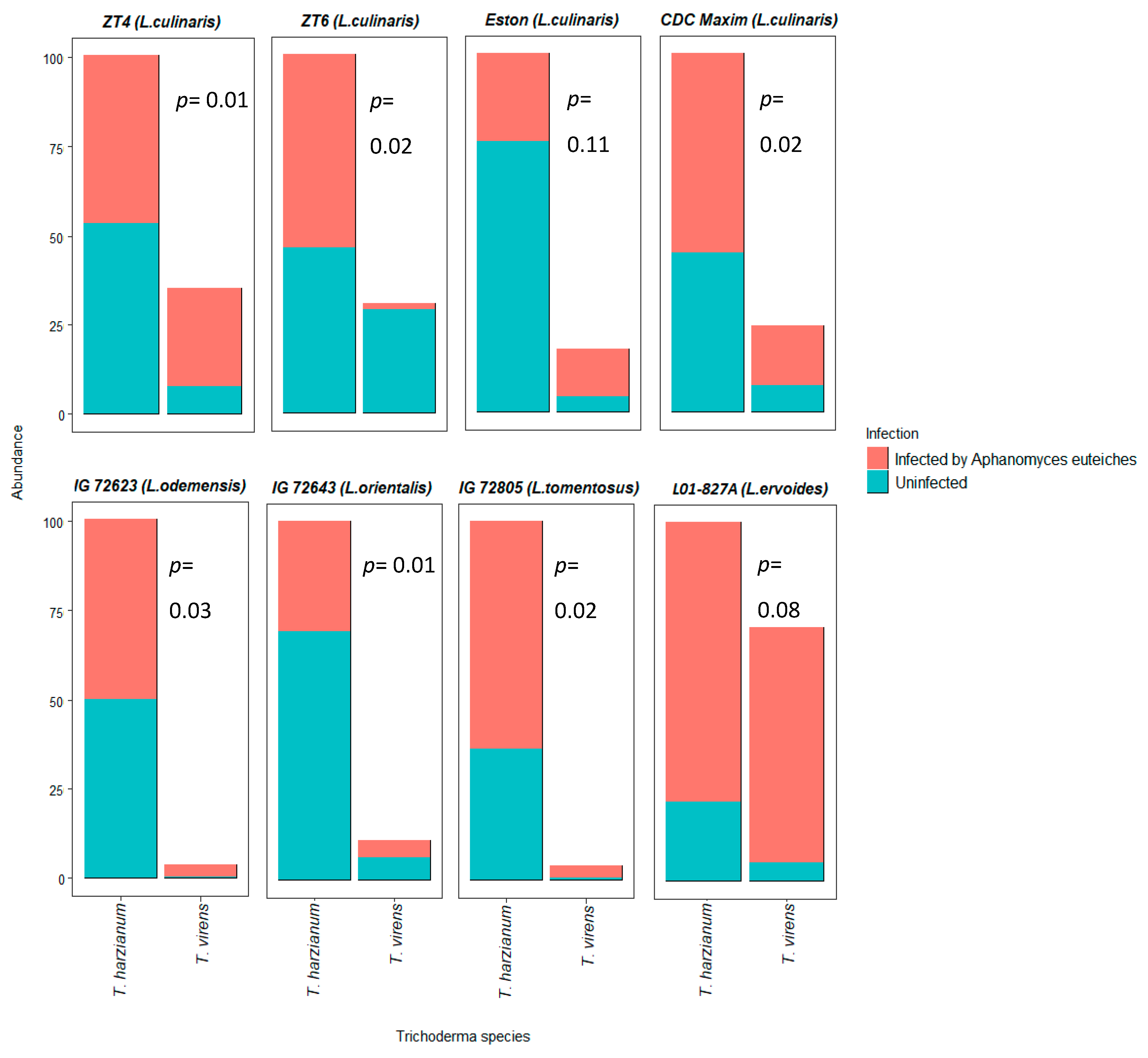
3.1. Composition of Fungal Communites and Trichoderma Abundance in the Roots
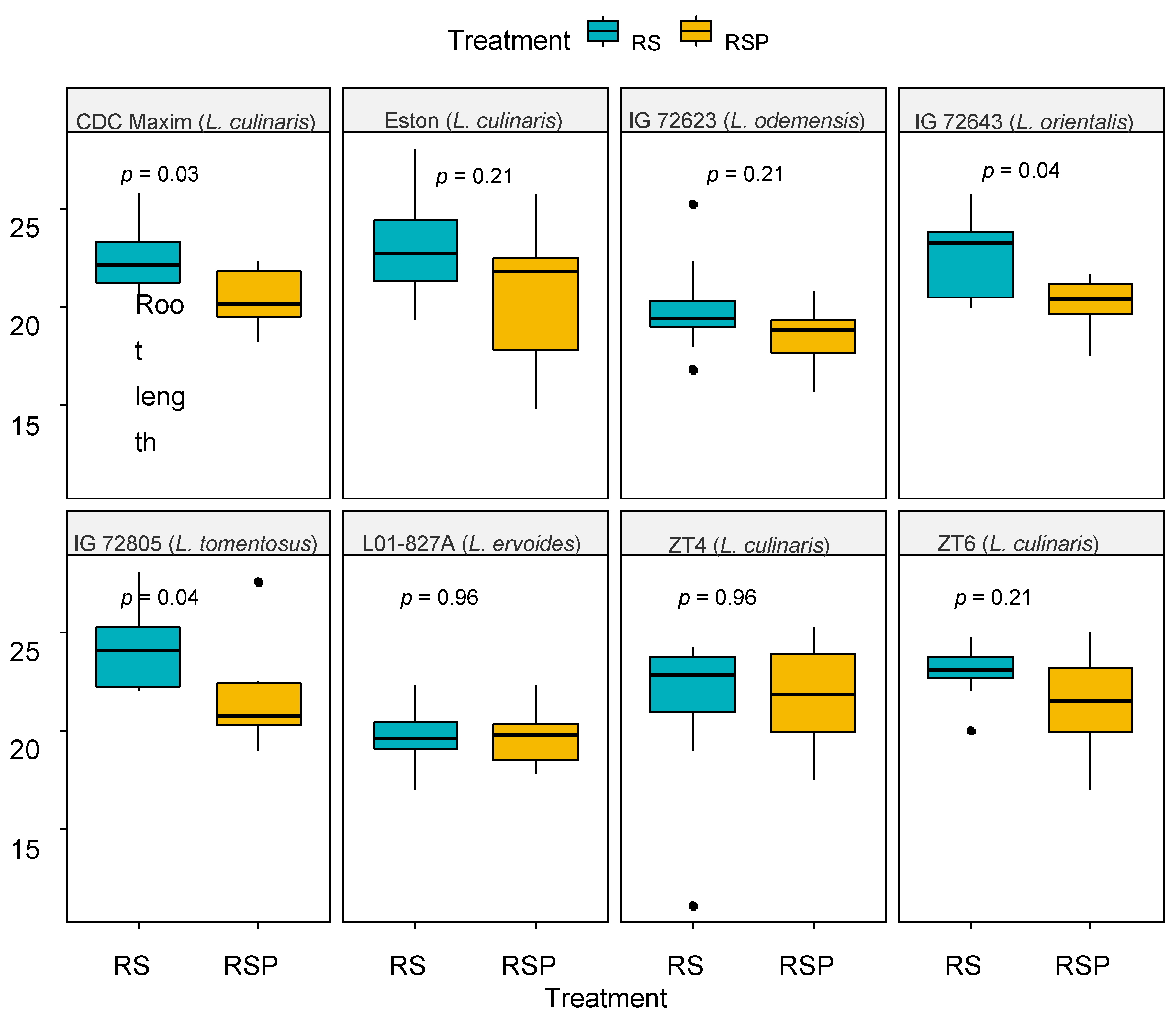
3.2. Relationship between Relative Abundance of Trichoderma spp. and Plant Growth
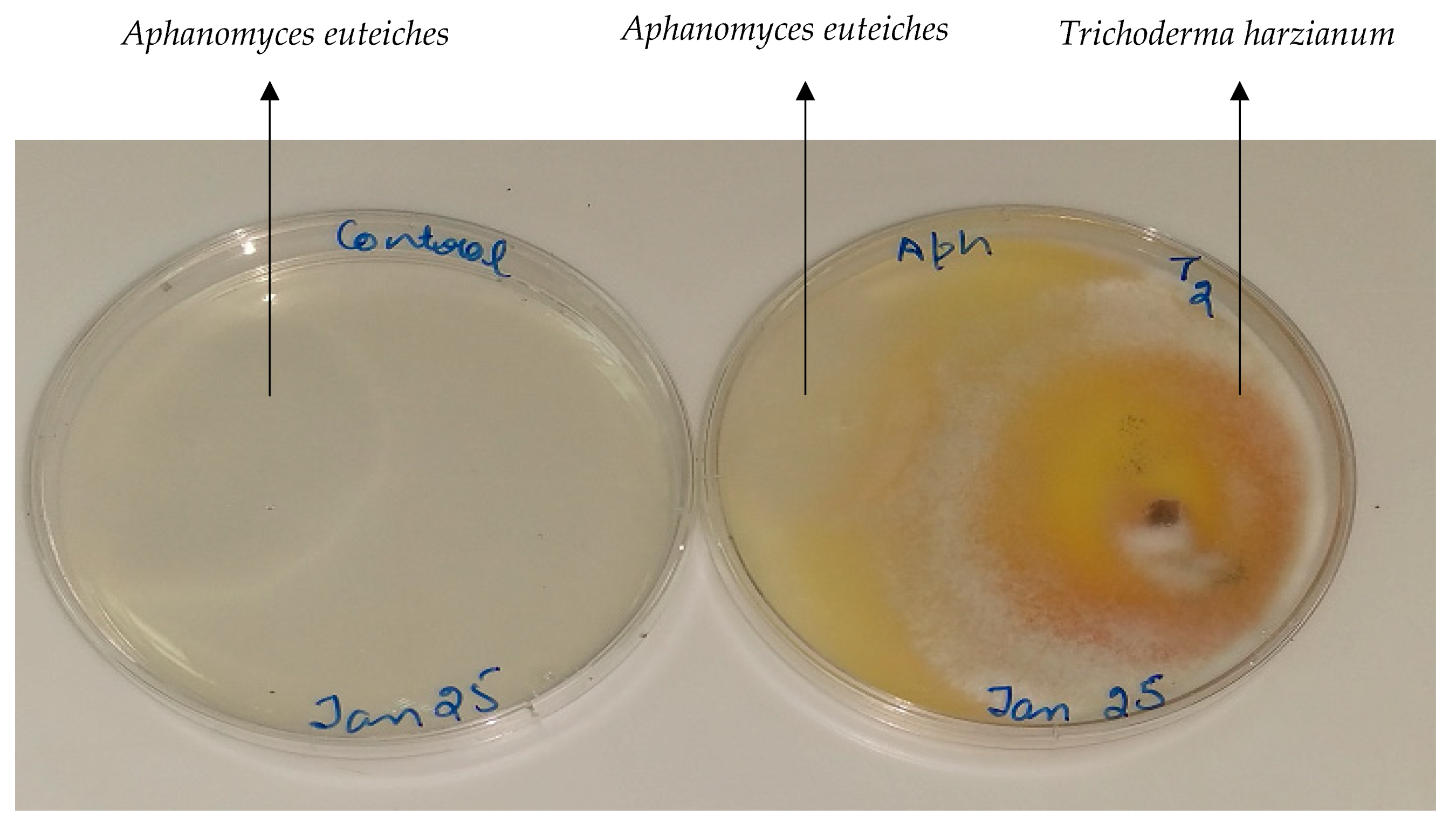
3.3. In Vitro Biocontrol Plate Assay
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barrow, J.R.; Lucero, M.E.; Reyes-Vera, I.; Havstad, K.M. Do symbiotic microbes have a role in regulating plant performance and response to stress? Commun. Integr. Biol. 2008, 1, 69–73. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 1–19. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.R.J. Ecology and evolution of plant mating. Trends Ecol. Evol. 1996, 11, 73–79. [Google Scholar]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Kemen, E. Microbe-microbe interactions determine oomycete and fungal host colonization. Curr. Opin. Plant Biol. 2014, 20, 75–81. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- van der Heijden, M.G.A.; Hartmann, M. Networking in the Plant Microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef]
- Sessitsch, A.; Pfaffenbichler, N.; Mitter, B. Microbiome Applications from Lab to Field: Facing Complexity. Trends Plant Sci. 2019, 24, 194–198. [Google Scholar] [CrossRef]
- Clúa, J.; Roda, C.; Zanetti, M.E.; Blanco, F.A. Compatibility between legumes and rhizobia for the establishment of a successful nitrogen-fixing symbiosis. Genes 2018, 9, 125. [Google Scholar]
- Granada, C.E.; Strochein, M.; Vargas, L.K.; Bruxel, M.; Saccol de Sá, E.L.; Passaglia, L.M.P. Genetic diversity and symbiotic compatibility among rhizobial strains and Desmodium incanumand Lotus spp. plants. Genet. Mol. Biol. 2014, 37, 396–405. [Google Scholar] [CrossRef]
- Bazghaleh, N.; Hamel, C.; Gan, Y.; Tar’an, B.; Knight, J.D. Genotype-specific variation in the structure of root fungal communities is related to chickpea plant productivity. Appl. Environ. Microbiol. 2015, 81, 2368–2377. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Inceoǧlu, Ö.; Salles, J.F.; Van Overbeek, L.; Van Elsas, J.D. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 2010, 76, 3675–3684. [Google Scholar] [CrossRef]
- Baetz, U.; Martinoia, E. Root exudates: The hidden part of plant defense. Trends Plant Sci. 2014, 19, 90–98. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef]
- Bazghaleh, N.; Hamel, C.; Gan, Y.; Knight, J.D.D.; Vujanovic, V.; Cruz, A.F.A.F.; Ishii, T. Phytochemicals induced in chickpea roots selectively and non-selectively stimulate and suppress fungal endophytes and pathogens. Plant Soil 2016, 409, 479–493. [Google Scholar] [CrossRef]
- de Werra, P.; Huser, A.; Tabacchi, R.; Keel, C.; Maurhofer, M. Plant-and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl. Environ. Microbiol. 2011, 77, 2807–2812. [Google Scholar] [CrossRef]
- Lombardi, N.; Vitale, S.; Turr, À.D.; Reverberi, M.; Fanelli, C.; Vinale, F.; Marra, R.; Ruocco, M.; Pascale, A.; D’Errico, G.; et al. Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol. Plant-Microbe Interact. 2018, 31, 982–994. [Google Scholar] [CrossRef]
- Savary, S.; Ficke, A.; Aubertot, J.N.; Hollier, C. Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Lobell, D.B.; Field, C.B. Global scale climate-crop yield relationships and the impacts of recent warming. Environ. Res. Lett. 2007, 2, 014002. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; Bataillon, T. A Population Genomics Perspective on the Emergence and Adaptation of New Plant Pathogens in Agro-Ecosystems. PLoS Pathog. 2012, 8, e1002893. [Google Scholar] [CrossRef]
- Fox, C.A.; MacDonald, K.B. Challenges related to soil biodiversity research in agroecosystems - Issues within the context of scale of observation. Can. J. Soil Sci. 2003, 83, 231–244. [Google Scholar] [CrossRef]
- Roy, F.; Boye, J.I.; Simpson, B.K. Bioactive proteins and peptides in pulse crops: Pea, chickpea and lentil. Food Res. Int. 2010, 43, 432–442. [Google Scholar] [CrossRef]
- Borrell, A.N.; Shi, Y.; Gan, Y.; Bainard, L.D.; Germida, J.J.; Hamel, C. Fungal diversity associated with pulses and its influence on the subsequent wheat crop in the Canadian prairies. Plant Soil 2017, 414, 13–31. [Google Scholar] [CrossRef]
- Karppinen, E.M.; Payment, J.; Chatterton, S.; Bainard, J.D.; Hubbard, M.; Gan, Y.; Bainard, L.D. Distribution and abundance of Aphanomyces euteiches in agricultural soils: Effect of land use type, soil properties, and crop management practices. Appl. Soil Ecol. 2020, 150, 103470. [Google Scholar] [CrossRef]
- Chatterton, S.; Harding, M.W.; Bowness, R.; Mclaren, D.L.; Banniza, S.; Gossen, B.D. Importance and causal agents of root rot on field pea and lentil on the Canadian prairies, 2014–2017. Can. J. Plant Pathol. 2019, 41, 98–114. [Google Scholar] [CrossRef]
- Wu, L.; Chang, K.F.; Conner, R.L.; Strelkov, S.; Fredua-Agyeman, R.; Hwang, S.F.; Feindel, D. Aphanomyces euteiches: A Threat to Canadian Field Pea Production. Engineering 2018, 4, 542–551. [Google Scholar] [CrossRef]
- Conner, R.L.; Chang, K.F.; Hwang, S.F.; Warkentin, T.D.; McRae, K.B. Assessment of tolerance for reducing yield losses in field pea caused by Aphanomyces root rot. Can. J. Plant Sci. 2013, 93, 473–482. [Google Scholar] [CrossRef]
- Wicker, E.; Hullé, M.; Rouxel, F. Pathogenic characteristics of isolates of Aphanomyces euteiches from pea in France. Plant Pathol. 2001, 50, 433–442. [Google Scholar] [CrossRef]
- Wu, L.; Chang, K.F.; Hwang, S.F.; Conner, R.; Fredua-Agyeman, R.; Feindel, D.; Strelkov, S.E. Evaluation of host resistance and fungicide application as tools for the management of root rot of field pea caused by Aphanomyces euteiches. Crop J. 2019, 7, 38–48. [Google Scholar] [CrossRef]
- Moussart, A.; Even, M.N.; Lesné, A.; Tivoli, B. Successive legumes tested in a greenhouse crop rotation experiment modify the inoculum potential of soils naturally infested by Aphanomyces euteiches. Plant Pathol. 2013, 62, 545–551. [Google Scholar] [CrossRef]
- Woo, S.L.; Ruocco, M.; Vinale, F.; Nigro, M.; Marra, R.; Lombardi, N.; Pascale, A.; Lanzuise, S.; Manganiello, G.; Lorito, M. Trichoderma-based Products and their Widespread Use in Agriculture. Open Mycol. J. 2014, 8, 71–126. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma-plant-pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Harman, G.E. Trichoderma-not just for biocontrol anymore. Phytoparasitica 2011, 39, 103–108. [Google Scholar] [CrossRef]
- Wong, M.M.L.; Gujaria-Verma, N.; Ramsay, L.; Yuan, H.Y.; Caron, C.; Diapari, M.; Vandenberg, A.; Bett, K.E. Classification and characterization of species within the genus Lens using genotyping-by-sequencing (GBS). PLoS ONE 2015, 10, e0122025. [Google Scholar] [CrossRef]
- Gupta, C.D.; Dubey, R.C.; Kang, S.C.; Maheshwari, D.K. Antibiotic Mediated Necrotrophic Effect of Pseudomonas GRC2 against Two Fungal Plant Pathogens. Curr. Sci. 2001, 81, 91–94. [Google Scholar]
- Baker, R. Trzchoderma SPP. as plant-growth stimulants. Crit. Rev. Biotechnol. 1988, 7, 97–106. [Google Scholar] [CrossRef]
- Mukherjee, M.; Mukherjee, P.K.; Horwitz, B.A.; Zachow, C.; Berg, G.; Zeilinger, S. Trichoderma-Plant-Pathogen Interactions: Advances in Genetics of Biological Control. Indian J. Microbiol. 2012, 52, 522–529. [Google Scholar] [CrossRef]
- Naseby, D.C.; Pascual, J.A.; Lynch, J.M. Effect of biocontrol strains of Trichoderma on plant growth, Pythium ultimum populations, soil microbial communities and soil enzyme activities. J. Appl. Microbiol. 2000, 88, 161–169. [Google Scholar] [CrossRef]
- Li, M.F.; Li, G.H.; Zhang, K.Q. Non-volatile metabolites from Trichoderma spp. Metabolites 2019, 9, 58. [Google Scholar] [CrossRef]
- Adams, P.; De-Leij, F.A.A.M.; Lynch, J.M. Trichoderma harzianum Rifai 1295-22 mediates growth promotion of crack willow (Salix fragilis) saplings in both clean and metal-contaminated soil. Microb. Ecol. 2007, 54, 306–313. [Google Scholar] [CrossRef]
- Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 2012, 158, 17–25. [Google Scholar] [CrossRef]
- Altomare, C.; Norvell, W.A.; Björkman, T.; Harman, G.E. Solubilization of phosphates and micronutrients by the plant-growth- promoting and biocontrol fungus Trichoderma harzianum Rifai 1295-22. Appl. Environ. Microbiol. 1999, 65, 2926–2933. [Google Scholar] [CrossRef]
- Dandurand, L.M.; Mosher, R.D.; Knudsen, G.R. Combined effects of Brassica napus seed meal and Trichoderma harzianum on two soilborne plant pathogens. Can. J. Microbiol. 2000, 46, 1051–1057. [Google Scholar] [CrossRef]
- Parnell, J.J.; Berka, R.; Young, H.A.; Sturino, J.M.; Kang, Y.; Barnhart, D.M.; Dileo, M.V. From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Howell, C.R. Mechanisms employed by Trichoderma species in the biological control of plant diseases: The history and evolution of current concepts. Plant Dis. 2003, 87, 4–10. [Google Scholar] [CrossRef]
- Hagn, A.; Pritsch, K.; Schloter, M.; Munch, J.C. Fungal diversity in agricultural soil under different farming management systems, with special reference to biocontrol strains of Trichoderma spp. Biol. Fertil. Soils 2003, 38, 236–244. [Google Scholar] [CrossRef]
- Harman, G.E. Overview of mechanisms and uses of Trichoderma spp. Phytopathology 2006, 96, 190–194. [Google Scholar] [CrossRef]
- Bremer, E.; Van Kessel, C.; Nelson, L.; Rennie, R.J.; Rennie, D.A. Selection of Rhizobium leguminosarum strains for lentil (Lens culinaris) under growth room and field conditions. Plant Soil 1990, 121, 47–56. [Google Scholar] [CrossRef]
- Saber, W.I.A.; Abd El-Hai, K.M.; Ghoneem, K.M. Synergistic effect of Trichoderma and Rhizobium on both biocontrol of chocolate spot disease and induction of nodulation, physiological activities and productivity of Vicia faba. Res. J. Microbiol. 2009, 4, 286–300. [Google Scholar] [CrossRef][Green Version]
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759. [Google Scholar] [CrossRef]
- Prashar, P.; Vandenberg, A. Genotype-specific responses to the effects of commercial Trichoderma formulations in lentil (Lens culinaris ssp. culinaris) in the presence and absence of the oomycete pathogen Aphanomyces euteiches. Biocontrol Sci. Technol. 2017, 27, 1123–1144. [Google Scholar] [CrossRef]
- Adam, E.; Bernhart, M.; Müller, H.; Winkler, J.; Berg, G. The Cucurbita pepo seed microbiome: Genotype-specific composition and implications for breeding. Plant Soil 2018, 422, 35–49. [Google Scholar] [CrossRef]
- Dakora, F.D.; Phillips, D.A. Root exudates as mediators of mineral acquisition in low-nutrient environments. In Food Security in Nutrient-Stressed Environments: Exploiting Plants’ Genetic Capabilities; Springer: Dordrecht, The Netherlands, 2002; pp. 201–213. [Google Scholar]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef]
- Tiago, O.; Maicon, N.; Ivan, R.C.; Diego, N.F.; Vinícius, J.S.; Mauricio, F.; Alan, J.d.P.; Velci, Q.d.S. Plant secondary metabolites and its dynamical systems of induction in response to environmental factors: A review. Afr. J. Agric. Res. 2017, 12, 71–84. [Google Scholar] [CrossRef]
- Badri, D.V.; Chaparro, J.M.; Zhang, R.; Shen, Q.; Vivanco, J.M. Application of natural blends of phytochemicals derived from the root exudates of arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 2013, 288, 4502–4512. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Bazghaleh, N.; Prashar, P.; Purves, R.W.; Vandenberg, A. Polyphenolic composition of lentil roots in response to infection by Aphanomyces euteiches. Front. Plant Sci. 2018, 9, 1131. [Google Scholar] [CrossRef]
- d’Arcy-Lameta, A. Study of soybean and lentil root exudates II. Identification of some polyphenolic compounds, relation with plantlet physiology In the framework of Leguminosae-Rhizobium symbiosis study, we were interested in determining the exact nature of root exud. Plant Soil 1986, 123, 113–123. [Google Scholar]
- Bazghaleh, N.; Hamel, C.; Gan, Y.; Tar’an, B.; Knight, J.D. Genotypic variation in the response of chickpea to arbuscular mycorrhizal fungi and non-mycorrhizal fungal endophytes. Can. J. Microbiol. 2018, 64, 265–275. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Tucci, M.; Ruocco, M.; De Masi, L.; De Palma, M.; Lorito, M. The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 2011, 12, 341–354. [Google Scholar] [CrossRef]
- Abi-Ghanem, R.; Carpenter-Boggs, L.; Smith, J.L. Cultivar effects on nitrogen fixation in peas and lentils. Biol. Fertil. Soils 2011, 47, 115–120. [Google Scholar] [CrossRef]
| Genotype | Species | Gene Pool * | Type |
|---|---|---|---|
| CDC Maxim | L. culinaris | Primary | Cultivated |
| Eston | L. culinaris | Primary | Cultivated |
| ZT-4 | L. culinaris | Primary | Cultivated |
| ZT-6 | L. culinaris | Primary | Cultivated |
| IG 72643 | L. orientalis | Primary | Wild |
| IG 72805 | L. tomentosus | Primary | Wild |
| IG 72623 | L. odemensis | Secondary | Wild |
| L01-827A | L. ervoides | Tertiary | Wild |
| DF | p | ||
|---|---|---|---|
| Fungal Community | Trichoderma Community | ||
| Genotype | 7 | 0.03 * | 0.02 * |
| Infection (A.s euteiches) | 1 | 0.09 | 0.79 |
| Treatment (Trichoderma) | 1 | 0.001 *** | 0.002 ** |
| Infection × Genotype × Treatment | 7 | 0.01 * | 0.35 |
| Residuals | 96 | ||
| DF | Sum Sq | Mean Sq | F | p | |
|---|---|---|---|---|---|
| Genotype | 7 | 167.2 | 23.89 | 3.66 | 0.001 ** |
| Infection (A. euteiches) | 1 | 0.1 | 0.07 | 0.01 | 0.91 |
| Treatment (Trichoderma) | 1 | 72.1 | 72.1 | 11.07 | 0.001 ** |
| Infection × Genotype × Treatment | 7 | 26.5 | 3.79 | 0.58 | 0.76 |
| Residuals | 96 |
| Factors | p Value | r |
|---|---|---|
| Root biomass: Trichoderma spp. | <0.05 | 0.2 |
| Shoot biomass: Trichoderma spp. | <0.05 | 0.19 |
| Root biomass: shoot biomass | <0.0001 | 0.51 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bazghaleh, N.; Prashar, P.; Woo, S.; Vandenberg, A. Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot. Microorganisms 2020, 8, 1290. https://doi.org/10.3390/microorganisms8091290
Bazghaleh N, Prashar P, Woo S, Vandenberg A. Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot. Microorganisms. 2020; 8(9):1290. https://doi.org/10.3390/microorganisms8091290
Chicago/Turabian StyleBazghaleh, Navid, Pratibha Prashar, Sheridan Woo, and Albert Vandenberg. 2020. "Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot" Microorganisms 8, no. 9: 1290. https://doi.org/10.3390/microorganisms8091290
APA StyleBazghaleh, N., Prashar, P., Woo, S., & Vandenberg, A. (2020). Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot. Microorganisms, 8(9), 1290. https://doi.org/10.3390/microorganisms8091290






