Gene Expression and Photophysiological Changes in Pocillopora acuta Coral Holobiont Following Heat Stress and Recovery
Abstract
:1. Introduction
2. Materials and Methods
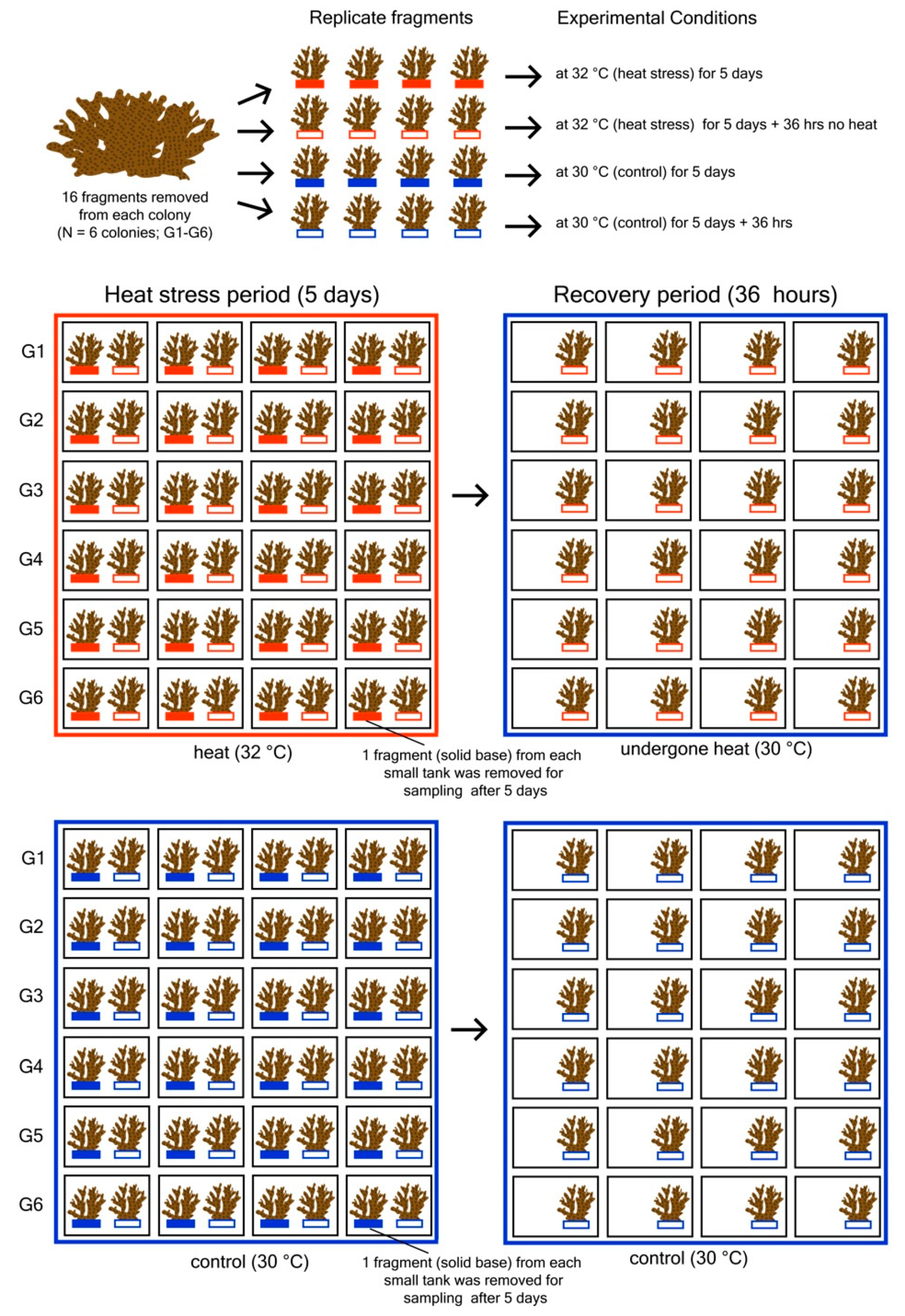
2.1. Experimental Design
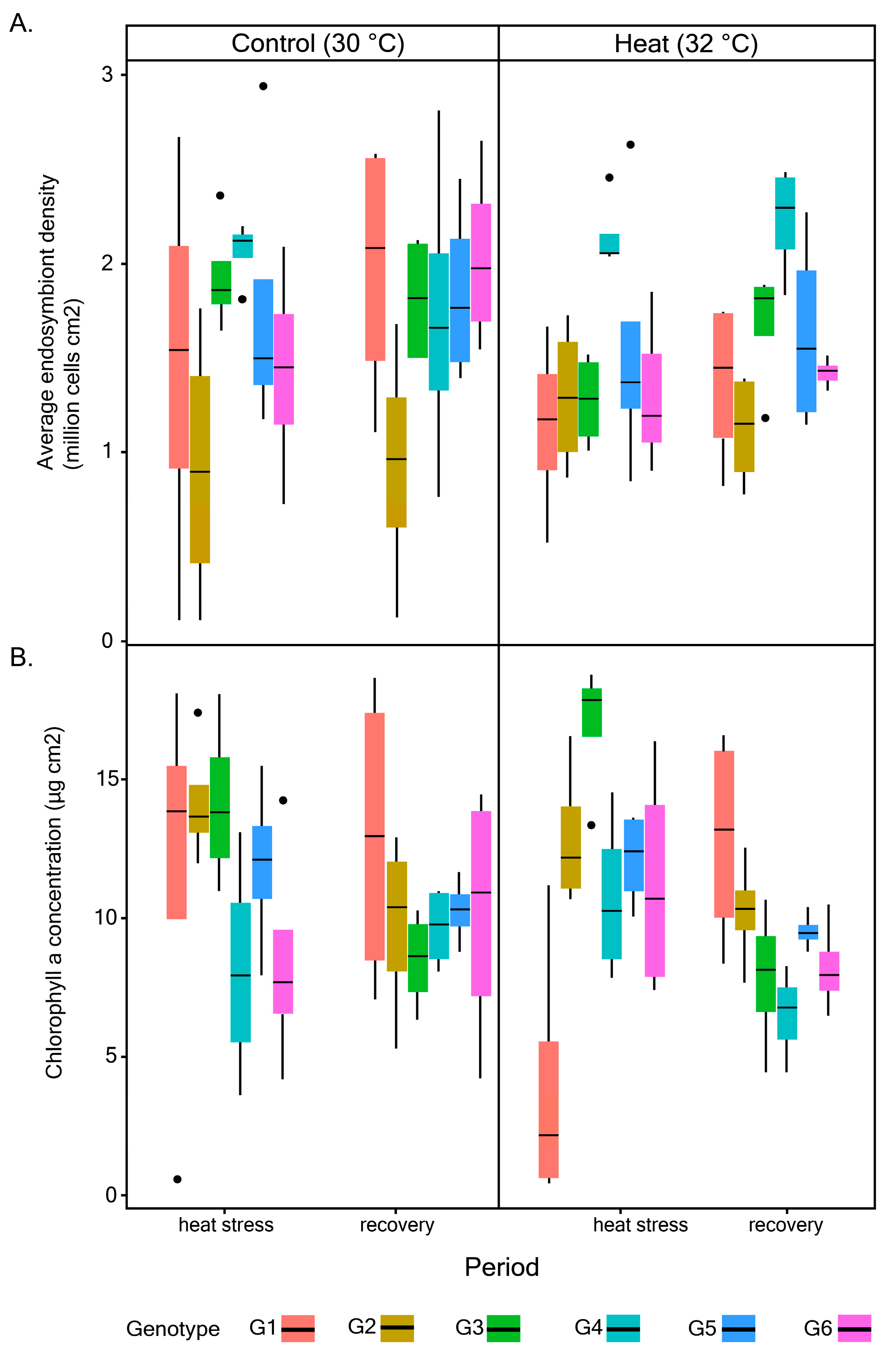
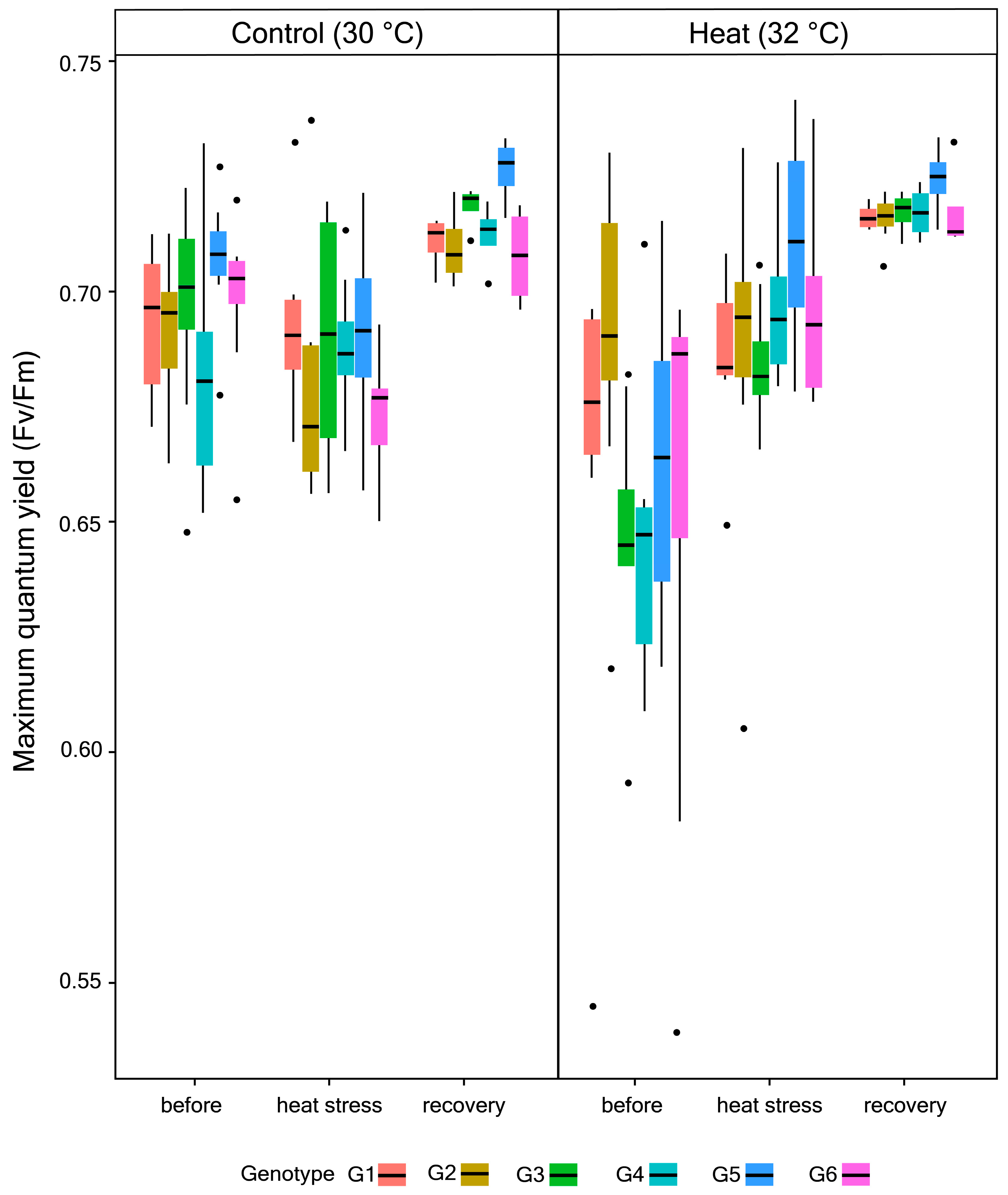
2.2. Quantification of Maximum Quantum Yield (Fv/Fm), Endosymbiont Density, and Chlorophyll (chl) a Concentration
2.3. RNA Extraction and Reverse Transcription (RT)
2.4. Primer Design and Validation
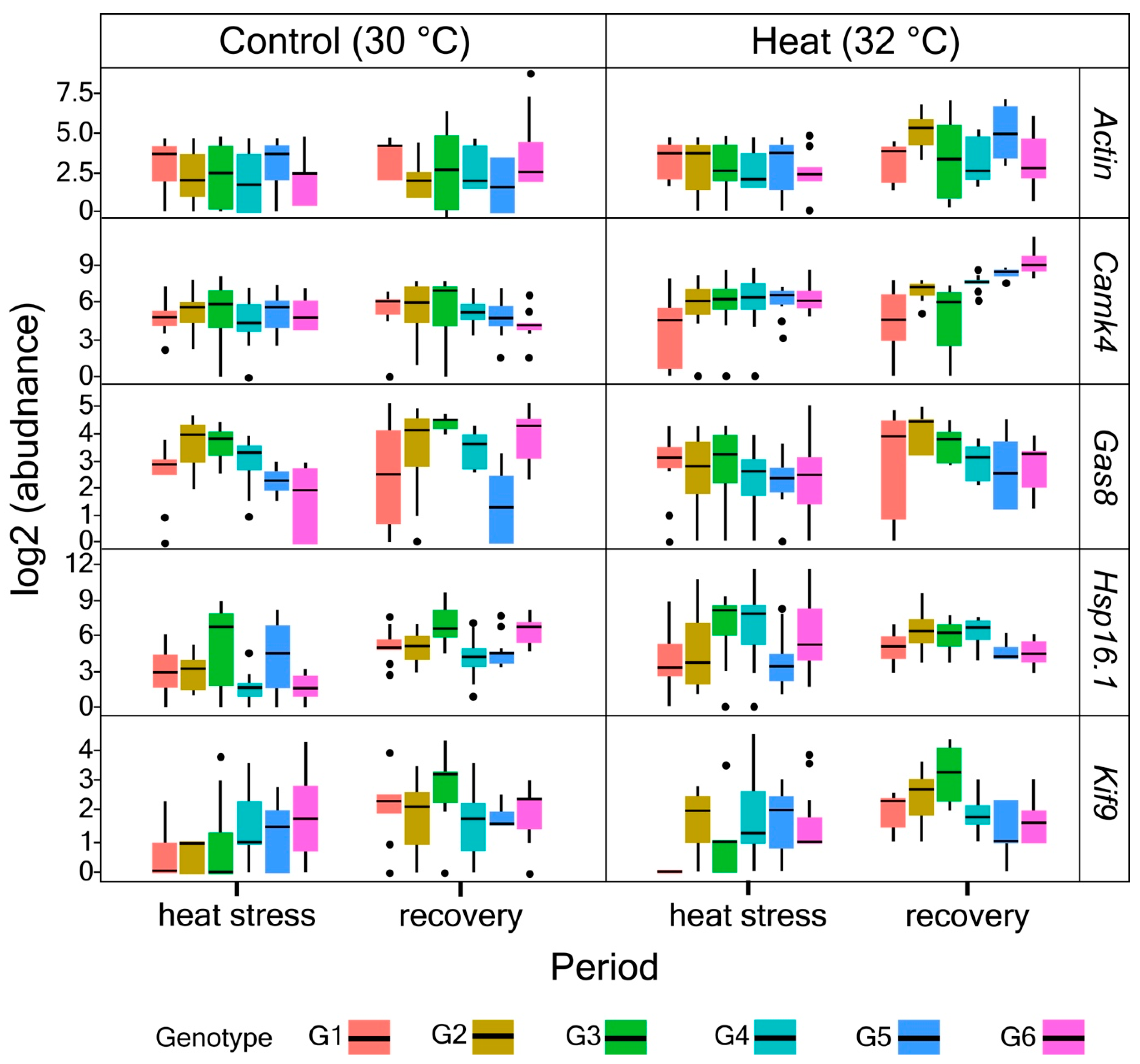
2.5. Quantification of Gene Expressions
2.6. Statistical Analyses
3. Results
3.1. Photophysiological Performance
3.2. Primer Validation and Gene Expression Profiles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral Reefs Under Rapid Climate Change and Ocean Acidification. Science 2007, 318, 1737–1742. [Google Scholar] [CrossRef] [Green Version]
- Lough, J.; Anderson, K.D.; Hughes, T.P. Increasing thermal stress for tropical coral reefs: 1871–2017. Sci. Rep. 2018, 8, 6079. [Google Scholar] [CrossRef] [Green Version]
- Hughes, T.P.; Barnes, M.; Bellwood, D.R.; Cinner, J.E.; Cumming, G.S.; Jackson, J.B.C.; Kleypas, J.; Van De Leemput, I.A.; Lough, J.; Morrison, T.H.; et al. Coral reefs in the Anthropocene. Nature 2017, 546, 82–90. [Google Scholar] [CrossRef]
- Heron, S.F.; Maynard, J.A.; Van Hooidonk, R.; Eakin, C.M. Warming Trends and Bleaching Stress of the World′s Coral Reefs 1985–2012. Sci. Rep. 2016, 6, 38402. [Google Scholar] [CrossRef] [Green Version]
- Somero, G. The physiology of climate change: How potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J. Exp. Biol. 2010, 213, 912–920. [Google Scholar] [CrossRef] [Green Version]
- Maor-Landaw, K.; Levy, O. Survey of Cnidarian Gene Expression Profiles in Response to Environmental Stressors: Summarizing 20 Years of Research, What Are We Heading for? Cnidaria Past Present Future 2016, 523–543. [Google Scholar] [CrossRef]
- Sweet, M.; Brown, B. Coral Responses to Anthropogenic Stress in the Twenty-First Century: An Ecophysiological Perspective. Oceanogr. Mar. Biol. 2016, 54, 271–314. [Google Scholar] [CrossRef]
- Venn, A.A.; Wilson, M.A.; Trapido-Rosenthal, H.G.; Keely, B.; Douglas, A.E. The impact of coral bleaching on the pigment profile of the symbiotic alga, Symbiodinium. Plant Cell Environ. 2006, 29, 2133–2142. [Google Scholar] [CrossRef] [PubMed]
- Buddemeier, R.W.; Fautin, D.G. Coral Bleaching as an Adaptive Mechanism. Bioscience 1993, 43, 320–326. [Google Scholar] [CrossRef]
- Loya, Y.; Sakai, K.; Yamazato, K.; Nakano, Y.; Sambali, H.; Van Woesik, R. Coral bleaching: The winners and the losers. Ecol. Lett. 2001, 4, 122–131. [Google Scholar] [CrossRef]
- Baker, A.C.; Glynn, P.W.; Riegl, B. Climate change and coral reef bleaching: An ecological assessment of long-term impacts, recovery trends and future outlook. Estuar. Coast. Shelf Sci. 2008, 80, 435–471. [Google Scholar] [CrossRef]
- Ng, C.S.L.; Huang, D.; Toh, K.B.; Sam, S.Q.; Kikuzawa, Y.P.; Toh, T.C.; Taira, D.; Chan, Y.K.S.; Hung, L.Z.T.; Sim, W.T.; et al. Responses of urban reef corals during the 2016 mass bleaching event. Mar. Pollut. Bull. 2020, 154, 111111. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E. Energy Signaling in the Regulation of Gene Expression during Stress. Mol. Plant 2010, 3, 300–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gates, R.D.; Edmunds, P.J. The Physiological Mechanisms of Acclimatization in Tropical Reef Corals. Am. Zool. 1999, 39, 30–43. [Google Scholar] [CrossRef]
- Wund, M.A. Assessing the Impacts of Phenotypic Plasticity on Evolution. Integr. Comp. Biol. 2012, 52, 5–15. [Google Scholar] [CrossRef]
- Stone, J.R. Phenotypic Evolution: A Reaction Norm Perspective; Carl, D., Ed.; Sinauer Associates Incorporated: Sunderland, MA, USA, 2000; Volume 75, pp. 55–56. [Google Scholar]
- West-Eberhard, M.J. Phenotypic plasticity and the origins of diversity. Annu. Rev. Ecol. Syst. 1989, 20, 249–278. [Google Scholar] [CrossRef]
- Wellband, K.; Heath, D.D. Plasticity in gene transcription explains the differential performance of two invasive fish species. Evol. Appl. 2017, 10, 563–576. [Google Scholar] [CrossRef]
- Grenier, S.; Barre, P.; Litrico, I. Phenotypic Plasticity and Selection: Nonexclusive Mechanisms of Adaptation. Science 2016, 2016, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Seneca, F.; Traylor-Knowles, N.; Palumbi, S.R. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. USA 2013, 110, 1387–1392. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.; Chen, Y.J.; Lu, C.Y.; Chen, C.S. The proteomic response of the reef coral Pocillopora acuta to experimentally elevated temperatures. PLoS ONE 2018, 13, e0192001. [Google Scholar] [CrossRef] [Green Version]
- Anthony, K.R.N.; E Fabricius, K. Shifting roles of heterotrophy and autotrophy in coral energetics under varying turbidity. J. Exp. Mar. Biol. Ecol. 2000, 252, 221–253. [Google Scholar] [CrossRef]
- Lesser, M.P.; Weis, V.M.; Patterson, M.R.; Jokiel, P.L. Effects of morphology and water motion on carbon delivery and productivity in the reef coral, Pocillopora damicornis (Linnaeus): Diffusion barriers, inorganic carbon limitation, and biochemical plasticity. J. Exp. Mar. Biol. Ecol. 1994, 178, 153–179. [Google Scholar] [CrossRef]
- Todd, P.A. Morphological plasticity in scleractinian corals. Biol. Rev. 2008, 83, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Ow, Y.X.; Todd, P.A. Light-induced morphological plasticity in the scleractinian coral Goniastrea pectinata and its functional significance. Coral Reefs 2010, 29, 797–808. [Google Scholar] [CrossRef]
- Aubin-Horth, N.; Renn, S.C.P. Genomic reaction norms: Using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 2009, 18, 3763–3780. [Google Scholar] [CrossRef]
- Maury, L.L.; Marguerat, S.; Bähler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef]
- Kenkel, C.D.; Aglyamova, G.; Alamaru, A.; Bhagooli, R.; Capper, R.; Cunning, R.; Devillers, A.; Haslun, J.A.; Hédouin, L.; Keshavmurthy, S.; et al. Development of Gene Expression Markers of Acute Heat-Light Stress in Reef-Building Corals of the Genus Porites. PLoS ONE 2011, 6, e26914. [Google Scholar] [CrossRef] [Green Version]
- Seneca, F.; Forêt, S.; Ball, E.; Smith-Keune, C.; Miller, D.J.; Van Oppen, M. Patterns of Gene Expression in a Scleractinian Coral Undergoing Natural Bleaching. Mar. Biotechnol. 2009, 12, 594–604. [Google Scholar] [CrossRef]
- Bay, L.K.; Ulstrup, K.E.; Nielsen, H.B.; Jarmer, H.; Goffard, N.; Willis, B.L.; Miller, D.J.; Van Oppen, M. Microarray analysis reveals transcriptional plasticity in the reef building coral Acropora millepora. Mol. Ecol. 2009, 18, 3062–3075. [Google Scholar] [CrossRef]
- Császár, N.; Seneca, F.; Van Oppen, M. Variation in antioxidant gene expression in the scleractinian coral Acropora millepora under laboratory thermal stress. Mar. Ecol. Prog. Ser. 2009, 392, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Stearns, S.C. The Evolutionary Significance of Phenotypic Plasticity. Bioscience 1989, 39, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Drury, C.; Manzello, D.; Lirman, D. Genotype and local environment dynamically influence growth, disturbance response and survivorship in the threatened coral, Acropora cervicornis. PLoS ONE 2017, 12, e0174000. [Google Scholar] [CrossRef] [Green Version]
- Kenkel, C.D.; Matz, M.V. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 2016, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Guest, J.R.; Baird, A.H.; Maynard, J.A.; Muttaqin, E.; Edwards, A.J.; Campbell, S.J.; Yewdall, K.; Affendi, Y.A.; Chou, L.M. Contrasting patterns of coral bleaching susceptibility in 2010 suggest an adaptive response to thermal stress. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Epstein, H.E.; Torda, G.; van Oppen, M.J.H. Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs 2019, 38, 373–386. [Google Scholar] [CrossRef]
- Smith, H.; Epstein, H.; Torda, G. The molecular basis of differential morphology and bleaching thresholds in two morphs of the coral Pocillopora acuta. Sci. Rep. 2017, 7, 10066. [Google Scholar] [CrossRef]
- Poquita-Du, R.C.; Huang, D.; Chou, L.M.; Todd, P.A. Mrinalini Short Term Exposure to Heat and Sediment Triggers Changes in Coral Gene Expression and Photo-Physiological Performance. Front. Mar. Sci. 2019, 6, 6. [Google Scholar] [CrossRef]
- Schmidt-Roach, S.; Miller, K.J.; Lundgren, P.; Andreakis, N. With eyes wide open: A revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia: Pocilloporidae) using morphology and genetics. Zool. J. Linnean. Soc. 2014, 170, 1–33. [Google Scholar] [CrossRef]
- Chou, L.M.; Toh, T.C.; Toh, K.B.; Ng, C.S.L.; Cabaitan, P.; Tun, K.; Goh, E.; Afiq-Rosli, L.; Taira, D.; Poquita-Du, R.C.; et al. Differential response of coral assemblages to thermal stress underscores the complexity in predicting bleaching susceptibility. PLoS ONE 2016, 11, e0159755. [Google Scholar] [CrossRef] [Green Version]
- Toh, T.C.; Huang, D.; Tun, K.; Chou, L.M. Summary of coral bleaching from 2014 to 2017 in Singapore. In Status of Coral Reefs in East Asian Seas Region: 2018; Kimura, T., Tun, K., Chou, L.M., Eds.; Ministry of the Environment of Japan and Japan Wildlife Research Center: Tokyo, Japan, 2018; pp. 21–23. [Google Scholar]
- Ralph, P.L.; Gademann, R.; Larkum, A.; Schreiber, U. In situ underwater measurements of photosynthetic activity of coral zooxanthellae and other reef-dwelling dinoflagellate endosymbionts. Mar. Ecol. Prog. Ser. 1999, 180, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Ben-Haim, Y.; Zicherman-Keren, M.; Rosenberg, E. Temperature-Regulated Bleaching and Lysis of the Coral Pocillopora damicornis by the Novel Pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 2003, 69, 4236–4242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffrey, S.; Humphrey, G. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 1975, 167, 191–194. [Google Scholar] [CrossRef]
- Matz, M.V.; Wright, R.M.; Scott, J.G. No Control Genes Required: Bayesian Analysis of qRT-PCR Data. PLoS ONE 2013, 8, e71448. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation of Statistical Computing: Vienna, Austria, 2018; Available online: http://www.R-project.org/ (accessed on 4 February 2018).
- Morrison, T.; Hurley, J.; Garcia, J.; Yoder, K.; Katz, A.; Roberts, D.; Cho, J.; Kanigan, T.; Ilyin, S.E.; Horowitz, D.; et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006, 34, e123. [Google Scholar] [CrossRef] [PubMed]
- Lalam, N. Statistical Inference for Quantitative Polymerase Chain Reaction Using a Hidden Markov Model: A Bayesian Approach. Stat. Appl. Genet. Mol. Biol. 2007, 6, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Sivaganesan, M.; Seifring, S.; Varma, M.; Haugland, R.A.; Shanks, O.C. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinform. 2008, 9, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Follestad, T.; Jørstad, T.S.; Erlandsen, S.E.; Sandvik, A.K.; Bones, A.M.; Langaas, M. A Bayesian hierarchical model for quantitative real-time PCR data. Stat. Appl. Genet. Mol. Biol. 2010, 9, 3. [Google Scholar] [CrossRef]
- Paradis, M.È.; Haine, D.; Gillespie, B.; Oliver, S.P.; Messier, S.; Comeau, J.; Scholl, D.T. Bayesian estimation of the diagnostic accuracy of a multiplex real-time PCR assay and bacteriological culture for 4 common bovine intramammary pathogens. J. Dairy Sci. 2012, 95, 6436–6448. [Google Scholar] [CrossRef]
- Warner, M.E.; Fitt, W.K.; Schmidt, G.W. The effects of elevated temperature on the photosynthetic efficiency of zooxanthellae in hospite from four different species of reef coral: A novel approach. Plant Cell Environ. 1996, 19, 291–299. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O. Climate change, coral bleaching and the future of the world′s coral reefs. Mar. Freshw. Res. 1999, 50, 839. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes, A.L.C.; Lozano-Cortés, D.F.; Zapata, F.A. Effect of short-term subaerial exposure on the cauliflower coral, Pocillopora damicornis, during a simulated extreme low-tide event. Coral Reefs 2017, 36, 401–414. [Google Scholar] [CrossRef] [Green Version]
- Wooldridge, S.A. Breakdown of the coral-algae symbiosis: Towards formalising a linkage between warm-water bleaching thresholds and the growth rate of the intracellular zooxanthellae. Biogeosciences 2013, 10, 1647–1658. [Google Scholar] [CrossRef] [Green Version]
- Hennige, S.J.; Suggett, D.J.; Warner, M.E.; McDougall, K.E.; Smith, D.J. Photobiology of Symbiodinium revisited: Bio-physical and bio-optical signatures. Coral Reefs 2009, 28, 179–195. [Google Scholar] [CrossRef]
- Roth, M.S. The engine of the reef: Photobiology of the coral-algal symbiosis. Front. Microbiol. 2014, 5, 422. [Google Scholar] [CrossRef] [PubMed]
- Tanzil, J.T.I.; Ng, A.P.K.; Tey, Y.Q.; Tan, B.H.Y.; Yun, E.Y.; Huang, D.; Abigayle, N.P.K.; Qing, T.Y.; Beverly, T.H.Y.; Eric, Y.Y.; et al. A preliminary characterisation of Symbiodinium diversity in some common corals from Singapore. Cosmos 2016, 12, 15–27. [Google Scholar] [CrossRef]
- Poquita-Du, R.C.; Huang, D.; Chou, L.M.; Todd, P.A. The contribution of stress-tolerant endosymbiotic dinoflagellate Durusdinium to Pocillopora acuta survival in a highly urbanized reef system. Coral Reefs 2020, 1–11. [Google Scholar] [CrossRef]
- Guest, J.R.; Low, J.; Tun, K.; Wilson, B.; Ng, C.; Raingeard, D.; Ulstrup, K.E.; Tanzil, J.T.I.; Todd, P.A.; Toh, T.C.; et al. Coral community response to bleaching on a highly disturbed reef. Sci. Rep. 2016, 6, 20717. [Google Scholar] [CrossRef] [Green Version]
- Lough, J.M.; Barnes, D.J. Several centuries of variation in skeletal extension, density and calcification in massive Porites colonies from the Great Barrier Reef: A proxy for seawater temperature and a background of variability against which to identify unnatural change. J. Exp. Mar. Biol. Ecol. 1997, 211, 29–67. [Google Scholar] [CrossRef]
- Marshall, A.T.; Clode, P. Calcification rate and the effect of temperature in a zooxanthellate and an azooxanthellate scleractinian reef coral. Coral Reefs 2004, 23, 218–224. [Google Scholar] [CrossRef]
- Kruzic, P.; Sr_sen, P.; Benkovic, L. The impact of seawater temperature on coral growth parameters of the colonial coral Cladocora caespitosa (Anthozoa, Scleractinia) in the eastern Adriatic Sea. Facies 2012, 58, 477–491. [Google Scholar] [CrossRef]
- Tanzil, J.T.I.; Brown, B.E.; Dunne, R.P.; Lee, J.N.; Kaandorp, J.A.; Todd, P.T. Regional decline in growth rates of massive Porites corals in Southeast Asia. Glob. Chang. Biol. 2013, 19, 30113023. [Google Scholar] [CrossRef] [PubMed]
- DeSalvo, M.K.; Voolstra, C.R.; Sunagawa, S.; Schwarz, J.A.; Stillman, J.H.; Coffroth, M.A.; Szmant, A.M.; Medina, M. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 2008, 17, 3952–3971. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.L.; McConnaughey, T.A. Geochemical perspectives on coral mineralization. Rev. Miner. Geochem. 2003, 54, 151–187. [Google Scholar] [CrossRef]
- Allemand, D.; Tambuttè, É.; Zoccola, D.; Tambuttè, S. Coral calcification, cells to reefs. In Coral Reefs: An Ecosystem in Transition; Dubinsky, Z., Stambler, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 119–150. [Google Scholar]
- Lough, J.M.; Barnes, D.J. Environmental controls on growth of the massive coral Porites. J. Exp. Mar. Biol. Ecol. 2000, 245, 225243. [Google Scholar] [CrossRef]
- Lough, J.M.; Cooper, T.F. New insights from coral growth band studies in an era of rapid environmental change. Earth Sci. Rev. 2011, 108, 170184. [Google Scholar] [CrossRef]
- Howe, S.A.; Marshall, A.T. Temperature effects on calcification rate and skeletal deposition in the temperate coral, Plesiastrea versipora (Lamarck). J. Exp. Mar. Biol. 2002, 275, 63–81. [Google Scholar] [CrossRef]
- Cole, C.; Finch, A.A.; Hintz, C.; Hintz, K.; Allison, N. Effects of seawater pCO2 and temperature on calcification and productivity in the coral genus Porites spp.: An exploration of potential interaction mechanisms. Coral Reefs 2018, 37, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Cunning, R.; Bay, R.A.; Gillette, P.; Baker, A.C.; Traylor-Knowles, N. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci. Rep. 2018, 8, 16134. [Google Scholar] [CrossRef] [Green Version]
- Taira, D.; Toh, T.C.; Ng, C.S.L.; Loke, H.X.; Afiq-Rosli, L.; Cabaitan, P.C.; Toh, K.B.; Poquita-Du, R.C.; Chou, L.M.; Song, T. Relocating bleached Platygyra sinensis facilitates recovery from thermal stress during a minor bleaching event. Mar. Freshw. Behav. Physiol. 2017, 50, 375–385. [Google Scholar] [CrossRef]
- Browne, N.; Precht, E.; Last, K.; Todd, P. Photo-physiological costs associated with acute sediment stress events in three near-shore turbid water corals. Mar. Ecol. Prog. Ser. 2014, 502, 129–143. [Google Scholar] [CrossRef] [Green Version]
- Fabricius, K.E. Effects of irradiance, flow, and colony pigmentation on the temperature microenvironment around corals: Implications for coral bleaching? Limnol. Oceanogr. 2006, 51, 30–37. [Google Scholar] [CrossRef]
- Hawley, S.A.; Pan, D.A.; Mustard, K.J.; Ross, L.; Bain, J.; Edelman, A.M.; Frenguelli, B.G.; Hardie, D.G. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005, 2, 9–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosic, N.; Kaniewska, P.; Chan, C.K.K.; Ling, E.Y.S.; Edwards, D.; Dove, S.G.; Hoegh-Guldberg, O. Early transcriptional changes in the reef-building coral Acropora aspera in response to thermal and nutrient stress. BMC Genom. 2014, 15, 1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSalvo, M.; Sunagawa, S.; Voolstra, C.; Medina, M. Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 2010, 402, 97–113. [Google Scholar] [CrossRef] [Green Version]
- Richier, S.; Rodriguez-Lanetty, M.; Schnitzler, C.E.; Weis, V.M. Response of the symbiotic cnidarian Anthopleura elegantissima transcriptome to temperature and UV increase. Comp. Biochem. Physiol. D Genom. Proteom. 2008, 3, 283–289. [Google Scholar] [CrossRef]
- Lesser, M.P. Oxidative stress causes coral bleaching during exposure to elevated temperature. Coral Reefs 1997, 16, 187–192. [Google Scholar] [CrossRef]
- Hou, J.; Xu, T.; Su, D.; Wu, Y.; Cheng, L.; Wang, J.; Zhou, Z.; Wang, Y. RNA-Seq reveals extensive transcriptional response to heat stress in the stony coral Galaxea fascicularis. Front. Genet. 2018, 9, 37. [Google Scholar] [CrossRef]
- Poquita-Du, R.C.; Ng, C.S.L.; Loo, J.; Afiq-Rosli, L.; Tay, Y.; Todd, P.A.; Chou, L.M.; Huang, D. New evidence shows that Pocillopora′damicornis-like′ corals in Singapore are actually Pocillopora acuta (Scleractinia: Pocilloporidae). Biodivers. Data J. 2017, 5, e11407. [Google Scholar] [CrossRef] [Green Version]
- Glynn, P.W.; Ault, J.S. A biogeographic analysis and review of the far eastern Pacific coral reef region. Coral Reefs 2000, 19, 1–23. [Google Scholar] [CrossRef]
- Paz-Garcia, D.A.; Hellberg, M.E.; de-Leon, F.J.G.; Balart, E.F. Switch between morphospecies of Pocillopora corals. Am. Nat. 2015, 186, 434–440. [Google Scholar] [CrossRef]
- Johnston, E.C.; Forsman, Z.H.; Flot, J.F.; Schmidt-Roach, S.; Pinzon, J.H.; Knapp, I.S.S.; Toonen, R.J. A genomic glance through the fog of plasticity and diversification in Pocillopora. Sci. Rep. 2017, 7, 5991. [Google Scholar] [CrossRef] [PubMed]
- Cziesielski, M.J.; Liew, Y.J.; Cui, G.; Schmidt-Roach, S.; Campana, S.; Marondedze, C.; Aranda, M. Multi-omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. Proc. R. Soc. B 2018, 285, 20172654. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, A.B.; Wang, Y.B.; Chen, C.S.; Chen, S.H.; Lin, C.Y. Dual-compartmental transcriptomic + proteomic analysis of a marine endosymbiosis exposed to environmental change. Mol. Ecol. 2016, 25, 5944–5958. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Carreto, S.; Vera-Estrella, R.; Portillo-Bobadilla, T.; Licea-Navarro, A.; Bernaldez-Sarabia, J.; Rudiño-Piñera, E.; Verleyen, J.J.; Rodríguez, E.; Rodríguez-Almazán, C. Transcriptomic and proteomic analysis of the tentacles and mucus of Anthopleura dowii Verrill, 1869. Mar. Drugs. 2019, 17, 436. [Google Scholar] [CrossRef] [Green Version]
- Baumann, J.; Grottoli, A.G.; Hughes, A.D.; Matsui, Y. Photoautotrophic and heterotrophic carbon in bleached and non-bleached coral lipid acquisition and storage. J. Exp. Mar. Biol. Ecol. 2014, 461, 469–478. [Google Scholar] [CrossRef]
- Yamashiro, H.; Oku, H.; Onaga, K. Effect of bleaching on lipid content and composition of Okinawan corals. Fish. Sci. 2005, 71, 448–453. [Google Scholar] [CrossRef]
| Gene Name (Abbreviation) | Biological Process | Forward Primer Reverse Primer |
|---|---|---|
| Actin * | Cytoskeleton | F: 5′-CAA GCA TCC TGT TCT CCT GAC-3′ |
| R: 5′-AGG TAG GCC GTC AAG TCC C-3′ | ||
| Acyl-CoA dehydrogenase family member 11 (ACAD-11) * | Fatty acid beta-oxidation | F: 5′-TAATCCAGCGACCCAGTGGA-3′ |
| R: 5′-AAGCCAGGCCTTCTTTTGCT-3′ | ||
| Heat shock protein 70 (HSP70) * | Stress response (heat) | F: 5′-TTTCGACAACAAGGCCACGG-3′ |
| R: 5′-TCTTCTTCGATCGTTAGGCGG-3′ | ||
| Calcium/calmodulin-dependent protein kinase type IV (Camk4) | Adaptive immune response | F: 5′-GGA ACC CCT GGA TAC TGT GC-3′ |
| R: 5′-ACA TCG CTT GAT CAC CTC GT-3′ | ||
| EF-hand domain-containing protein 1 (EFH1) | Cell cycle | F: 5′-GTTCTACACCCCAGCTGACT-3′ |
| R: 5′-CTTGTGGAACAGATGCCACC-3′ | ||
| Growth arrest-specific protein 8 (GAS8) | Cilia motility | F: 5′-TGG AGA AGA AGG AAG CGC AG-3′ |
| R: 5′-GTA CTC CGA ACG ACT GGA GC-3′ | ||
| Small heat shock protein 16.1 (Hsp16.1) | Stress response (heat) | F: 5′-TGG TCA ACC CTT ACT GCC AT-3′ |
| R: 5′-TCT CTC TCT GAG CGA TGC TG-3′ | ||
| Kinesin-like protein (KIF9) | Extracellular matrix disassembly | F: 5′-CAA CGG AAC GAT TTT GGC GT-3′ |
| R: 5′-GAT CCG TAC AGT CAC AGC GT-3′ | ||
| XIAP-associated factor 1 (XAF1) | Apoptosis | F: 5′-TGCGAAAACTGCAACAGACG-3′ |
| R: 5′-CACAAGGCAAAGACACAGGC-3′ |
| Final model = Average Endosymbiont Density~Treatment × Period × Site/Colony | |||
|---|---|---|---|
| Fixed effects | Chisq | Df | p |
| Treatment | 3.14 × 105 | 1 | <2.2 × 10−16 |
| Period | 2.07 × 105 | 1 | <2.2 × 10−16 |
| Site | 7.33 × 10 | 1 | 0.007 |
| Treatment × period | 1.34 × 103 | 1 | <2.2 × 10−16 |
| Treatment × site | 5.34 × 104 | 1 | <2.2 × 10−16 |
| Period × site | 7.59 × 103 | 1 | <2.2 × 10−16 |
| Treatment × period × site | 3.56 × 103 | 1 | <2.2 × 10−16 |
| Treatment × period × site × colony | 2.58 × 106 | 16 | <2.2 × 10−16 |
| Final model = Average chl a Concentration~Treatment × Period × Site/Colony | |||
|---|---|---|---|
| Fixed effects | Chisq | Df | p |
| Treatment | 0.46 | 1 | 0.498 |
| Period | 5.04 | 1 | 0.025 |
| Site | 4.50 | 1 | 0.034 |
| Treatment × period | 0.33 | 1 | 0.564 |
| Treatment × site | 0.62 | 1 | 0.430 |
| Period × site | 0.22 | 1 | 0.639 |
| Treatment × period × site | 3.77 | 1 | 0.052 |
| Treatment × period × site × colony | 41.57 | 16 | <2.2 × 10−16 |
| Final model = Average Fv/Fm~Treatment × Period × Site/Colony | |||
|---|---|---|---|
| Fixed effects | Chisq | Df | p |
| Treatment | 0.116 | 1 | 0.733 |
| Period | 95.529 | 1 | <2.2 × 10−16 |
| Site | 1.531 | 1 | 0.216 |
| Treatment × period | 5.753 | 1 | 0.016 |
| Treatment × site | 0.183 | 1 | 0.669 |
| Period × site | 0.184 | 1 | 0.668 |
| Treatment × period × site | 0.659 | 1 | 0.417 |
| Treatment × period × site × colony | 21.918 | 16 | 0.146 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poquita-Du, R.C.; Goh, Y.L.; Huang, D.; Chou, L.M.; Todd, P.A. Gene Expression and Photophysiological Changes in Pocillopora acuta Coral Holobiont Following Heat Stress and Recovery. Microorganisms 2020, 8, 1227. https://doi.org/10.3390/microorganisms8081227
Poquita-Du RC, Goh YL, Huang D, Chou LM, Todd PA. Gene Expression and Photophysiological Changes in Pocillopora acuta Coral Holobiont Following Heat Stress and Recovery. Microorganisms. 2020; 8(8):1227. https://doi.org/10.3390/microorganisms8081227
Chicago/Turabian StylePoquita-Du, Rosa Celia, Yi Le Goh, Danwei Huang, Loke Ming Chou, and Peter A. Todd. 2020. "Gene Expression and Photophysiological Changes in Pocillopora acuta Coral Holobiont Following Heat Stress and Recovery" Microorganisms 8, no. 8: 1227. https://doi.org/10.3390/microorganisms8081227
APA StylePoquita-Du, R. C., Goh, Y. L., Huang, D., Chou, L. M., & Todd, P. A. (2020). Gene Expression and Photophysiological Changes in Pocillopora acuta Coral Holobiont Following Heat Stress and Recovery. Microorganisms, 8(8), 1227. https://doi.org/10.3390/microorganisms8081227







