COVID-19 Is a Multifaceted Challenging Pandemic Which Needs Urgent Public Health Interventions
Abstract
:1. Background
2. Impact of Coronaviruses in Animals and Humans
3. Emergent Zoonotic Coronaviruses of the Century Responsible for Coronavirus Epidemics and Pandemics
4. New SARS-CoV-2 Insights
5. The Mechanism of SARS-CoV-2 Entry
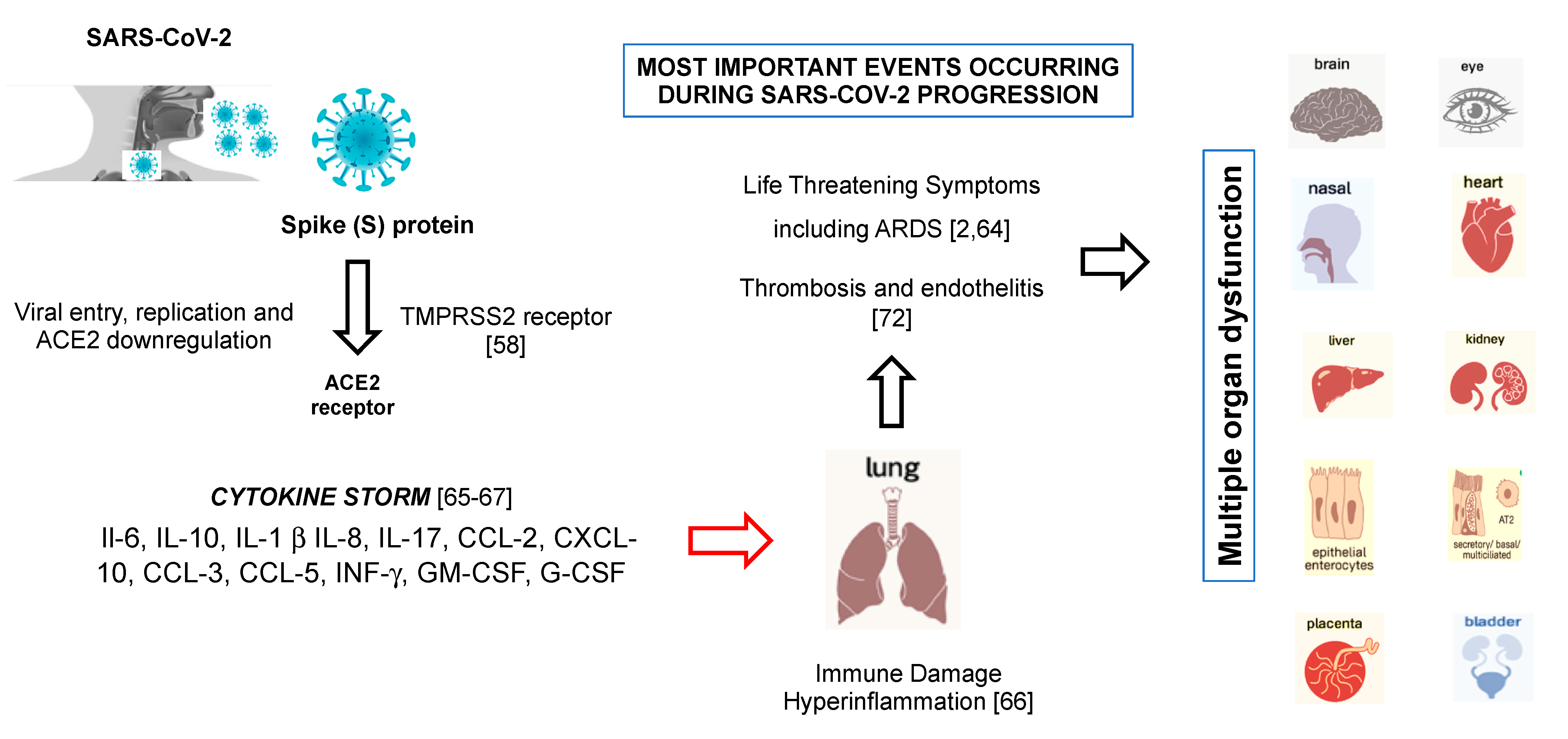
6. SARS-CoV-2 and the Cytokine Storm Syndrome
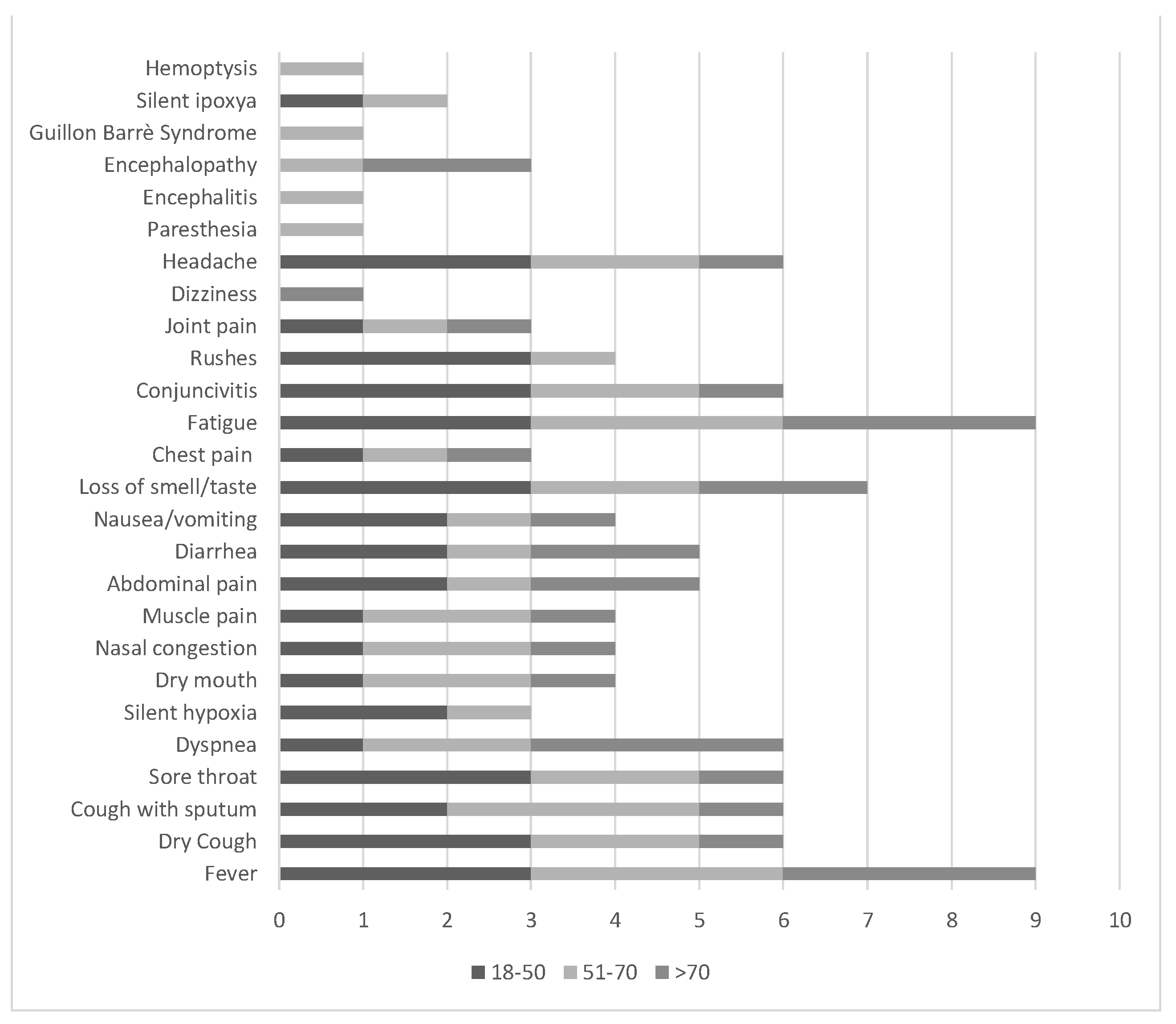
7. Clinical Manifestations: The “COVID-19 Planet”
8. Diagnosis of SARS-CoV-2 Infection
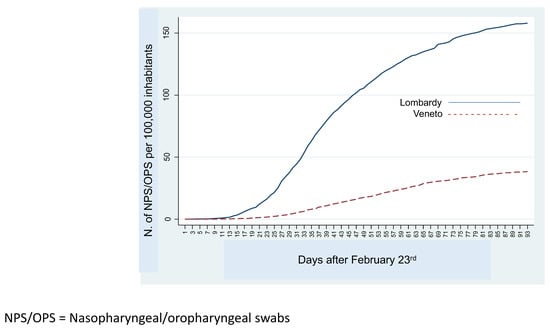
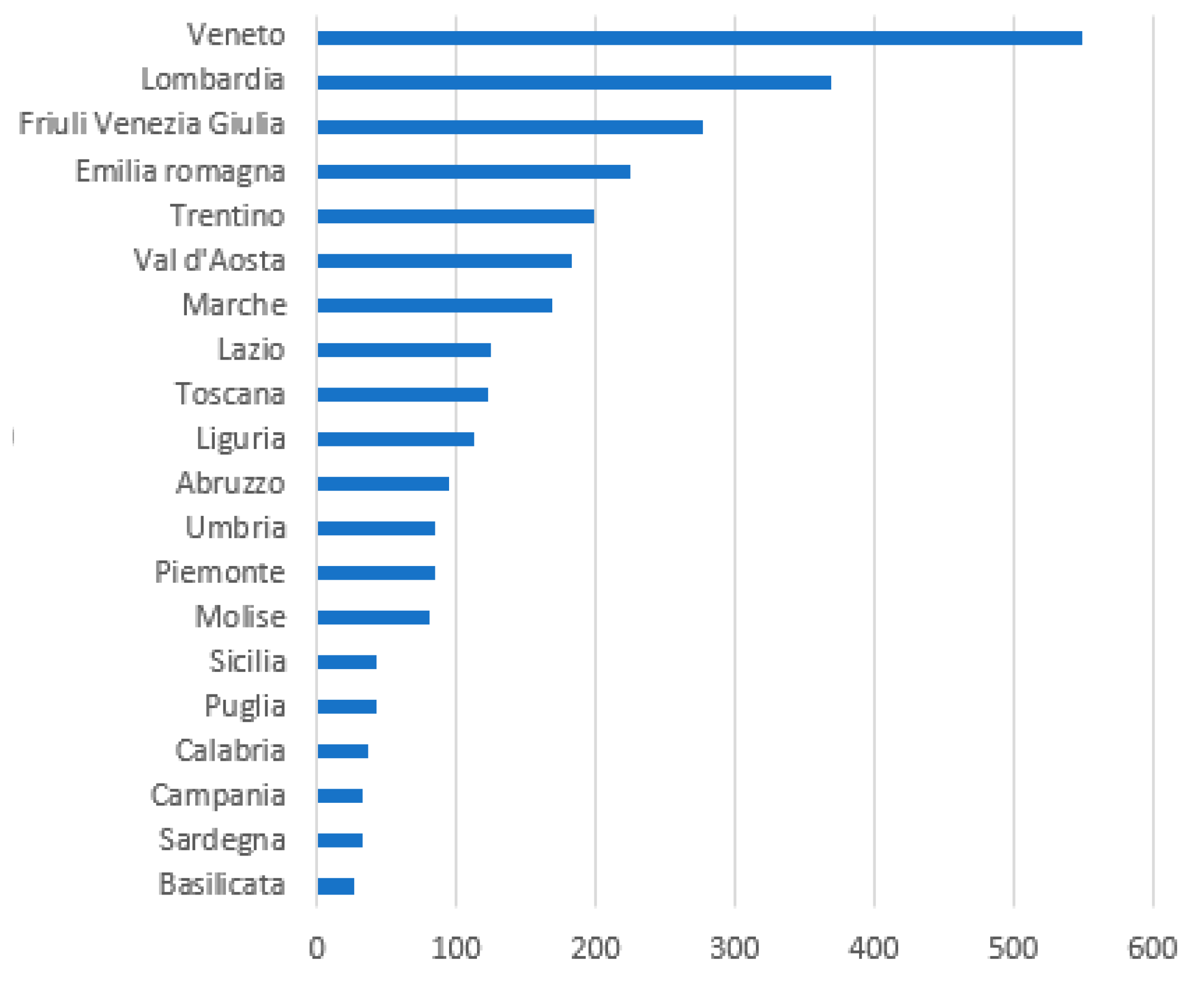
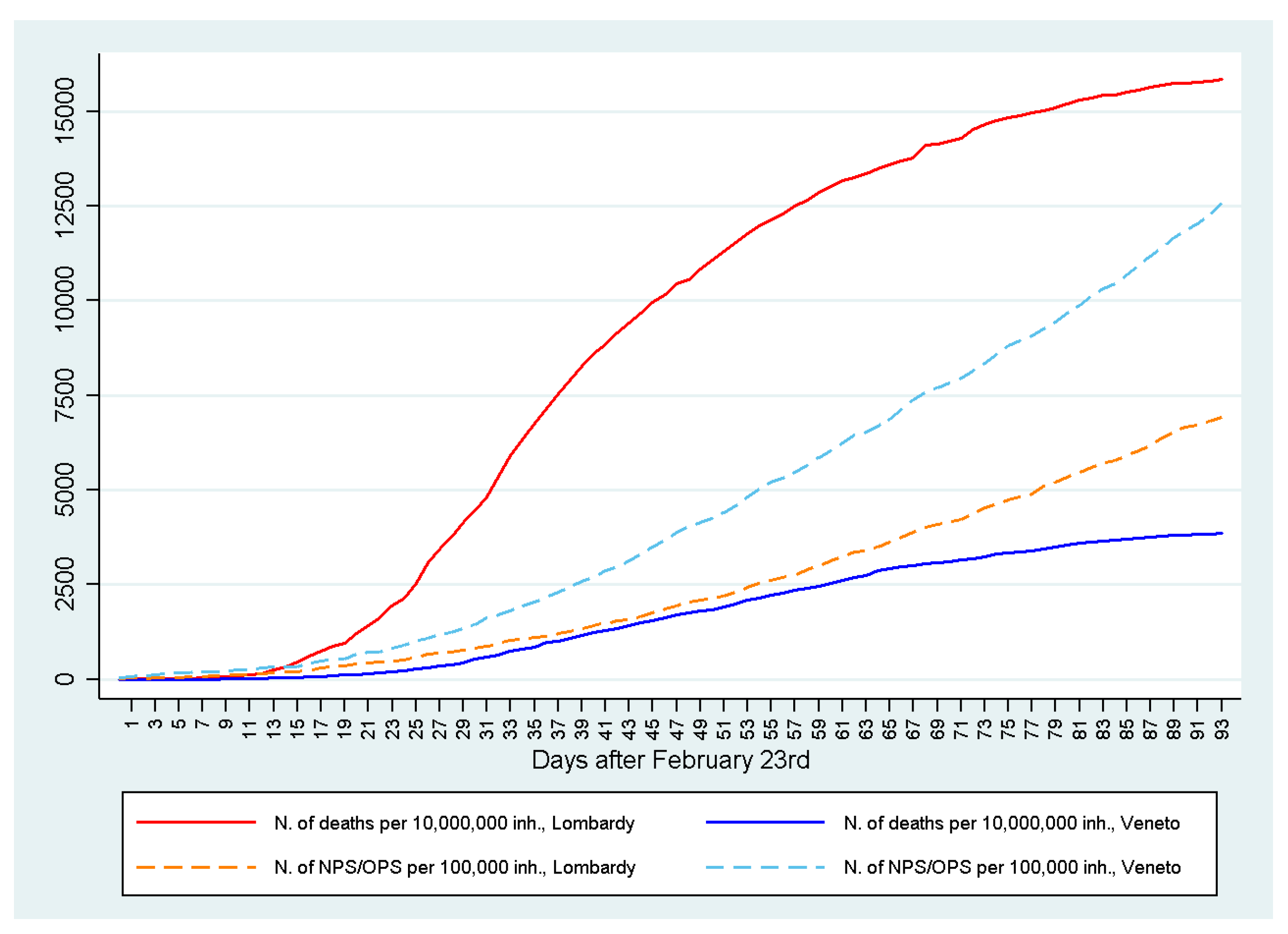
9. The Fight Against COVID-19 in Northern Italy
10. Conclusions
11. Perspectives
- RNA-Dependent RNA Polymerase Inhibitors (RdRp).Among RNA-Dependent RNA Polymerase Inhibitors (RdRp), there is Remdesivir (RDV), an adenosine analog previously used as anti-Ebola virus drug recently approved by Food and Drug Administration (FDA), to employ in emergency for the treatment of COVID-19 in adults and children hospitalized with severe disease [96]. The mechanism of action of RDV makes it potentially useful in the treatment of COVID-19. At least 23 studies of RDV are currently listed on various trial registers, intending to study 23,500 patients, but fewer than a quarter are double-blind, and some are uncontrolled observational studies [97]. Although measurement of efficacy will require ongoing randomized, placebo-controlled trials of RDV therapy, recently, in a cohort of patients hospitalized for severe COVID-19 who were treated with compassionate-use RDV, clinical improvement was observed in 36 of 53 patients (68%) [97]. More recently, our research team proposed to administer RDV by aerosol to combat SARS-CoV-2 in the upper respiratory tract in patients with severe clinical conditions and in ICUs in order to avoid possible side effects of systemic intravenous RDV administration [98]. Other recent potential effective antivirals include Favipiravir (Avigan), in the same class of the RDV, whose antiviral activity is exhibited through selectively targeting RdRp, are attractive targets for antiviral therapies, which interrupt the nucleotide incorporation process during viral RNA replication [90]. Favipiravir is a guanine analog approved for treatment against influenza virus infection in Japan and also can effectively inhibit replication of Ebola, yellow fever, chikungunya, norovirus, and enterovirus and could potentially exhibit effects against SARS-CoV-2 [99]. Neuraminidase and protease inhibitors have not proven to be particularly effective against SARS-CoV-2, although additional studies are necessary.
- Inhibitors of TMPRSS2 serine protease.The serine protease TMPRSS2 required for SARS- CoV-2 entry into host cells, highly conserved among the MERS-CoV, SARS-CoV-1, and SARS-CoV-2 viruses, has been identified as a promising target for treatment of COVID-19 [59]. The TMPRSS2 cell entry of SARS-CoV-2 inhibitors camostat mesylate, nafamostat, and bromhexine may treat COVID-19 [60].
- Anti-inflammatory molecules potentially efficacy against SARS-CoV-2.For severe COVID-19 illness, many experimental studies are underway to test the validity of anti-inflammatory molecules used successfully for other diseases that may have a potential anti-SARS-CoV-2 effect [100]. In this context, the use of corticosteroids (dexamethasone) as well as the immunomodulators sarilumab, tocilizumab, meplazumab, bevacizumab, baricitinib, avipatadil, may ameliorate the cytokine storm that contributes to mortality [101]. Tocilizumab has been widely used in rheumatic diseases, such as rheumatoid arthritis. It is an IL-6 receptor (IL-6R) blocker that can effectively block the IL-6 signal transduction pathway and thus is likely to become an effective drug for patients with severe COVID-19 and to reduce the mortality [102]. To block IL-17 pathway by biological drugs that are already available and used to treat different pathologies could also be a novel, additional strategy to treat patients infected by SARS-CoV-2 [71]. COVID-19 was initially underestimated especially in Lombardy and other regions of Northern Italy, in which hospitals have become amplifiers of SARS-CoV-2 infection. If these patients were treated early with anti-inflammatory drugs and anticoagulants, they could have avoided being hospitalized, where many of them died.
- Antagonists of Proteinases.The growing recognition of endothelitis and thrombosis in COVID-19 patients provides a strong incentive to determine the potential utility of antagonists of PAR1 (Proteinase-activated receptor 1) inhibitors to improve the outcome of such patients. PAR1 is widely expressed in cell types relevant to COVID-19 pathobiology, which include pneumocytes, endothelial cells, fibroblasts, and platelets. Activation of PAR1 by the serine protease thrombin is a critical element in platelet aggregation and coagulation. In particular, serine protease inhibitor Nafamostat, a serine protease inhibitor that works as an anticoagulant, has demonstrated satisfactory results in inhibiting the action of MERS-CoV and has been shown to be effective against SARS-CoV-2 infection, preventing membrane fusion [103].
- Hyperimmune plasma.The use of hyperimmune plasma obtained from convalescent patients recovered from the disease has shown to be a very promising and specific approach for the treatment of SARS-COV-2 infection [104]. This could represent a promising specific approach in the treatment of COVID-19 also on the basis of experience gained in other countries on a limited number of patients [105]. Convalescent plasma therapy also appears to be characterized by a high level of safety, as documented on all occasions in which it has been used in recent years, including COVID-19 itself. However, no randomized controlled trials or controlled non-randomized studies evaluating benefits and harms of convalescent plasma have been completed [106]. The European Commission in its recent Guide to Member States has pointed out that hyperimmune plasma would be a low-risk therapy immediately usable in selected categories of patients and a bridge alternative to be used while waiting for the production of a vaccine or the availability of drugs of proven efficacy [107]. So far, there is general consensus on the importance of combating uncontrolled inflammation, and the resulting ARDS or CSS, caused by this infection.
12. Highlights
- COVID-19 is a global pandemic that has currently emerged as one of the most intense and overwhelming viral infection for the humankind to manage.
- As the number of individuals infected with SARS-CoV-2 continues to rise globally, rapid diagnostics at earlier stages, therapeutics, and vaccines will become crucial for the management of the COVID-19 pandemic.
- There is some scientific evidence of efficacy of particular drugs such as antiviral (i.e., RDV), antiparasitic (i.e., hydroxychloroquine), and anti-inflammatory approaches (i.e., tocilizumab) for treatment of COVID-19.
- In the light of the exuberance of the host’s inflammatory response, a potential cause of lung damage and subsequent mortality, the priority should be to identify drugs with potent and specific antiviral, anti-inflammatory, and anticoagulant properties.
- A rational therapy would require careful evaluation of the cytokine profile of selected cohorts of subjects, which include SARS-CoV-2 positive patients with pneumonia or admitted to ICU. COVID-19 clinical evidence has shown that the first seven days of illness are crucial. To initiate therapeutic trials based on the rational use of anti-inflammatory drugs that directly inhibit the synthesis process of inflammatory cytokines including IL-16 and IL-17, would be desirable in the near future.
Author Contributions
Funding
Conflicts of Interest
References
- McIntosh, K. Coronaviruses: A comparative review. Curr. Top. Microbiol. Immunol. 1974, 63, 85–129. [Google Scholar]
- Weiss, S.R.; Navas-Martin, S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meulen, V. Biology of coronaviruses. Adv. Exp. Med. Biol. 1984, 173, 227–235. [Google Scholar] [PubMed] [Green Version]
- Wenzel, R.P.; Hendley, J.O.; Davies, J.A.; Gwaltney, J.M., Jr. Coronavirus infections in military recruits. Three-year study with coronavirus strains OC43 and 229E. Am. Rev. Respir. Dis. 1974, 109, 621–624. [Google Scholar] [PubMed]
- Van Bever, H.P.; Chng, S.Y.; Goh, D.Y. Childhood severe acute respiratory syndrome, coronavirus infections and asthma. Pediatr. Allergy. Immunol. 2004, 15, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Pfefferle, S.; Oppong, S.; Drexler, J.F.; Gloza-Rausch, F.; Ipsen, A.; Seebens, A.; Muller, M.A.; Annan, A.; Vallo, P.; Adu-Sarkodie, Y.; et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009, 15, 1377–1384. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef] [Green Version]
- Eccles, R. Understanding the symptoms of the common cold and influenza. Lancet Infect. Dis. 2005, 5, 718–725. [Google Scholar] [CrossRef]
- Messacar, K.; Abzug, M.J.; Dominguez, S.R. The Emergence of Enterovirus-D68. Microbiol. Spectr. 2016, 4, 1–12. [Google Scholar]
- Contini, C. International conference on chlamydial and Mycoplasma human infections. Future Microbiol. 2007, 2, 373–376. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS); World Health Organization: Geneva, Switzerland, 2003. Available online: http://tdr.who.int/csr/sars/WHOconsensus.pdf (accessed on 10 June 2020).
- Burks, J.S.; DeVald, B.L.; Jankovsky, L.D.; Gerdes, J. Two coronaviruses isolated from central nervous system tissue of two multiple sclerosis patients. Science 1980, 209, 933–934. [Google Scholar] [CrossRef] [PubMed]
- Resta, S.; Luby, J.P.; Rosenfiled, C.R.; Siegel, J.D. Isolation and propagation of a human enteric coronavirus. Science 1985, 229, 978–981. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- WHO. Coronavirus Disease (COVID-19) Outbreak. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 29 February 2020).
- Zhai, S.L.; Wei, W.K.; Lv, D.H.; Xu, Z.H.; Chen, Q.L.; Sun, M.F.; Li, F.; Wang, D. Where did SARS-CoV-2 come from? Vet. Rec. 2020, 186, 254. [Google Scholar] [CrossRef] [Green Version]
- Benvenuto, D.; Giovanetti, M.; Ciccozzi, A.; Spoto, S.; Angeletti, S.; Ciccozzi, M. The 2019-new coronavirus epidemic: Evidence for virus evolution. J. Med. Virol. 2020, 92, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Contini, C.; Di Nuzzo, M.; Barp, N.; Bonazza, A.; De Giorgio, R.; Tognon, M.; Rubino, S. The novel zoonotic COVID-19 pandemic: An expected global health concern. J. Infect. Dev. Ctries. 2020, 14, 254–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [Green Version]
- Riordan, J.F. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003, 4, 225. [Google Scholar] [CrossRef] [Green Version]
- Nicholls, J.; Dong, X.P.; Jiang, G.; Peiris, M. SARS: Clinical virology and pathogenesis. Respirology 2003, 8, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Goubar, A.; Bitar, D.; Cao, W.C.; Feng, D.; Fang, L.Q.; Desenclos, J.C. An approach to estimate the number of SARS cases imported by international air travel. Epidemiol. Infect. 2009, 137, 1019–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; He, L.; Zhang, Q.; Huang, Z.; Che, X.; Hou, J.; Wang, H.; Shen, H.; Qiu, L.; Li, Z.; et al. Organ distribution of severe acute respiratory syndrome(SARS) associated coronavirus (SARS-CoV) in SARS patients: Implications for pathogenesis and virus transmission pathways. J. Pathol. 2004, 203, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Enjuanes, L.; Zuñiga, S.; Castaño-Rodriguez, C.; Gutierrez-Alvarez, J.; Canton, J.; Sola, I. Molecular basis of coronavirus virulence and vaccine development. Adv. Virus Res. 2016, 96, 245–286. [Google Scholar] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Alnaeem, A.; Kasem, S.; Qasim, I.; Al-Doweriej, A.; Refaat, M.; Al-Shabebi, A.; Hemida, M.G. The dipeptidyl peptidase-4 expression in some MERS-CoV naturally infected dromedary camels in Saudi Arabia 2018–2019. Virus Dis. 2020, 6, 1–4. [Google Scholar] [CrossRef]
- Ramadan, N.; Shaib, H. Middle East respiratory syndrome coronavirus (MERS-CoV). Germs 2019, 9, 35–42. [Google Scholar] [CrossRef]
- Islam, A.; Epstein, J.H.; Rostal, M.K.; Islam, S.; Rahman, M.Z.; Hossain, M.E.; Uzzaman, M.S.; Munster, V.J.; Peiris, M.; Flora, M.S.; et al. Middle East respiratory syndrome coronavirus in dromedary camels: An outbreak investigation. Lancet Infect. Dis. 2014, 14, 140–145. [Google Scholar]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Deslandes, A.; Berti, V.; Tandjaoui-Lambotte, Y.; Alloui, C.; Carbonnelle, E.; Zahar, J.R.; Brichler, S.; Cohen, Y. SARS-CoV-2 was already spreading in France in late December 2019. Int. J. Antimicrob. Agents 2020, 55, 106006. [Google Scholar] [CrossRef]
- Capobianchi, M.R.; Rueca, M.; Messina, F.; Giombini, E.; Carletti, F.; Colavita, F.; Castilletti, C.; Lalle, E.; Bordi, L.; Vairo, F.; et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin. Microbiol. Infect. 2020, 26, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H. Genomic characterization and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Xiao, X.; Wei, X.; Li, J.; Yang, J.; Tan, H.; Zhu, J.; Zhang, Q.; Wu, J.; Liu, L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020, 92, 595–601. [Google Scholar] [CrossRef] [Green Version]
- Lam, T.T.; Shum, M.H.; Zhu, H.C.; Tong, Y.G.; Ni, X.B.; Liao, Y.S.; Wei, W.; Cheung, W.Y.M.; Li, W.J.; Li, L.F.; et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. Nature 2020. [Google Scholar] [CrossRef] [Green Version]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.M.; Raj, S.V.; Zaki, A.M.; Osterhaus, A.D.M.E.; Haagmans, B.L.; Gorbalenya, A.E.; Snijder, E.J.; et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 2012, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Lescure, F.X.; Bouadma, L.; Nguyen, D.; Parisey, M.; Wicky, P.H.; Behillil, S.; Gaymard, A.; Bouscambert-Duchamp, M.; Donati, F.; Le Hingrat, Q.; et al. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020, 20, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Hoehl, S.; Rabenau, H.; Berger, A.; Kortenbusch, M.; Cinatl, J.; Bojkova, D.; Behrens, P.; Böddinghaus, B.; Götsch, U.; Naujoks, F.; et al. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N. Engl. J. Med. 2020, 382, 1278–1280. [Google Scholar] [CrossRef]
- Rothe, C.; Schunk, M.; Sothmann, P.; Gisela Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef] [Green Version]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.C.; Bai, W.Z.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Qing, H.; Li, Z.; Yang, Z.; Shi, M.; Huang, Z.; Song, J.; Song, Z. The possibility of COVID-19 transmission from eye to nose. Acta Ophthalmol. 2020, 98, e388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozkurt, B.; Egrilmez, S.; Şengör, T.; Yıldırım, O.; İrkeç, M. The COVID-19 Pandemic: Clinical Information for Ophthalmologists. Turk. J. Ophthalmol. 2020, 50, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Tong, J.; Liu, M.; Shen, Y.; Guo, D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, W.J.; Liang, W.H.; Zhao, Y.; Liang, H.R.; Chen, Z.S.; Li, Y.M.; Liu, X.Q.; Chen, R.C.; Tang, C.L.; Wang, T.; et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: A nationwide analysis. Eur. Respir. J. 2020, 55, 2000547. [Google Scholar] [CrossRef] [Green Version]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Ling, Y.; Xu, S.B.; Lin, Y.X.; Tian, D.; Zhu, Z.Q.; Dai, F.H.; Wu, F.; Song, Z.G.; Huang, W.; Chen, J.; et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020, 133, 1039–1043. [Google Scholar] [CrossRef]
- Wang, W.; Xum, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Mim, M.A.; Naznin Rakhi, N.; Saha, O.; Rahaman, M.M. Recommendation of fecal specimen for routine molecular detection of SARS-CoV-2 and for COVID-19 discharge criteria. Pathog. Glob. Health 2020, 114, 168–169. [Google Scholar]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; Do Cao, J.; Benachi, A.; De Luca, D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef]
- Kasraeian, M.; Zare, M.; Vafaei, H.; Asadi, N.; Faraji, A.; Bazrafshan, K.; Roozmeh, S. COVID-19 Pneumonia and Pregnancy; A Systematic Review and Meta-Analysis. J. Matern. Fetal Neonatal Med. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.A. An analysis of 38 pregnant women with COVID-19, their newborn infants, and maternal-fetal transmisssion of SARS-CoV-2: Maternal coronavirus infections and pregnancy outcomes. Arch. Pathol. Lab. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Calderaro, A.; Arcangelettim, M.C.; De Conto, F.; Buttrini, M.; Montagna, P.; Montecchini, S.; Ferraglia, F.; Pinardi, F.; Chezzi, C. SARS-CoV-2 infection diagnosed only by cell culture isolation before the local outbreak in an Italian seven-week-old suckling baby. Int. J. Infect. Dis. 2020, 96, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramesh, N.; Siddaiah, A.; Joseph, B. Tackling corona virus disease 2019 (COVID 19) in workplaces. Indian J. Occup. Environ. Med. 2020, 24, 16–18. [Google Scholar] [PubMed]
- Brenner, S.R. Covid-19, TMPRSS2, and whether android regulation affects pandemic virus gender incidence and age distribution of disease. Med. Hypotheses 2020, 140, 109773. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Zhou, B.; Zhang, L.; Balaji, K.S.; Wei, C.; Liu, X.; Chen, H.; Peng, J.; Fu, J. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol. Biol. Rep. 2020, 1–10. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Lui, R.N.; Sung, J.J. Covid-19 and the digestive system. J. Gastroenterol. Hepatol. 2020, 35, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Matthai, J.; Shanmugam, N.; Sobhan, P.; Indian Society of pediatric gastroenterology, hepatology and nutrition; Pediatric Gastroenterology Chapter Of Indian Academy Of Pediatrics Coronavirus Disease (COVID-19) and the Gastrointestinal System in Children. Coronavirus Disease (COVID-19) and the Gastrointestinal System in Children. Indian Pediatr. 2020, 57, 533–535. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Ruscitti, P.; Berardicurti, O.; Iagnocco, A.; Giacomelli, R. Cytokine storm syndrome in severe COVID-19. Autoimmun. Rev. 2020, 19, 102562. [Google Scholar] [CrossRef]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef]
- Zhu, J.; Zhong, Z.; Ji, P.; Li, H.; Li, B.; Pang, J.; Zhang, J.; Zhao, C. Clinicopathological characteristics of 8697 patients with COVID-19 in China: A meta-analysis. Fam. Med. Community Health 2020, 8, e000488. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Badawi, A.; Ryoo, S.G. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): A systematic review and meta-analysis. Int. J. Infect. Dis. 2016, 49, 129–133. [Google Scholar] [CrossRef] [Green Version]
- Wichmann, D.; Sperhake, J.P.; Lütgehetmann, M.; Steurer, S.; Edler, C.; Heinemann, A.; Heinrich, F.; Mushumba, H.; Kniep, I.; Schröder, A.S.; et al. Autopsy findings and venous thromboembolism in patients with COVID-19. Ann. Intern. Med. 2020, M20-2003. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes. Infect. 2020, 22, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020, 109, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Englund, K. Clinical presentation and course of COVID-19. Cleve. Clin. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Scangas, G.A.; Bleier, B.S. Anosmia: Differential diagnosis, evaluation, and management. Am. J. Rhinol. Allergy 2017, 31, 3–7. [Google Scholar] [CrossRef]
- Carod-Artal, F.J. Neurological complications of coronavirus and COVID-19. Rev. Neurol. 2020, 70, 311–322. [Google Scholar]
- Clerkin, K.J.; Fried, J.A.; Raikhelkar, J.; Sayer, G.; Griffin, J.M.; Masoumi, A.; Jain, S.S.; Burkhoff, D.; Kumaraiah, D.; Rabbani, L.; et al. COVID-19 and Cardiovascular Disease. Circulation 2020, 141, 1648–1655. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, Y.; Miao, X.; Streithorst, Y.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.W.; Kim, N.G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv 2020. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. Version 2. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Int. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance (accessed on 3 March 2020).
- Wang, H.; Li, X.; Li, T.; Zhang, S.; Wang, L.; Wu, X.; Liu, J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.; Guarner, J. Role of serology in the COVID-19 pandemic. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Elaborazione GIMBE Dati Protezione Civile. Available online: https://coronavirus.gimbe.org/ (accessed on 19 March 2020).
- Contini, C.; Migali, G.; Rizzo, L.; Secomandi, R. A Cosa Servono I Tamponi. 2020. Available online: https://www.lavoce.info/archives/65587/a-cosa-servono-i-tamponi/ (accessed on 15 April 2020).
- Cao, G.W.; Zhang, B.X.; Chen, X.P. Strategy and Policy Working Group for NCIP Epidemic Response. Urgent research agenda for the novel coronavirus epidemic: Transmission and non-pharmaceutical mitigation strategies. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 1–6. [Google Scholar]
- Jin, Y.; Cai, L.; Cheng, Z.; Cheng, H.; Deng, T.; Fan, Y.P.; Fang, C.; Huang, D.; Huang, L.Q.; Huang, Q.; et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil. Med. Res. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavezzo, E.; Franchin, E.; Ciavarella, C.; Cuomo-Dannenburg, G.; Barzon, L.; Del Vecchio, C.; Rossi, L.; Manganelli, R.; Loregian, A.; Navarin, N.; et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature 2020. [Google Scholar] [CrossRef]
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [Green Version]
- Ahn, D.G.; Shin, H.J.; Kim, M.H.; Lee, S.; Kim, H.S.; Myoung, J.; Kim, B.T.; Kim, S.J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef]
- Ferner, R.E.; Aronson, J.K. Remdesivir in covid-19. BMJ 2020, 369, m1610. [Google Scholar] [CrossRef] [Green Version]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.X.; et al. Compassionate use of Remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Contini, C.; Gallenga, C.E.; Neri, G.P.; Maritati, M.; Conti, P. A new pharmacological approach based on Remdesivir aerosolized administration on SARS-CoV-2 pulmonary inflammation: A possible and rational therapeutic application. Med. Hypotheses 2020, 144, 109876. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, P.; Ciurtin, C.; Scully, M.; Levi, M.; Chambers, R.C. JAK inhibitors in COVID-19: Need for vigilance regarding increased inherent thrombotic risk. Eur. Respir. J. 2020, 2001919. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Phelan, A.; Griffin, I.; Tucker, C.; Oechsle, O.; Smith, D.; Richardson, P. COVID-19: Combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020, 20, 400–402. [Google Scholar] [CrossRef]
- Zhang, S.; Li, L.; Shen, A.; Chen, Y.; Qi, Z. Rational use of Tocilizumab in the treatment of novel coronavirus pneumonia. Clin. Drug Investig. 2020, 40, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, H.C.; Roback, J.D. Convalescent plasma: Therapeutic hope or hopeless strategy in the SARS-CoV-2 pandemic. Transfus. Med. Rev. 2020. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Valk, S.J.; Piechotta, V.; Chai, K.L.; Doree, C.; Monsef, I.; Wood, E.M.; Lamikanra, A.; Kimber, C.; McQuilten, Z.; So-Osman, C.; et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: A rapid review. Cochrane Database Syst. Rev. 2020, 5. [Google Scholar] [CrossRef]
- European Commission Directorate General for Health and Food Safety. An EU Programme of COVID-19 Convalescent Plasma Collection and Transfusion: Guidance on Collection, Testing, Processing, Storage, Distribution and Monitored Use. Available online: https://ec.europa.eu/health/sites/health/files/blood_tissues_organs/docs/guidance_plasma_covid19_e (accessed on 8 April 2020).
| Virus and Viral Diseases |
| Orthomyxovirus (Influenza) |
| Paramyxovirus (Parainfluenza) PIV-1, -2, -3, and 4 |
| Human metapneumovirus * |
| Human rhinovirus § |
| Adenovirus |
| Coronavirus ^ |
| Enterovirus ^^ |
| Respiratory syncytial virus ° |
| Bacteria and Bacterial Infections |
| Haemophilus influenzae pneumonia |
| Streptococcus pneumoniae pneumonia |
| Moraxella catarrhalis pneumonia |
| Bordetella pertussis and Bordetella parapertussis |
| Legionella pneumophila °° |
| Mycoplasma pneumoniae °° |
| Chlamydia pneumoniae °° |
| SARS-CoV-1 | MERS-CoV | SARS-CoV-2 | |||
|---|---|---|---|---|---|
| Origin | Guangdong Province, China | Saudi Arabia | Wuhan, China | Rome, Italy | Paris, France |
| Potential reservoir | Bat * | Bat * | Bat * | ||
| Intermediate host | Palm-civet | Camel/dromedary | Pangolin (to be established yet) | ||
| Final host | Humans | Humans | Humans | ||
| NCBI GenBank No. | AY278741.1 | NC_019843.3 | MN908947.3 | MT077125.1 | EPI_ISL_406596 (GISAID No.) |
| Reference | [37] | [38] | [39] | [33] | [40] |
| Complete genome length (nt) | 29,727 | 30,119 | 29,903 | 29,785 | 29,853 |
| Spike gene location (nt) | 21,492–25,259 | 21,456–25,517 | 21,563–25,384 | 21,507–25,328 | 21,563–25,384 |
| Spike gene length (nt) | 3768 | 4062 | 3822 | ||
| Spike genomic sequence homology with Bat * # | 74.21% (Query cover 97 %) | 75.47% (Query cover 8%) | 92.89% (Query cover 100%) | 92.83% (Query cover 100%) | |
| Spike protein length (aa) | 1255 | 1353 | 1273 | ||
| Spike amino acid sequence homology with Bat * # | 76.67% (Query cover 100%) | 34.22% (Query cover 87%) | 97.41% (Query cover 100%) | 97.25% (Query cover 100%) | |
| The predominant receptor | Human angiotensin-converting enzyme-2 (ACE2) | Human dipeptidyl peptidase 4 (DPP4 or CD26) | Human angiotensin-converting enzyme-2 (ACE2) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contini, C.; Caselli, E.; Martini, F.; Maritati, M.; Torreggiani, E.; Seraceni, S.; Vesce, F.; Perri, P.; Rizzo, L.; Tognon, M. COVID-19 Is a Multifaceted Challenging Pandemic Which Needs Urgent Public Health Interventions. Microorganisms 2020, 8, 1228. https://doi.org/10.3390/microorganisms8081228
Contini C, Caselli E, Martini F, Maritati M, Torreggiani E, Seraceni S, Vesce F, Perri P, Rizzo L, Tognon M. COVID-19 Is a Multifaceted Challenging Pandemic Which Needs Urgent Public Health Interventions. Microorganisms. 2020; 8(8):1228. https://doi.org/10.3390/microorganisms8081228
Chicago/Turabian StyleContini, Carlo, Elisabetta Caselli, Fernanda Martini, Martina Maritati, Elena Torreggiani, Silva Seraceni, Fortunato Vesce, Paolo Perri, Leonzio Rizzo, and Mauro Tognon. 2020. "COVID-19 Is a Multifaceted Challenging Pandemic Which Needs Urgent Public Health Interventions" Microorganisms 8, no. 8: 1228. https://doi.org/10.3390/microorganisms8081228
APA StyleContini, C., Caselli, E., Martini, F., Maritati, M., Torreggiani, E., Seraceni, S., Vesce, F., Perri, P., Rizzo, L., & Tognon, M. (2020). COVID-19 Is a Multifaceted Challenging Pandemic Which Needs Urgent Public Health Interventions. Microorganisms, 8(8), 1228. https://doi.org/10.3390/microorganisms8081228