Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus
Abstract
:1. Introduction
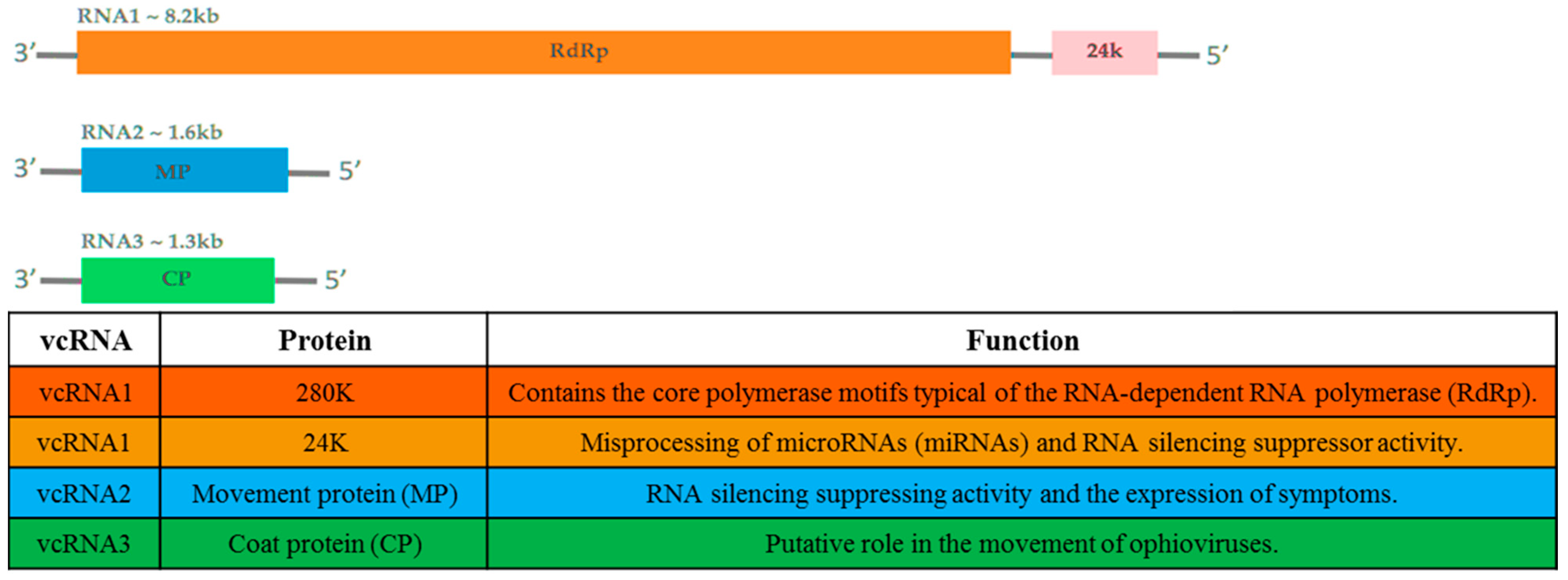
2. Taxonomy, Genome Structure, and Organization
3. Symptoms and Economical Impact
4. Transmission and Epidemiology
5. Signaling Pathways in Citrus Psorosis Pathogenesis
6. Methods to Detect the Disease
6.1. Biological Indexing and Cross Protection
6.2. Antibodies–Antigen-Based Methods
6.3. Nucleic Acid-Based Methods
| Tested Methods Main Findings | References |
|---|---|
| DAS- and TAS-ELISA | [82] |
| TAS-ELISA is five times more sensitive than DAS-ELISA. Threshold of sensitivity: CPsV is still detectable in a leaf extract dilution 1/31,250 for TAS-ELISA and only 1/6,250 for DAS-ELISA, with optical density (OD) of 0.036 and 0.027, respectively. High specificity of TAS-ELISA in non-infected samples. It always gives low OD values (0.006). | |
| RT- and Heminested RT-PCR Heminested RT-PCR allows the detection of eight CPsV isolates, whereas no signal was detected with the conventional RT-PCR using only two primers. Threshold of sensitivity: CPsV is still detectable until a dilution of 10−5 with heminested RT-PCR. | [78] |
| DAS-, TAS-ELISA, and DTBIA DTBIA for CPsV detection in the same trees is reliable only when using young shoots. It is less consistent in old leaves. TAS-ELISA sensitivity is two to eight times higher in young shoots compared to old ones. High specificity of ELISA tests: the ELISA readings of positive controls were within the range 0.300–0.550, and those of the negative controls were within the range 0.002–0.013. | [46] |
| TAS-ELISA-HRP and TAS-ELISA-AP TAS-ELISA-HRP readings are at least two times higher than those of TAS-ELISA-AP after two hours of incubation. TAS-ELISA-HRP is highly sensitive and comparable to the level of the RT-PCR. It gives the same results as RT-PCR using primers CPV1 and CPV2 (Table 2). High specificity: TAS-ELISA in non-infected samples always gives low OD values. | [33] |
| One-Step RT-PCR and DAS-ELISA One-step RT-PCR is more sensitive than DAS-ELISA. (One-step RT-PCR allows the detection of 22/30 positive samples, whereas DAS-ELISA allows the detection of only 7/30 positive samples and gives 15/30 false negatives.) The primer pair Ps66/Ps65 is more sensitive than two other primer pairs (Table 2). Ps66/Ps65 allows the detection of 22/30 positive samples, whereas CPV1/CPV2 and CPsV-f/CPsV-r allow the detection of only 11/22 and 12/22, respectively. | [84] |
| Real-Time RT-PCR and RT-PCR Real-time RT-PCR is 100 times more sensitive than conventional RT-PCR. High specificity: no amplification was obtained with healthy controls or with plant tissue infected with other citrus viruses and viroid. | [34] |
| Real-Time RT-PCR and TAS-ELISA Real time RT-PCR sensitivity is three orders of magnitude higher than that of TAS-ELISA. TAS-ELISA gives eight false-negative field samples. Two of these eight samples showed symptoms of psorosis; the remaining samples were asymptomatic. Threshold of sensitivity: positive TAS-ELISA readings were only obtained with undiluted samples or samples diluted by 10−1. In contrast, real-time RT-PCR was positive up to the 10−4 dilution. | [75] |
| Singleplex and Triplex Real-Time RT-PCR | [86] |
| No significant differences in detection between singleplex and triplex assays in CPsV detection. The specificity was not affected by the inclusion of the three singleplex assays in a multiplex real-time RT-PCR reaction. No amplification was detected with samples from healthy citrus or water control. |
| Name of RT-PCR Test Primer/Probe Name | Sequence | Tm (°C) | Targeted RNA/Region (Genomic Coordinates) | Size of the Expected Product | References |
|---|---|---|---|---|---|
| RT-PCR | |||||
| Primer 1 | 5′-ACAATAAGCAAGACAAC-3′ | 45 | RNA1 (DN a) | 218 bp | [44] |
| DN | 5′-CCATGTCACTTCTATTC-3′ | ||||
| CPV1 | 5′-GCTTCCTGGAAAAGCTGATG-3′ | 50 | RNA3/CP (665–684 a) | 600 bp | [25] |
| CPV2 | 5′-TCTGTTTTTGTCAACACACTCC-3′ | RNA3/CP (1243–1264 a) | |||
| Ps65 | 5′-TGCCATCTGGAGTGAGGCT-3′ | 45 | RNA3/CP (1182–1200 b) | 430 bp | [49] |
| Ps66 | 5′-TCGAAGCTGTATGATGGTGA-3′ | RNA3/CP (768–787 b) | |||
| ConsF | 5′-ACAAAGAAATTCCCTGCAAGGG-3′ | 58 and 59–60 for singleplex and multiplex test, respectively. | RNA3/ CP (766–1200 c) | 411 bp | [85] |
| ConsR | 5′-AAGTTTCTATCATTCTGAAACCC-3′ | ||||
| CPsV-f | 5′-TGAGGAA/GTTGAGCCATGC-3′ | 58 | RNA3/CP d | 390 bp | [42] |
| CPsV-r | 5′-CCATCTGGAGTGAGGCTGTA-3′ | ||||
| Heminested RT-PCR | |||||
| Primer 1 | 46 and 47 for the first PCR done with primers 1–7 and the heminested PCR done with primers 6–7, respectively. | RNA1 (DN a) | 195 bp | [78] | |
| Primer 6 | 5′-GAGGAAGGTATTTCCATAGG-3′ | ||||
| Primer 7 | 5′-CCTATTAATGATAATTGCAC-3′ | ||||
| Real-time RT-PCR | |||||
| CPV200f | 5′-GCWGGWAATCGRTCTGTGAGRTAT-3′ | 58 and 60 for singleplex and multiplex tests, respectively. | RNA3/CP (200–223 e) | 106 bp | [34] |
| CPV287r | 5′-AGCAAWGGCATCARGGAYTC-3′ | RNA3/CP (287–306 e) | |||
| CPVp | 5′-Cy5b-TCYCCTGCTGTTGGWGCAACTYC-BHQ1-3′ | RNA3/CP (263–285 e) | |||
| CP1cf | 5′-GTTCAAGATGGAGCAAGTTGATGG-3′ | 56 | RNA3/CP (738–850 f) | 113 bp | [75] |
| CP3r | 5′-GAGACCCTTGTGTAAAAACCAGCAC-3′ | ||||
| CPsV-792 F1 | 5′-TCACAAATCAGTGAGGAATTGAGC-3′ | 60 for singleplex tests. For multiplex tests, the Tm was defined according to the manufacturer’s recommendation. | RNA3/CP (792–816 g) | 154 bp | [86] |
| CPsV-791 F2 | 5′-CACAAATCAGTGATGAATTGAGCC-3′ | RNA3/CP (793–817 g) | |||
| CPsV-946 R1 | 5′-GCAAACCCAGCATATCTCACAG-3′ | RNA3/CP (947–925 g) | |||
| CPsV-946 R2 | 5′-CGCAAACCCAGCATATCTTACAG-3′ | RNA3/CP (948–925 g) | |||
| CPsV-851 p-VIC | 5′-TCTCAAGATTGATATAGACAAC-3′ | RNA3/CP (851–873 g) | |||
7. Strategies to Control the Disease
7.1. CPsV Sanitation
7.2. Plant Biotechnology and Genetic Engineering for Resistance
7.3. Breeding for Resistance
8. Disease Situation in the Mediterranean Region: Focus on Morocco
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harvey, H.L. Citrus psorosis. J. Dep. Agric. West. Aust. 1961, 2, 493–494. [Google Scholar]
- Roistacher, C.N. Diagnosis and management of virus and virus like diseases of citrus. In Diseases of Fruits and Vegetables; Naqvi, S.A.M.H., Ed.; Springer: Dordrecht, The Netherlands, 2004; Volume I, pp. 109–189. [Google Scholar]
- Roistacher, C.N. Psorosis complex: Psorosis-A, psorosis-B and ringspot. In Graft-Transmissible Diseases of Citrus (Handbook for Detection and Diagnosis); FAO: Rome, Italy, 1991; pp. 115–126. [Google Scholar]
- Vaira, A.M.; Milne, R.G. Ophiovirus. In Desk Encyclopedia of Plant and Fungal Virology; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 243–250. ISBN 9780123751485. [Google Scholar]
- Hernández-Rodríguez, L.; Bertalmio, A.; Rubio, L.; Rolón, R.; Maeso, D.; Rivas, F. Inability of the brown citrus aphid (Toxoptera citricida) to transmit citrus psorosis virus under controlled conditions. J. Citrus Pathol. 2020, 7, 1–6. [Google Scholar]
- Gomez, C.A. Metodologías de Diagnóstico de CPsV (psorosis) en Cítricos; Seretaria de Agroindustria: Buenos Aires, Argentina, 2019; Volume 3200, pp. 1–6. [Google Scholar]
- Chambers, G.; Englezou, A. Protecting Australian Citrus Germplasm through Improved Diagnostic Tools; Hort Innovation: Sydney, Australia, 2018. [Google Scholar]
- Bové, J.M. Maladies à virus des citrus dans les pays du Bassin Méditerranéen. Fruits 1967, 22, 125–140. [Google Scholar]
- Derrick, K.S.; Timmer, L.W. Citrus blight and other diseases of recalcitrant etiology. Annu. Rev. Phytopathol. 2000, 38, 181–205. [Google Scholar] [CrossRef]
- Wang, N. The citrus huanglongbing crisis and potential solutions. Mol. Plant. 2019, 12, 607–609. [Google Scholar] [CrossRef]
- Albrecht, U.; Fiehn, O.; Bowman, K.D. Metabolic variations in different citrus rootstock cultivars associated with different responses to Huanglongbing. Plant. Physiol. Biochem. 2016, 107, 33–44. [Google Scholar] [CrossRef]
- Moreno, P.; Guerri, J.; García, M.L. The psorosis disease of citrus: A pale light at the end of the tunnel. J. Citrus Pathol. 2015, 2, 1–18. [Google Scholar]
- Epstein, A.H. Root graft transmission of tree pathogens. Annu. Rev. Phytopathol. 1978, 16, 181–192. [Google Scholar] [CrossRef]
- García, M.L.; Dal Bó, E.; da Graça, J.V.; Gago-Zachert, S.; Hammond, J.; Moreno, P.; Natsuaki, T.; Pallás, V.; Navarro, J.A.; Reyes, C.A.; et al. ICTV virus taxonomy profile: Ophioviridae. J. Gen. Virol. 2017, 98, 1161–1162. [Google Scholar] [CrossRef] [Green Version]
- Naum-Onganía, G.; Gago-Zachert, S.; Peña, E.; Grau, O.; Garcia, M.L. Citrus psorosis virus RNA 1 is of negative polarity and potentially encodes in its complementary strand a 24K protein of unknown function and 280K putative RNA dependent RNA polymerase. Virus Res. 2003, 96, 49–61. [Google Scholar] [CrossRef]
- Reyes, C.A.; Ocolotobiche, E.E.; Marmisollé, F.E.; Robles Luna, G.; Borniego, M.B.; Bazzini, A.A.; Asurmendi, S.; García, M.L. Citrus psorosis virus 24K protein interacts with citrus miRNA precursors, affects their processing and subsequent miRNA accumulation and target expression. Mol. Plant. Pathol. 2016, 17, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Robles Luna, G.; Reyes, C.A.; Peña, E.J.; Ocolotobiche, E.; Baeza, C.; Borniego, M.B.; Kormelink, R.; García, M.L. Identification and characterization of two RNA silencing suppressors encoded by ophioviruses. Virus Res. 2017, 235, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Sánchez de la Torre, M.E.; García, M.L.; Riva, O.; Bó, E.D.; Jones, L.; Zandomeni, R.; Grau, O. Genome Organization of the top component of citrus psorosis virus and identification of the coat protein gene. Int. Organ. Citrus Virol. Conf. Proc. 2000, 14, 345–346. [Google Scholar]
- Sánchez de la Torre, M.E.; López, C.; Grau, O.; García, M.L. RNA 2 of Citrus psorosis virus is of negative polarity and has a single open reading frame in its complementary strand. J. Gen. Virol. 2002, 83, 1777–1781. [Google Scholar] [CrossRef] [PubMed]
- Robles Luna, G.; Peña, E.J.; Borniego, M.B.; Heinlein, M.; Garcia, M.L. Ophioviruses CPsV and MiLBVV movement protein is encoded in RNA 2 and interacts with the coat protein. Virology 2013, 441, 152–161. [Google Scholar] [CrossRef]
- Borniego, M.B.; Karlin, D.; Peña, E.J.; Robles Luna, G.; García, M.L. Bioinformatic and mutational analysis of ophiovirus movement proteins, belonging to the 30K superfamily. Virology 2016, 498, 172–180. [Google Scholar] [CrossRef]
- Robles Luna, G.; Peña, E.J.; Borniego, M.B.; Heinlein, M.; García, M.L. Citrus psorosis virus movement protein contains an aspartic protease required for autocleavage and the formation of tubule-like structures at plasmodesmata. J. Virol. 2018, 92, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Velázquez, K.; Pina, J.A.; Navarro, L.; Moreno, P.; Guerri, J. Association of citrus psorosis B symptoms with a sequence variant of the Citrus psorosis virus RNA 2. Plant. Pathol. 2012, 61, 448–456. [Google Scholar] [CrossRef]
- da Graca, J.V.; Lee, R.F.; Moreno, P.; Civerolo, E.L.; Derrick, K.S. Comparison of isolates of citrus ringspot, psorosis, and other viruslike agents of citrus. Plant. Dis. 1991, 75, 613–616. [Google Scholar] [CrossRef]
- Barthe, G.A.; Ceccardi, T.L.; Manjunath, K.L.; Derrick, K.S. Citrus psorosis virus: Nucleotide sequencing of the coat protein gene and detection by hybridization and RT-PCR. J. Gen. Virol. 1998, 79, 1531–1537. [Google Scholar] [CrossRef]
- Peña, E.J.; Robles Luna, G.; Zanek, M.C.; Borniego, M.B.; Reyes, C.A.; Heinlein, M.; García, M.L. Citrus psorosis and Mirafiori lettuce big-vein ophiovirus coat proteins localize to the cytoplasm and self interact in vivo. Virus Res. 2012, 170, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Alioto, D.; Malfitano, M.; Troisi, A.; Peluso, A.; Martin, S.; Milne, R.G.; Guerri, J.; Moreno, P. Variability of the coat protein gene of Citrus psorosis virus in Campania, southern Italy. Arch. Virol. 2003, 148, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Martín, S.; López, C.; García, M.L.; Naum-Onganía, G.; Grau, O.; Flores, R.; Moreno, P.; Guerri, J. The complete nucleotide sequence of a Spanish isolate of Citrus psorosis virus: Comparative analysis with other ophioviruses. Arch. Virol. 2005, 150, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Martín, S.; García, M.L.; Troisi, A.; Rubio, L.; Legarreta, G.; Grau, O.; Alioto, D.; Moreno, P.; Guerri, J. Genetic variation of populations of Citrus psorosis virus. J. Gen. Virol. 2006, 87, 3097–3102. [Google Scholar] [CrossRef] [PubMed]
- Achachi, A.; Curk, F.; Jijakli, M.H.; Gaboun, F.; El Fahime, E.; Soulaymani, A.; El Guilli, M.; Ibriz, M. Variability and genetic structure of a natural population of Citrus psorosis virus. Ann. Microbiol. 2015, 65, 1195–1199. [Google Scholar] [CrossRef]
- Djelouah, K.; Potere, O.; Boscia, D.; D’Onghia, A.M.; Savino, V. Production of monoclonal antibodies to citrus psorosis virus. Int. Organ. Citrus Virol. Conf. Proc. 2000, 14, 152–158. [Google Scholar]
- Roistacher, C.N. Psorosis—A review. Int. Organ. Citrus Virol. Conf. Proc. 1993, 15, 139–154. [Google Scholar]
- Zanek, M.C.; Peña, E.; Reyes, C.A.; Figueroa, J.; Stein, B.; Grau, O.; Garcia, M.L. Detection of Citrus psorosis virus in the northwestern citrus production area of Argentina by using an improved TAS-ELISA. J. Virol. Methods 2006, 137, 245–251. [Google Scholar] [CrossRef]
- Loconsole, G.; Saponari, M.; Savino, V. Development of real-time PCR based assays for simultaneous and improved detection of citrus viruses. Eur. J. Plant. Pathol. 2010, 128, 251–259. [Google Scholar] [CrossRef]
- Kayim, M. Biological and molecular detection of citrus psorosis virus in citrus in the Eastern Mediterrenean Region of Turkey. J. Plant. Biochem. Biotechnol. 2010, 19, 259–262. [Google Scholar] [CrossRef]
- Moore, P.W.; Nauer, E.; Yendol, W. California scaly bark disease of citrus. Calif. Agric. 1957, 11, 8–9. [Google Scholar]
- Wallace, J.M. Virus-strain interference in relation to symptoms of psorosis disease of citrus. J. Agric. Sci. 1957, 27, 223–246. [Google Scholar] [CrossRef] [Green Version]
- Wallace, J.M. A Half Century of Research on Psorosis. Int. Organ. Citrus Virol. Conf. Proc. 1957, 15, 5–21. [Google Scholar]
- Moony, P. Growing Citrus in New Zealand: A Practical Guide; New Zealand Citrus Growers Incorporated: Wellington, New Zealand, 2001. [Google Scholar]
- Achachi, A.; Aït Barka, E.; Ibriz, M. Recent advances in Citrus psorosis virus. Virus Dis. 2014, 25, 261–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tennant, P.F.; Robinson, D.; Fisher, L.; Bennett, S.M.; Hutton, D.; Coates-Beckford, P.; Mc Laughlin, W. Diseases and Pests of Citrus (Citrus spp.). Tree For. Sci. Biotechnol. 2009, 3, 81–107. [Google Scholar]
- Rosa, C.; Polek, M.; Falk, B.W.; Rowhani, A. Improved efficiency for quantitative and qualitative indexing for Citrus tristeza virus and Citrus psorosis virus. Plant. Dis. 2007, 91, 1089–1095. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Moreno, P. Biological diversity of citrus ringspot isolates in Spain. Plant. Pathol. 1993, 42, 347–357. [Google Scholar] [CrossRef]
- Garcia, M.L.; Sanchez De La Torre, M.E.; Dal Bo, E.; Djelouah, K.; Rouag, N.; Luisoni, E.; Milne, R.G.; Grau, O. Detection of citrus psorosis-ringspot virus using RT-PCR and DAS-ELISA. Plant. Pathol. 1997, 46, 830–836. [Google Scholar] [CrossRef]
- de Zubrzycki, A.D.; Zubrzycki, H.M.; Correa, M. Determination of the distribution of psorosis in commercial plantings. Int. Organ. Citrus Virol. Conf. Proc. 1984, 9, 165–170. [Google Scholar]
- Martín, S.; Alioto, D.; Milne, R.G.; Guerri, J.; Moreno, P. Detection of Citrus psorosis virus in field trees by direct tissue blot immunoassay in comparison with ELISA, symptomatology, biological indexing and cross-protection tests. Plant. Pathol. 2002, 51, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Martín, S.; Milne, R.G.; Alioto, D.; Guerri, J.; Moreno, P. Psorosis-like symptoms induced by causes other than citrus psorosis virus. Int. Organ. Citrus Virol. Conf. Proc. 2002, 15, 197–204. [Google Scholar]
- Roistacher, C.N.; D’Onghia, A.M.; Djelouah, K. Defining psorosis by biological indexing and ELISA. Int. Organ. Citrus Virol. Conf. Proc. 2000, 14, 144–151. [Google Scholar]
- Martín, S.; Alioto, D.; Milne, R.G.; Garnsey, S.M.; Laura Garcia, M.; Grau, O.; Guerri, J.; Moreno, P. Detection of citrus psorosis virus by ELISA, Molecular hybridization, RT-PCR and immunosorbent electron microscopy and its association with citrus psorosis disease. Eur. J. Plant. Pathol. 2004, 110, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Velázquez, K.; Pérez, J.M.; Alonso, M.; Batista, L.; Rodríguez, J.; Legarreta, G.; Grau, O.; García, M.L. Detection of citrus psorosis virus in Cuba. Int. Organ. Citrus Virol. Conf. Proc. 2005, 16, 427–428. [Google Scholar]
- Golino, D.A.; Sim, S.T.; Cunningham, M.; Rowhani, A. Transmission of rose mosaic viruses. In Proceedings of the IVth International Symposium on Rose Research and Cultivation, Hannover, Germany, 25–30 August 2007; pp. 217–224. [Google Scholar]
- Nienhaus, F.; Saad, A.T. First report on plant virus diseases in Lebanon, Jordan, and Syria. Z. Pflanzenkrankh. Pflanzenschutz 1967, 74, 459–471. [Google Scholar]
- Wallace, J.M. Chapter 2- Virus and viruslike diseases. In The Citrus Industry; Reuther, W., Calavan, E.C., Carman, G.E., Eds.; Univeristy of California: Oakland, CA, USA, 1978; Volume IV, p. 362. ISBN 9789896540821. [Google Scholar]
- D’Onghia, A.M.; Djelouah, K.; Savino, V. Serological detection of Citrus psorosis virus in seeds but not in seedlings of infected mandarin and sour orange. J. Plant. Pathol. 2000, 82, 233–235. [Google Scholar] [CrossRef]
- Yarwood, C.E. Mechanical transmission of plant viruses. Adv. Virus Res. 1957, 4, 243–278. [Google Scholar]
- Garnsey, S.M.; Timmer, L.W. Mechanical transmissibility of citrus ringspot virus isolates from Florida, Texas, and California. Int. Organ. Citrus Virol. Conf. Proc. 1980, 8, 174–179. [Google Scholar]
- El-Dougdoug, K.A.; Ghazal, S.A.; Mousa, A.A.; Fahmy, H.; Sofy, A.R. Differentiation among three Egyptian isolates of citrus psorosis virus. Int. J. Virol. 2009, 5, 49–63. [Google Scholar] [CrossRef]
- Skaria, M.; Miao, H.; Avila, E. Post-freeze Status of Citrus psorosis virus. Int. Organ. Citrus Virol. Conf. Proc. 2002, 15, 366–367. [Google Scholar]
- Palle, S.R.; Miao, H.; Seyran, M.; Louzada, E.S.; da Graça, J.V.; Skaria, M. Evidence for association of citrus psorosis virus with symptomatic trees and an olpidium like fungus in Texas. Int. Organ. Citrus Virol. Conf. Proc. 2005, 15, 423–426. [Google Scholar]
- Gottwald, T.R.; Palle, S.R.; Miao, H.; Seyran, M.; Skaria, M.; da Graça, J.V. Assessment of the possibility of natural spread of citrus psorosis disease in Texas. Int. Organ. Citrus Virol. Conf. Proc. 2005, 16, 240–250. [Google Scholar]
- Lot, H.; Campbell, R.N.; Souche, S.; Milne, R.G.; Roggero, P. Transmission by olpidium brassicae of Mirafiori lettuce virus and Lettuce big-vein virus, and their roles in lettuce big-vein etiology. Phytopathology 2002, 92, 288–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benatena, H.N.; Portillo, M.M. Natural spread of psorosis in sweet orange seedlings. Int. Organ. Citrus Virol. Conf. Proc. 1984, 19, 159–164. [Google Scholar]
- Poritllo, M.M.; Benatena, H.N. Transmission of PSOROSIS from citrus to citrus by Aphids. Rev. Soc. Entomol. Argentina 1989, 45, 299–305. [Google Scholar]
- Carvalho, S.A.; Santos, F.A.; Machado, M.A. Eliminação de vírus do complexo sorose dos citros por microenxertia associada a termoterapia. Fitopatol. Bras. 2002, 27, 306–308. [Google Scholar] [CrossRef]
- Delaunois, B.; Jeandet, P.; Clément, C.; Baillieul, F.; Dorey, S.; Cordelier, S. Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front. Plant. Sci. 2014, 5, 1–50. [Google Scholar] [CrossRef] [Green Version]
- Kachroo, A.; Vincelli, P.; Kachroo, P. Signaling mechanisms underlying resistance responses: What have we learned, and how is it being applied? Phytopathology 2017, 107, 1452–1461. [Google Scholar] [CrossRef] [Green Version]
- van Bel, A.J.E.; Helariutta, Y.; Thompson, G.A.; Ton, J.; Dinant, S.; Ding, B.; Patrick, J.W. Phloem: The integrative avenue for resource distribution, signaling, and defense. Front. Plant. Sci. 2013, 4, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant. Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Marmisolle, F.E.; Arizmendi, A.; Ribone, A.; Rivarola, M.; García, M.L.; Reyes, C.A. Up-regulation of microRNA targets correlates with symptom severity in Citrus sinensis plants infected with two different isolates of citrus psorosis virus. Planta 2020, 251, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dalio, R.J.D.; Magalhaes, D.M.; Rodrigues, C.M.; Arena, G.D.; Oliveira, T.S.; Souza-Neto, R.R.; Picchi, S.C.; Martins, P.M.M.; Santos, P.J.C.; Maximo, H.J.; et al. PAMPs, PRRs, effectors and R-genes associated with citrus-pathogen interactions. Ann. Bot. 2017, 119, 749–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, P.M.; Kazan, K.; Wilson, I.; Anderson, J.P.; Richmond, T.; Somerville, S.C.; Manners, J.M. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 2000, 97, 11655–11660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant. Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.Y.; Park, J.M. Cross-talk in viral defense signaling in plants. Front. Microbiol. 2016, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Souza, P.F.N.; Carvalho, F.E.L. Killing two birds with one stone: How do plant viruses break down plant defenses and manipulate cellular processes to replicate themselves? J. Plant. Biol. 2019, 62, 170–180. [Google Scholar] [CrossRef] [Green Version]
- De Francesco, A.; Costa, N.; Plata, M.I.; García, M.L. Improved detection of citrus psorosis virus and coat protein-derived transgenes in citrus plants: Comparison between RT-qPCR and TAS-ELISA. J. Phytopathol. 2015, 163, 915–925. [Google Scholar] [CrossRef]
- Velázquez, K.; Renovell, A.; Comellas, M.; Serra, P.; García, M.L.; Pina, J.A.; Navarro, L.; Moreno, P.; Guerri, J. Effect of temperature on RNA silencing of a negative-stranded RNA plant virus: Citrus psorosis virus. Plant. Pathol. 2010, 59, 982–990. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Zhou, Y. Strategies for viral cross protection in plants. Methods Mol. Biol. 2012, 894, 69–81. [Google Scholar] [CrossRef]
- Legarreta, G.G.; García, M.L.; Costa, N.; Grau, O. A highly sensitive heminested RT-PCR assay for the detection of citrus psorosis virus targeted to a conserved region of the genome. J. Virol. Methods 2000, 84, 15–22. [Google Scholar] [CrossRef]
- D’Onghia, A.M.; Djelouah, K.; Alioto, D.; Castellano, M.A.; Savino, V. ELISA correlates with biological indexing for the detection of citrus psorosis-associated virus. J. Plant. Pathol. 1998, 80, 157–163. [Google Scholar]
- D’Onghia, A.M.; Djelouah, K.; Frasheri, D.; Potere, O. Detection of citrus psorosis virus by direct tissue blot immunoassay. J. Plant. Pathol. 2001, 83, 139–142. [Google Scholar]
- Potere, O.; Boscia, D.; Djelouah, K.; Elicio, V.; Savino, V. Use of monoclonal antibodies to citrus psorosis virus for diagnosis. J. Plant. Pathol. 1999, 81, 209–212. [Google Scholar]
- Alioto, D.; Gangemi, M.; Deaglio, S.; Sposato, P.; Noris, E.; Luisoni, E.; Milne, R.G. Improved detection of citrus psorosis virus using polyclonal and monoclonal antibodies. Plant. Pathol. 1999, 48, 735–741. [Google Scholar] [CrossRef] [Green Version]
- Mrani, N.; D’Onghia, A.M.; Djelouah, K.; Zemzami, M.; Frasheri, D.; Martelli, G.P. Distribution of citrus psorosis virus in Morocco. Int. Organ. Citrus Virol. Conf. Proc. 2002, 15, 358–362. [Google Scholar]
- Achachi, A.; Jijakli, M.H.; El Fahime, E.; Soulaymani, A.; Ibriz, M. Detection of citrus psorosis virus using an improved one-step RT-PCR. Arab. J. Sci. Eng. 2015, 40, 7–13. [Google Scholar] [CrossRef]
- Roy, A.; Fayad, A.; Barthe, G.; Brlansky, R.H. A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J. Virol. Methods 2005, 129, 47–55. [Google Scholar] [CrossRef]
- Osman, F.; Hodzic, E.; Kwon, S.J.; Wang, J.; Vidalakis, G. Development and validation of a multiplex reverse transcription quantitative PCR (RT-qPCR) assay for the rapid detection of Citrus tristeza virus, Citrus psorosis virus, and Citrus leaf blotch virus. J. Virol. Methods 2015, 220, 64–75. [Google Scholar] [CrossRef] [Green Version]
- Loconsole, G.; Fatone, M.T.; Savino, V. Specific digoxigenin-labelled riboprobes for detection of Citrus psorosis virus and Citrus variegation virus by molecular hybridization. J. Plant. Pathol. 2009, 91, 311–319. [Google Scholar]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High throughput sequencing for plant virus detection and discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.B.; Glasa, M.; Katis, N.; Olmos, A. Recent advances on detection and characterization of fruit tree viruses using high-throughput sequencing technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, E.E.; Coletta-Filho, H.D.; Nourin, S.; Falk, B.W.; Nerva, L.; Oliveira, T.S.; Dorta, S.O.; Machado, M.A. Deep sequencing analysis of RNAs from citrus plants grown in a citrus sudden death–affected area reveals diverse known and putative novel viruses. Viruses 2017, 9, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.H.; Osman, F.; Bodaghi, S.; Dang, T.; Greer, G.; Huang, A.; Hammado, S.; Abu-Hajar, S.; Campos, R.; Vidalakis, G. Full genome characterization of 12 citrus tatter leaf virus isolates for the development of a detection assay. PLoS ONE 2019, 14, e0223958. [Google Scholar] [CrossRef] [PubMed]
- Jooste, T.L.; Visser, M.; Cook, G.; Burger, J.T.; Maree, H.J. In Silico probe-based detection of Citrus viruses in NGS data. Phytopathology 2017, 107, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L. Citrus sanitation, quarantine and certification programs. Int. Organ. Citrus Virol. Conf. Proc. 1993, 20, 383–391. [Google Scholar]
- D’Onghia, A.M.; Carimi, F.; De Pasquale, F.; Djelouah, K.; Martelli, G.P. Elimination of citrus psorosis virus by somatic embryogenesis from stigma and style cultures. Plant. Pathol. 2001, 50, 266–269. [Google Scholar] [CrossRef]
- El-Sawy, A.; Gomaa, A.; Abd-El-Zaher, M.H.; Reda, A.; Danial, N. Production of somatic embryogenesis via in vitro culture of stigma and style for elimination of citrus psorosisvirus ( CpsV ) from some citrus genotypes. J. Hortic. Sci. Ornam. Plants 2013, 5, 110–117. [Google Scholar] [CrossRef]
- Carimi, F.; De Pasquale, F.; Fiore, S.; D’Onghia, A.M. Sanitation of citrus germplasm by somatic embryogenesis and shoot-tip grafting. In Improvement of the Citrus Sector by the Setting up of the Common Conservation Strategies for the Free Exchange of Healthy Citrus Genetic Resources; D’Onghia, A.M., Menini, U., Martelli, G.P., Eds.; CIHEAM-IAMB: Montpellier, France, 2001; pp. 61–65. [Google Scholar]
- Zanek, M.C.; Reyes, C.A.; Cervera, M.; Peña, E.J.; Velázquez, K.; Costa, N.; Plata, M.I.; Grau, O.; Peña, L.; García, M.L. Genetic transformation of sweet orange with the coat protein gene of Citrus psorosis virus and evaluation of resistance against the virus. Plant. Cell Rep. 2008, 27, 57–66. [Google Scholar] [CrossRef]
- Reyes, C.A.; Peña, E.J.; Zanek, M.C.; Sanchez, D.V.; Grau, O.; García, M.L. Differential resistance to Citrus psorosis virus in transgenic nicotiana benthamiana plants expressing hairpin RNA derived from the coat protein and 54K protein genes. Plant Cell Rep. 2009, 28, 1817–1825. [Google Scholar] [CrossRef]
- Reyes, C.A.; De Francesco, A.; Peña, E.J.; Costa, N.; Plata, M.I.; Sendin, L.; Castagnaro, A.P.; García, M.L. Resistance to Citrus psorosis virus in transgenic sweet orange plants is triggered by coat protein-RNA silencing. J. Biotechnol. 2011, 151, 151–158. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, A.; Costa, N.; García, M.L. Citrus psorosis virus coat protein-derived hairpin construct confers stable transgenic resistance in citrus against psorosis A and B syndromes. Transgenic Res. 2017, 26, 225–235. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, A.; Simeone, M.; Gómez, C.; Costa, N.; García, M.L. Transgenic sweet orange expressing hairpin CP-mRNA in the interstock confers tolerance to citrus psorosis virus in the non-transgenic scion. Transgenic Res. 2020, 29, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, K.; Alba, L.; Zarza, O.; Vives, M.C.; Pina, J.A.; Juárez, J.; Navarro, L.; Moreno, P.; Guerri, J. The response of different genotypes of citrus and relatives to Citrus psorosis virus inoculation. Eur. J. Plant. Pathol. 2016, 144, 73–81. [Google Scholar] [CrossRef]
- Jarrar, S.; Djelouah, K.; D’Onghia, A.M.; Martelli, G.P. A preliminary survey of virus and virus-like diseases of citrus in palestine. Int. Organ. Citrus Virol. Conf. Proc. 2002, 15, 423–426. [Google Scholar]
- Najar, A.; Duran-Vila, N.; Khlij, A.; Bové, M. Virus and virus-like diseases of citrus in Tunisia. Int. Organ. Citrus Virol. Conf. Proc. 2005, 16, 484–486. [Google Scholar]
- Hamdi, I.; Najar, A. Incidence and molecular characterization of citrus psorosis virus in Tunisia. Tunis. J. Plant. Prot. 2017, 12, 135–147. [Google Scholar]
- Yilmaz, M.A.; Baloglu, S. Elisa detection of Citrus Psorosis Virus (CPsV) in the Eastern Mediterranean region of Turkey. Options Méditerranéennes Série B Etudes Rech. 2002, 43, 85–87. [Google Scholar]
- Barbarossa, L.; Loconsole, G.; Vovlas, C. Virus and virus-like diseases of citrus in Epirus. J. Plant. Pathol. 2007, 89, 273–276. [Google Scholar]
- Stamo, B.; D’Onghia, A.M. Investigations on Citrus Psorosis Virus (CPsV) and Citrus infectious Variegation Virus (CVV) in Albania. Options Méditerranéennes Sér. B Etudes Rech. 2002, 43, 89–91. [Google Scholar]
- Kyriakou, A.P. Citrus infectious Variegation Virus (CVV) and Citrus Psorosis Virus (CPsV) in Cyprus. Options Méditerranéennes Sér. B Etudes Rech. 2002, 43, 93–95. [Google Scholar]
- Gatt, M.B.; D’Onghia, A.M. Serological Investigations on the Main Citrus Viruses in Malta. Options Méditerranéennes Sér. B Etudes Rech. 2002, 43, 97–99. [Google Scholar]
- Felkai, K. Etude du Virus de la Psorose: Identification Biologique et Immuno-Enzymatique DAS-ELISA de Citrus Psorosis Ophiovirus (CPsV) sur Agrumes, Saad Dahlab de Blida. Ph.D. Thesis, University Saad Dahlab de Blida, Ouled Yaich, Algeria, 2008. [Google Scholar]
- da Graça, J.V.; Bar-Joseph, M.; Derrick, K.S. Immunoblot detection of citrus psorosis in Israel using citrus ringspot antiserum. Int. Organ. Citrus Virol. Conf. 1993, 12, 432–434. [Google Scholar]
- El-Otmani, M.; Coggins, J.C.W.; Duymovic, A. Citrus cultivars and production in Morocco. HortScience 1990, 25, 1343–1346. [Google Scholar] [CrossRef]
- Bibi, I.; Afechtal, M.; Chafik, Z.; Bamouh, A.; Benyazid, J.; Bousamid, A.; Kharmach, E. Occurrence and distribution of virus and virus-like diseases of citrus in North-Est of Morocco—Moulouya perimeter. In Proceedings of the Onzième Congrès de l’Association Marocaine de Protection des Plantes, Rabat, Maroc, 26–27 March 2019; pp. 119–134. [Google Scholar]
- Belabess, Z.; Afechtal, M.; Khfif, K.; Benyazid, J. Prévalence des phytvirus et virus-like associés aux agrumes au Nord-Est du Maroc. In Proceedings of the Onzième Congrès de l’Association Marocaine de Protection des Plantes, Rabat, Maroc, 26–27 March 2019; pp. 135–149. [Google Scholar]
- Bekki, L. Etude préliminaire de la psorose des agrumes dans la région de Moulouya. Mémoire troisième cycle pour l’obtention du Diplôme d’Ingénieur d’Etat en Agron. Option Prot. Plantes 2007, 16, 1–88. [Google Scholar]
- Achachi, A.; Ibriz, M. Biological and molecular detection of citrus psorosis virus in the northwest region of Morocco. In Proceedings of the XIIth International Citrus Congress, Valencia, Spain, 20 January 2015; pp. 831–836. [Google Scholar]
- Afechtal, M. Present status of virus and virus-like diseases of citrus in Morocco. Integr. Control. Citrus Fruit Crop. IOBC WPRS Bull. 2018, 132, 215–220. [Google Scholar]
- Adams, I.P.; Fox, A.; Boonham, N.; Massart, S.; De Jonghe, K. The impact of high throughput sequencing on plant health diagnostics. Eur. J. Plant. Pathol. 2018, 152, 909–919. [Google Scholar] [CrossRef]
- Nouri, S.; Salem, N.; Nigg, J.C.; Falk, B.W. Diverse array of new viral sequences identified in worldwide populations of the Asian Citrus Psyllid (Diaphorina citri) using viral metagenomics. J. Virol. 2016, 90, 2434–2445. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Gao, S.; Hernandez, A.G.; Wechter, W.P.; Fei, Z.; Ling, K.S. Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PLoS ONE 2012, 7, e37127. [Google Scholar] [CrossRef]
- Sun, L.; Nasrullah; Ke, F.; Nie, Z.; Wang, P.; Xu, J. Citrus genetic engineering for disease resistance: Past, present and future. Int. J. Mol. Sci. 2019, 20, 5256. [Google Scholar] [CrossRef] [Green Version]


| Date (Collection Period) | Locality (Number of Visited Groves) | Plant Material | Symptoms | Diagnostic Techniques (Tested Tissue) | Tested Varieties | CPsV * Prevalence (Positive/Total) | Positive Varieties (Percent (%) of Infection) | References |
|---|---|---|---|---|---|---|---|---|
| 2000 (March) | Gharb (2) | Five flowers, or six mature leaves from trees that were not in bloom. | Bark scaling and psorosis-like symptoms on the leaves (mottling, flecking, and ringspots). | DTBIA (ovary tissue) and ELISA (mature leaves from non-flowering trees). | Maroc-late and Salustiana orange trees. | 50% (172/346) | Maroc-late (71%) and Salustiana (36%). | [83] |
| Souss (7) | Nour clementine, Ortanique mandarin, Washington Navel, Maroc-late and Salustiana orange, Eureka lemon, and Star ruby grapefruit trees. | 9% (9/100) | Ortanique (20%), Washington Navel (20%), Maroc-late (14%) and Salustiana (20%). | |||||
| Haouz (7) | 28% (28/100) | Ortanique (50%), Washington Navel (49%), Maroc-late (14%), Salustiana (20%), Eureka (4%), and Star ruby (8%). | ||||||
| 2007 (June) | Moulouya (8) | Young leaves from the four cardinal orientations. | Bark scaling. | ELISA. | Berkane clementine and Washington Navel orange trees. | 77% (50/65) | Berkane clementine (22%) and Washington Navel (41%). | [117] |
| Vein clearing on leaves. | 8% (3/39) | |||||||
| Leaves showing oak leaf pattern. | 7% (1/15) | |||||||
| No symptoms. | 6% (5/81) | |||||||
| 2011 (January) | Gharb (DN) | Four young shoots. | Typical psorosis bark scaling. | RT-PCR (leaves) and/or biological indexing. | Maroc-late, Washington sanguine, Washington Navel orange, Sidi Aissa clementine, and sour orange trees. | 100% (8/8) | DN | [118] |
| Non-scaled trees with young-leaf symptoms. | 100% (4/4) | |||||||
| Atypical bark scaling. | 0% (0/2) | |||||||
| Concave gum symptoms. | 0% (0/1) | |||||||
| Non-scaled trees showing young leaf symptoms. | 0% (0/5) | |||||||
| No symptoms. | 0% (0/4) | |||||||
| 2008–2013 (DN) | Gharb, Haouz, Loukkos, Moulouya, Souss, and Tadla (102) | DN | Randomly and plants showing virus-like symptoms were included in the sampling. | ELISA and RT-PCR. | Navelina, Salustiana, Hamlin, Cadenera, Jaffa (Shamouti), Maroc-late, Vernia, Grosse Sanguine, Sanguinelli, and Tarocco orange trees. | 33% (1854/5620) | DN | [119] |
| DN | Gharb (DN) | Four young shoots. | Bark scaling. | ELISA, RT-PCR (leaves for both techniques), and/or biological indexing. | Maroc-late, Salustiana, and Washington Navel sweet orange and Nules clementine trees. | 100% (2/2) | Maroc-late (50%) | [84] |
| No symptoms. | 0% (0/2) | |||||||
| Bark scaling. | 100% (2/2) | Salustiana (100%) | ||||||
| No symptoms. | 100% (1/1) | |||||||
| No symptoms. | 0% (0/1) | Washington Navel (0%) | ||||||
| Moulouya (DN) | Bark scaling. | 100% (11/11) | Washington Navel (78%) | |||||
| No symptoms. | 43% (3/7) | |||||||
| No symptoms. | 75% (3/4) | Nules (75%) | ||||||
| Tadla (DN) | Bark scaling. | 100% (11/11) | DN | |||||
| No symptoms. | 0% (0/1) | |||||||
| 2018 (May–July) | Moulouya (37) | Four young shoots. | Bark scaling. | ELISA and RT-PCR. | Berkane, Nour, Muska, Nules, Orograndi clementine and Maroc-late, Navel, and Navelina orange trees. | 25% (16/65) | Nour (13%), Berkane (44%), Navel (25%), Navelina (6%), unknown orange cultivar (12%) | [116] |
| Virus-like symptoms. | 17% (/29/170) | Nour (14%), Berkane (21%), Nules (7%), Navel (10%), Navelina (17%), Maroc late (17%), unknown orange cultivar (14%) | ||||||
| 2018 (July–September) | Moulouya (3) | DN | Bark scaling in the trunk and main branches and internal staining in the underlying wood in citrus cultivars. | ELISA and RT-PCR. | Berkane, Nules, and Orogrande clementine trees. | 39% (309/790) | Berkane (DN), Nules (DN) and Orogrande (DN) | [115] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belabess, Z.; Sagouti, T.; Rhallabi, N.; Tahiri, A.; Massart, S.; Tahzima, R.; Lahlali, R.; Jijakli, M.H. Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus. Microorganisms 2020, 8, 1197. https://doi.org/10.3390/microorganisms8081197
Belabess Z, Sagouti T, Rhallabi N, Tahiri A, Massart S, Tahzima R, Lahlali R, Jijakli MH. Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus. Microorganisms. 2020; 8(8):1197. https://doi.org/10.3390/microorganisms8081197
Chicago/Turabian StyleBelabess, Zineb, Tourya Sagouti, Naima Rhallabi, Abdessalem Tahiri, Sébastien Massart, Rachid Tahzima, Rachid Lahlali, and M. Haissam Jijakli. 2020. "Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus" Microorganisms 8, no. 8: 1197. https://doi.org/10.3390/microorganisms8081197
APA StyleBelabess, Z., Sagouti, T., Rhallabi, N., Tahiri, A., Massart, S., Tahzima, R., Lahlali, R., & Jijakli, M. H. (2020). Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus. Microorganisms, 8(8), 1197. https://doi.org/10.3390/microorganisms8081197








