Gastric Microbiome Diversities in Gastric Cancer Patients from Europe and Asia Mimic the Human Population Structure and Are Partly Driven by Microbiome Quantitative Trait Loci
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Microbiome Inference
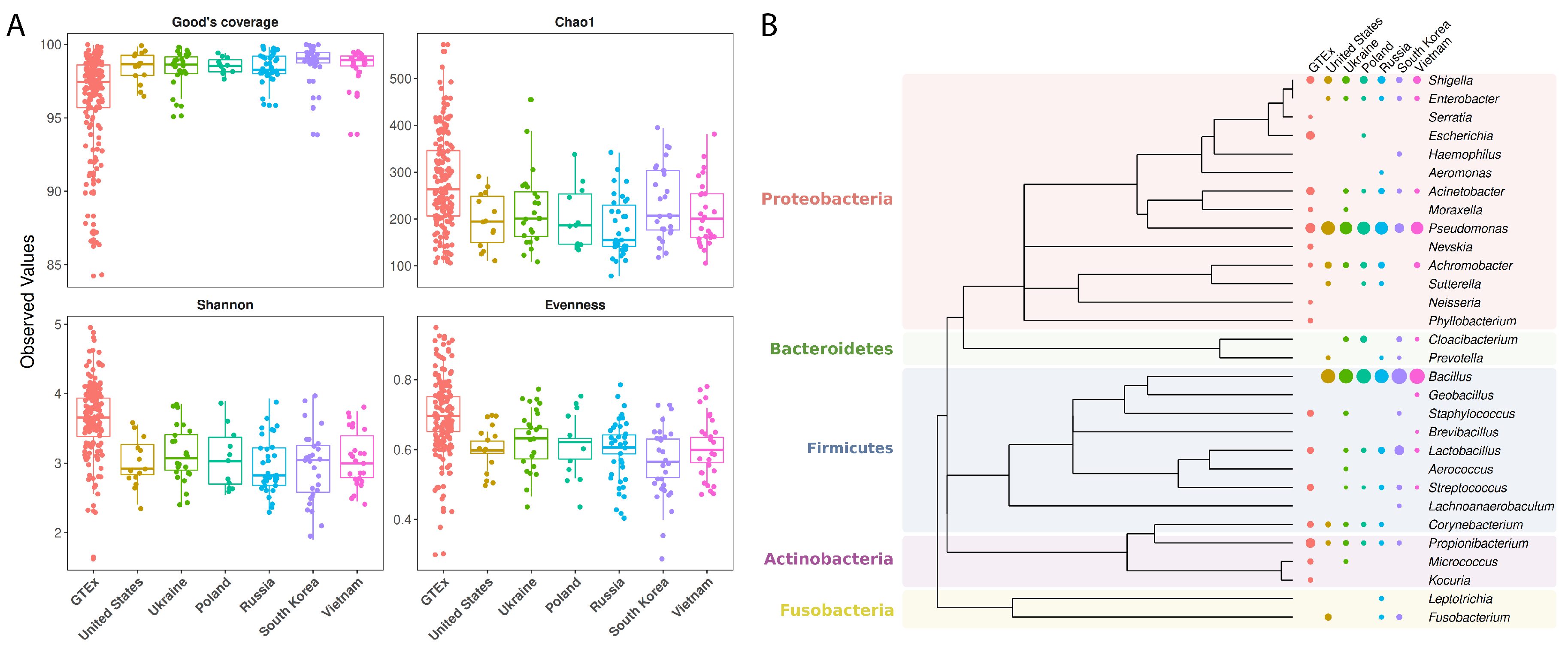
2.3. Checking for Genus Coverage, Richness and Evenness
2.4. Phylogenetic Analyses
2.5. Evaluating Microbiota Composition in Gastric Tumorigenesis
2.6. Host-Genome and Microbiome Associations
3. Results
3.1. Quality Control of RNASeq Data for Microbiome Inference
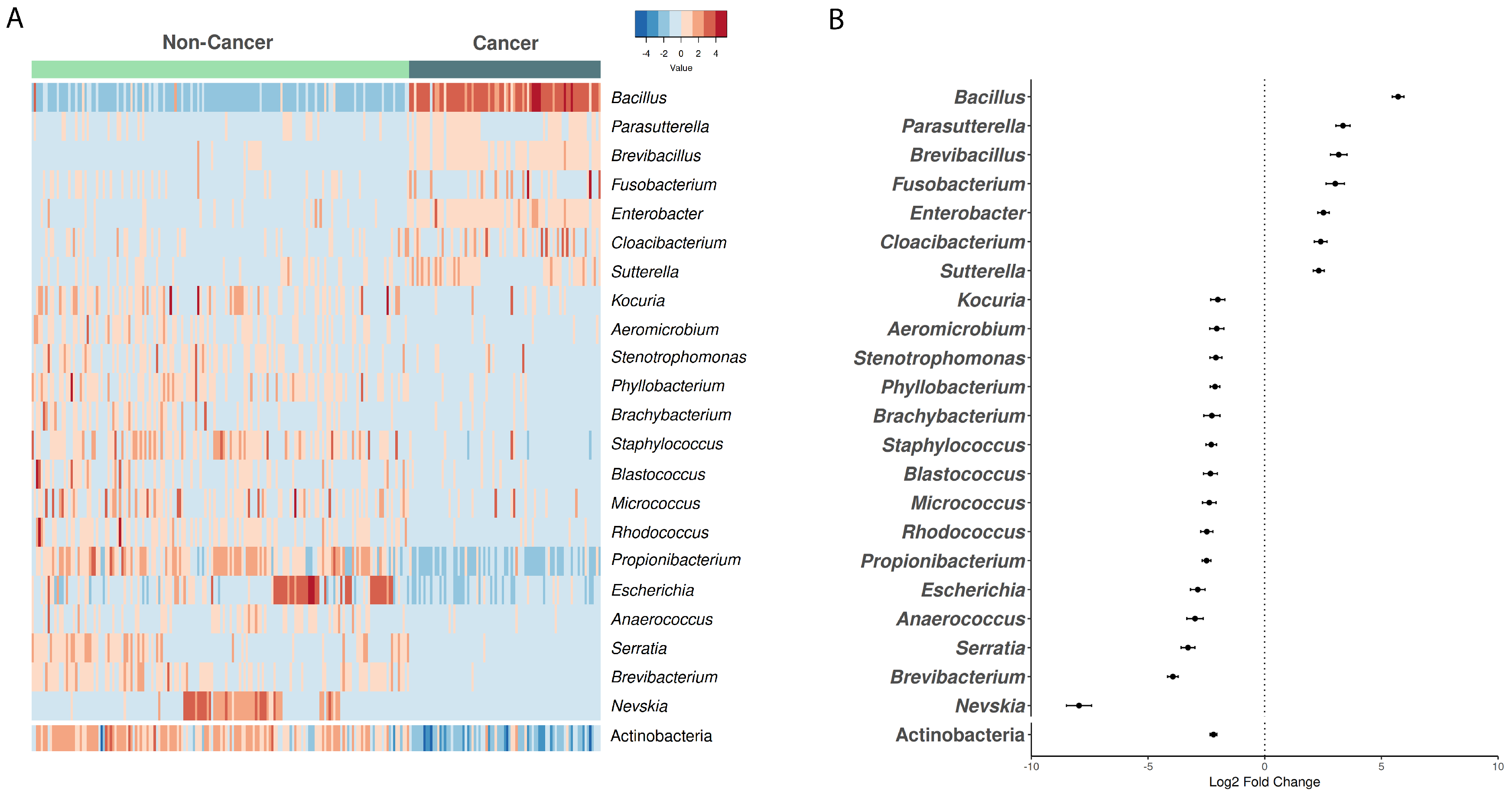
3.2. Microbiome Composition in Cancer vs. Non-Cancer Samples from European Ancestry
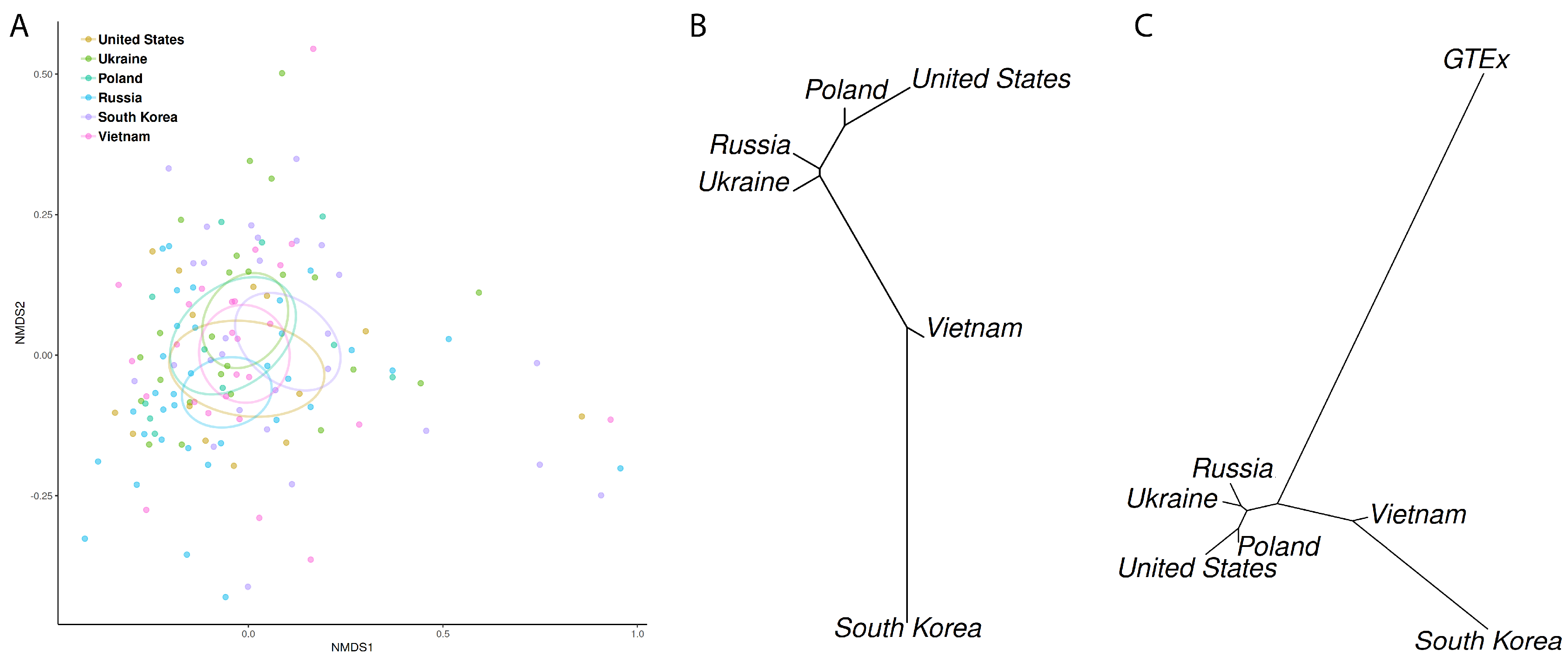
3.3. Microbiome Profiling in Function of Host Geographic Origin
3.4. Identification of Host Genetic Variation Associated with the Microbiome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moeller, A.H.; Caro-Quintero, A.; Mjungu, D.; Georgiev, A.V.; Lonsdorf, E.V.; Muller, M.N.; Pusey, A.E.; Peeters, M.; Hahn, B.H.; Ochman, H. Cospeciation of gut microbiota with hominids. Science 2016, 353, 380–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Gomez, A.; Petrzelkova, K.J.; Burns, M.B.; Yeoman, C.J.; Amato, K.R.; Vlckova, K.; Modry, D.; Todd, A.; Jost Robinson, C.A.; Remis, M.J.; et al. Gut Microbiome of Coexisting BaAka Pygmies and Bantu Reflects Gradients of Traditional Subsistence Patterns. Cell Rep. 2016, 14, 2142–2153. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Wang, J.; Thingholm, L.B.; Skiecevičienė, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef]
- Hughes, D.A.; Bacigalupe, R.; Wang, J.; Rühlemann, M.C.; Tito, R.Y.; Falony, G.; Joossens, M.; Vieira-Silva, S.; Henckaerts, L.; Rymenans, L.; et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Parekh, P.J.; Balart, L.A.; Johnson, D.A. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin. Transl. Gastroenterol. 2015, 6, e91. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Bobryshev, Y.V.; Kozarov, E.; Sobenin, I.A.; Orekhov, A.N. Role of gut microbiota in the modulation of atherosclerosis-associated immune response. Front. Microbiol. 2015, 6, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribaldone, D.G.; Caviglia, G.P.; Abdulle, A.; Pellicano, R.; Ditto, M.C.; Morino, M.; Fusaro, E.; Saracco, G.M.; Bugianesi, E.; Astegiano, M. Adalimumab Therapy Improves Intestinal Dysbiosis in Crohn’s Disease. J. Clin. Med. 2019, 8, 1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Li, B.; Sun, D.; Chen, S. Gut Microbiota: Implications in Alzheimer’s Disease. J. Clin. Med. 2020, 9, 2042. [Google Scholar] [CrossRef]
- Thomas, R.M.; Jobin, C. The Microbiome and Cancer: Is the ‘Oncobiome’ Mirage Real? Trends Cancer 2015, 1, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Dicksved, J.; Lindberg, M.; Rosenquist, M.; Enroth, H.; Jansson, J.K.; Engstrand, L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 2009, 58, 509–516. [Google Scholar] [CrossRef]
- Aviles-Jimenez, F.; Vazquez-Jimenez, F.; Medrano-Guzman, R.; Mantilla, A.; Torres, J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014, 4, 4202. [Google Scholar] [CrossRef] [Green Version]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Meehan, J.; Su, Z.; Ng, H.W.; Shu, M.; Luo, H.; Ge, W.; Perkins, R.; Tong, W.; Hong, H. Whole genome sequencing of 35 individuals provides insights into the genetic architecture of Korean population. BMC Bioinform. 2014, 15 (Suppl. 11), S6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [Green Version]
- Rajilić-Stojanović, M.; de Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef]
- Cavadas, B.; Ferreira, J.; Camacho, R.; Fonseca, N.; Pereira, L. QmihR: Pipeline for Quantification of Microbiome in Human RNA-seq. In Proceedings of the 11th International Conference on Practical Applications of Computational Biology & Bioinformatics, PACBB 2017, Porto, Portugal, 21–23 June 2017; pp. 173–179. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M. A new method for non-parametric multivariate analysis of variance. Aust. J. Ecol. 2001, 26, 2–46. [Google Scholar] [CrossRef]
- Krijthe, J.H. Rtsne: T-Distributed Stochastic Neighbor Embedding Using Barnes-Hut Implementation. 2015. Available online: https://github.com/jkrijthe/Rtsne (accessed on 28 August 2019).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Cantalupo, P.G.; Katz, J.P.; Pipas, J.M. HeLa nucleic acid contamination in the cancer genome atlas leads to the misidentification of human papillomavirus 18. J. Virol. 2015, 89, 4051–4057. [Google Scholar] [CrossRef] [Green Version]
- Robinson, K.M.; Crabtree, J.; Mattick, J.S.; Anderson, K.E.; Dunning Hotopp, J.C. Distinguishing potential bacteria-tumor associations from contamination in a secondary data analysis of public cancer genome sequence data. Microbiome 2017, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef] [Green Version]
- Delgado, S.; Cabrera-Rubio, R.; Mira, A.; Suárez, A.; Mayo, B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb. Ecol. 2013, 65, 763–772. [Google Scholar] [CrossRef]
- Tsuda-Sakurai, K.; Miura, M. The hidden nature of protein translational control by diphthamide: The secrets under the leather. J. Biochem. 2019, 165, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Takeuchi, O.; Standley, D.M.; Kumagai, Y.; Kawagoe, T.; Miyake, T.; Satoh, T.; Kato, H.; Tsujimura, T.; Nakamura, H.; et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 2009, 458, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Ra, E.A.; Lee, T.A.; Won Kim, S.; Park, A.; Choi, H.J.; Jang, I.; Kang, S.; Hee Cheon, J.; Cho, J.W.; Eun Lee, J.; et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat. Commun. 2016, 7, 11726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaniuk, N.A.; Canadien, V.; Bagshaw, R.D.; Bakowski, M.; Braun, V.; Landekic, M.; Mitra, S.; Huang, J.; Heo, W.D.; Meyer, T.; et al. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell Microbiol. 2011, 13, 1812–1823. [Google Scholar] [CrossRef]
- Daniels, J.; Kadlubar, S. Sulfotransferase genetic variation: From cancer risk to treatment response. Drug Metab. Rev. 2013, 45, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Abdel-El-Haleem, D. Acinetobacter: Environmental and biotechnological applications. Afr. J. Biotechnol. 2003, 2, 71–74. [Google Scholar]
- Dreyfus, M.; Régnier, P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell 2002, 111, 611–613. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Pandit, S.; Deutscher, M.P. Polyadenylation of stable RNA precursors in vivo. Proc. Natl. Acad. Sci. USA 1998, 95, 12158–12162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, B.K.; Kushner, S.R. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999, 34, 1094–1108. [Google Scholar] [CrossRef]

| SNP | Genus | GeneID | p-Value | q-Value | Genotype Frequency | Fst (p-Values) | |||||
| MAF_EUR | MAF_EAS | MAF_AFR | EUR_AFR | EUR_EAS | EAS_AFR | ||||||
| TCGA-EUROPE | rs1131600 | Corynebacterium | DPH1 | 7.97104 × 10−8 | 0.01501 | G|G: 0.028 A|G: 0.252 A|A: 0.720 | G|G: 0.004 A|G: 0.105 A|A: 0.891 | A|G: 0.020 A|A: 0.980 | 0.14323 (0.00000 ± 0.0000) | 0.04831 (0.00000 ± 0.0000) | 0.0359 (0.00000 ± 0.0000) |
| TCGA-EUROPE | rs61997220 | Lactobacillus | ZC3H12D | 3.07635 × 10−7 | 0.02414 | T|T: 0.907 C|T: 0.093 | T|T: 1.000 | T|T: 0.995 C|T: 0.005 | 0.04507 (0.00000 ± 0.0000) | 0.04582 (0.00000 ± 0.0000) | 0.00113 (0.25978 + 0.0048) |
| TCGA-EUROPE | rs9262143 | Acinetobacter | PPP1R18 | 5.68970 × 10−7 | 0.02414 | C|C: 0.845 C|T: 0.151 T|T: 0.004 | C|C: 1.000 | C|C: 0.995 C|T: 0.005 | 0.08232 (0.00000 ± 0.0000) | 0.07868 (0.00000 ± 0.0000) | 0.00113 (0.26255 ± 0.0046) |
| TCGA-EUROPE | rs17350674 | Cloacibacterium | KIF24 | 5.97391 × 10−7 | 0.02414 | C|C: 0.614 A|A: 0.050 A|C: 0.336 | C|C: 0.988 A|A: 0.002 A|C: 0.010 | C|C: 0.980 A|C: 0.020 | 0.21379 (0.00000 ± 0.0000) | 0.19977 (0.00000 ± 0.0000) | 0 (0.52688 ± 0.0052) |
| TCGA-EUROPE | rs2219078 | Acinetobacter | SULT1C3 | 6.40940 × 10−7 | 0.02414 | G|G: 0.658 A|G: 0.300 A|A: 0.042 | G|G: 0.097 A|G: 0.452 A|A: 0.450 | G|G: 0.472 A|G: 0.443 A|A: 0.085 | 0.03291 (0.00000 ± 0.0000) | 0.38529 (0.00000 ± 0.0000) | 0.24086 (0.00000 ± 0.0000) |
| TCGA-EUROPE | rs2523989 | Acinetobacter | TRIM31 | 1.03982 × 10−6 | 0.03263 | C|C: 0.775 C|T: 0.209 T|T: 0.016 | C|C: 0.905 C|T: 0.093 T|T: 0.002 | C|C: 0.903 C|T: 0.095 T|T: 0.002 | 0.03296 (0.00000 ± 0.0000) | 0.03172 (0.00000 ± 0.0000) | 0 (0.99990 ± 0.0000) |
| TCGA-EUROPE | rs7198494 | Achromobacter | C16orf46 | 1.75767 × 10−6 | 0.04728 | A|A: 0.648 A|G: 0.294 G|G: 0.058 | A|A: 0.962 A|G: 0.036 G|G: 0.002 | A|A: 0.526 A|G: 0.389 G|G: 0.085 | 0.01389 (0.00010 ± 0.0001) | 0.15724 (0.00000 ± 0.0000) | 0.21452 (0.00000 ± 0.0000) |
| TCGA-EUROPE | rs61997220 | Fusobacterium | ZC3H12D | 2.66174 × 10−6 | 0.05857 | T|T: 0.907 C|T: 0.093 | T|T: 1.000 | T|T: 0.995 C|T: 0.005 | 0.04507 (0.00000 ± 0.0000) | 0.04582 (0.00000 ± 0.0000) | 0.00113 (0.25978 ± 0.0048) |
| TCGA-EUROPE | rs62572859 | Lactobacillus | C9orf129 | 2.79953 × 10−6 | 0.05857 | C|C: 0.750 C|T: 0.221 T|T: 0.030 | C|C: 0.919 C|T: 0.081 | C|C: 0.433 C|T: 0.475 T|T: 0.092 | 0.09115 (0.00000 ± 0.0000) | 0.05751 (0.00000 ± 0.0000) | 0.22638 (0.00000 ± 0.0000) |
| TCGA-EUROPE | rs1014867 | Lactobacillus | FAT4 | 5.53680 × 10−6 | 0.09807 | C|C: 0.895 C|T: 0.103 T|T: 0.002 | C|C: 0.885 C|T: 0.109 T|T: 0.006 | C|C: 0.865 C|T: 0.126 T|T: 0.009 | 0.00188 (0.09425 ± 0.0026) | 0 (0.56796 ± 0.0055) | 0.00016 (0.33403 ± 0.0052) |
| TCGA-EUROPE | rs1782360 | Lactobacillus | LRBA | 6.08310 × 10−6 | 0.09807 | G|G: 0.855 C|C: 0.012 C|G: 0.133 | G|G: 0.754 C|C: 0.022 C|G: 0.224 | G|G: 0.495 C|C: 0.077 C|G: 0.428 | 0.13144 (0.00000 ± 0.0000) | 0.01505 (0.00040 ± 0.0002) | 0.06804 (0.00000 ± 0.0000) |
| TCGA-EUROPE | rs4963198 | Corynebacterium | LRRC56 | 6.84488 × 10−6 | 0.09914 | G|G: 0.109 A|A: 0.437 A|G: 0.453 | G|G: 0.022 A|A: 0.768 A|G: 0.210 | G|G: 0.319 A|A: 0.194 A|G: 0.487 | 0.09743 (0.00000 ± 0.0000) | 0.11482 (0.00000 ± 0.0000) | 0.33553 (0.00000 ± 0.0000) |
| GTEx | rs61733127 | Streptococcus | PHLPP2 | 3.22625 × 10−11 | 0.00001 | G|G: 0.024 A|G: 0.249 A|A: 0.728 | G|G: 0.012 A|G: 0.109 A|A: 0.879 | A|G: 0.056 A|A: 0.944 | 0.09268 (0.00000 ± 0.0000) | 0.03325 (0.00000 ± 0.0000) | 0.01635 (0.00000 ± 0.0000) |
| GTEx | rs74344827 | Streptococcus | TAT | 9.87962 × 10−8 | 0.01160 | G|G: 0.700 A|G: 0.274 A|A: 0.026 | G|G: 0.905 A|G: 0.087 A|A: 0.008 | G|G: 0.663 A|G: 0.309 A|A: 0.029 | 0.00052 (0.22265 ± 0.0035) | 0.06188 (0.00000 ± 0.0000) | 0.07456 (0.00000 ± 0.0000) |
| GTEx | rs73229817 | Corynebacterium | PDLIM2 | 2.60383 × 10−7 | 0.02039 | C|C: 0.901 C|T: 0.097 T|T: 0.002 | C|C: 1.000 | C|C: 0.998 C|T: 0.002 | 0.05466 (0.00000 ± 0.0000) | 0.0498 (0.00000 ± 0.0000) | 0 (0.99990 ± 0.0000) |
| GTEx | rs12807209 | Nevskia | MUC6 | 5.79755 × 10−7 | 0.03405 | G|G: 0.998 C|G: 0.002 | G|G: 1.000 | G|G: 0.750 C|C: 0.018 C|G: 0.231 | 0.11679 (0.00000 ± 0.0000) | 0 (0.50579 ± 0.0054) | 0.11913 (0.00000 ± 0.0000) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavadas, B.; Camacho, R.; Ferreira, J.C.; Ferreira, R.M.; Figueiredo, C.; Brazma, A.; Fonseca, N.A.; Pereira, L. Gastric Microbiome Diversities in Gastric Cancer Patients from Europe and Asia Mimic the Human Population Structure and Are Partly Driven by Microbiome Quantitative Trait Loci. Microorganisms 2020, 8, 1196. https://doi.org/10.3390/microorganisms8081196
Cavadas B, Camacho R, Ferreira JC, Ferreira RM, Figueiredo C, Brazma A, Fonseca NA, Pereira L. Gastric Microbiome Diversities in Gastric Cancer Patients from Europe and Asia Mimic the Human Population Structure and Are Partly Driven by Microbiome Quantitative Trait Loci. Microorganisms. 2020; 8(8):1196. https://doi.org/10.3390/microorganisms8081196
Chicago/Turabian StyleCavadas, Bruno, Rui Camacho, Joana C. Ferreira, Rui M. Ferreira, Ceu Figueiredo, Alvis Brazma, Nuno A. Fonseca, and Luísa Pereira. 2020. "Gastric Microbiome Diversities in Gastric Cancer Patients from Europe and Asia Mimic the Human Population Structure and Are Partly Driven by Microbiome Quantitative Trait Loci" Microorganisms 8, no. 8: 1196. https://doi.org/10.3390/microorganisms8081196
APA StyleCavadas, B., Camacho, R., Ferreira, J. C., Ferreira, R. M., Figueiredo, C., Brazma, A., Fonseca, N. A., & Pereira, L. (2020). Gastric Microbiome Diversities in Gastric Cancer Patients from Europe and Asia Mimic the Human Population Structure and Are Partly Driven by Microbiome Quantitative Trait Loci. Microorganisms, 8(8), 1196. https://doi.org/10.3390/microorganisms8081196





