Seasonal Variations in the Culturable Mycobiome of Acropora loripes along a Depth Gradient
Abstract
1. Introduction
2. Methods
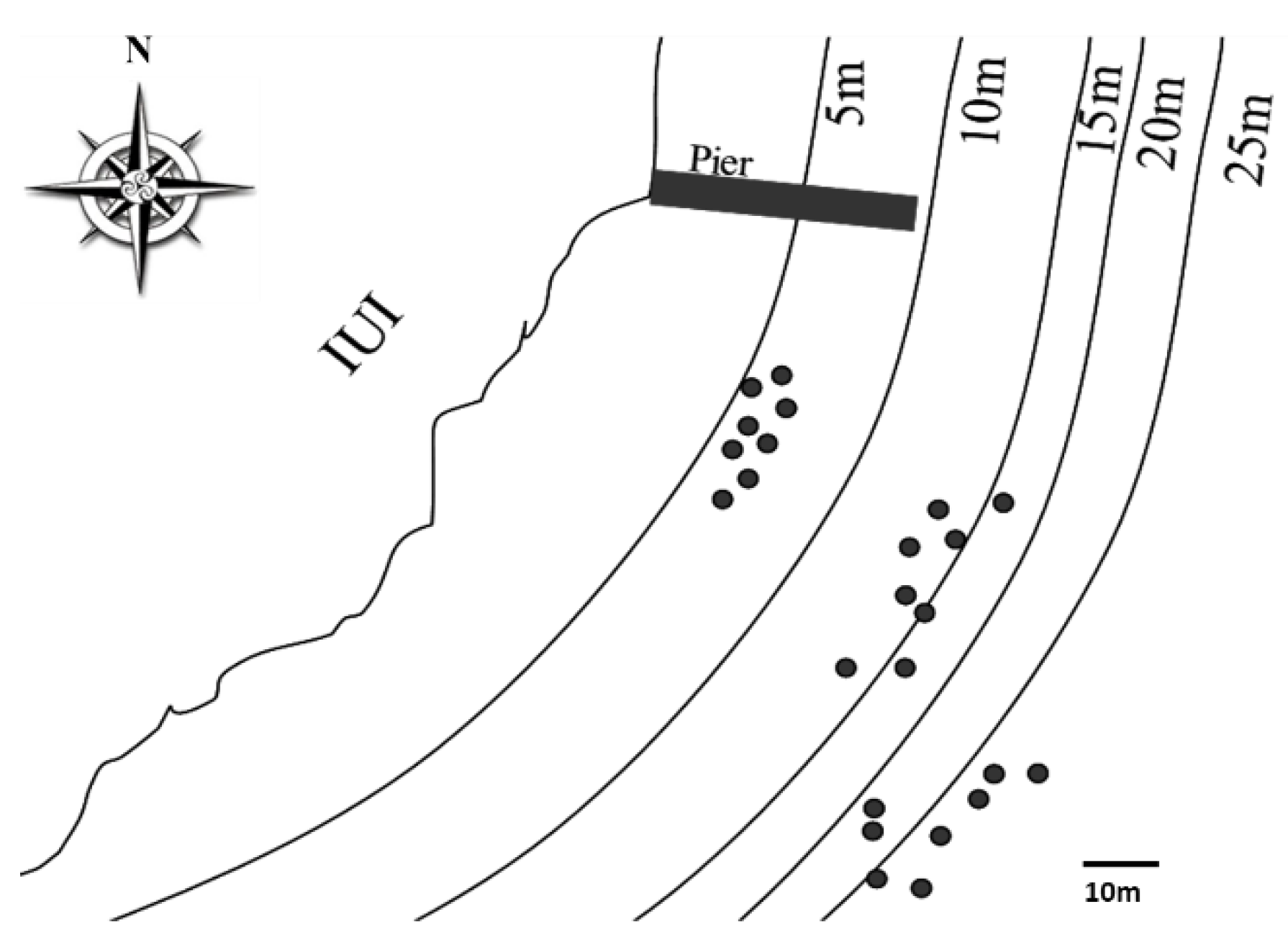
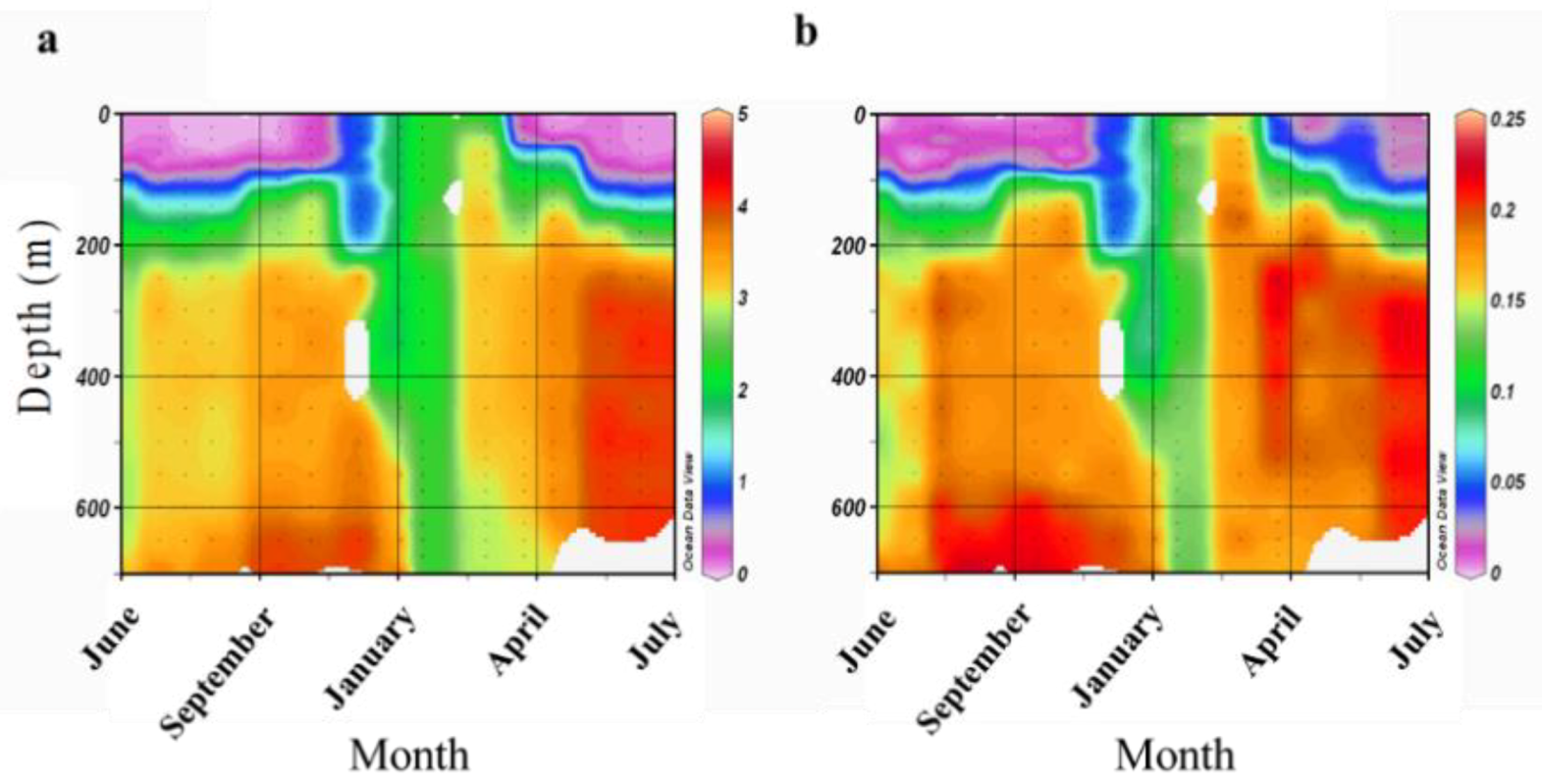
2.1. Sampling Site and Coral Collection
2.2. Sample Processing
2.3. DNA Extraction and PCR Amplification
2.4. Phylogenetic and Diversity Analysis
3. Results
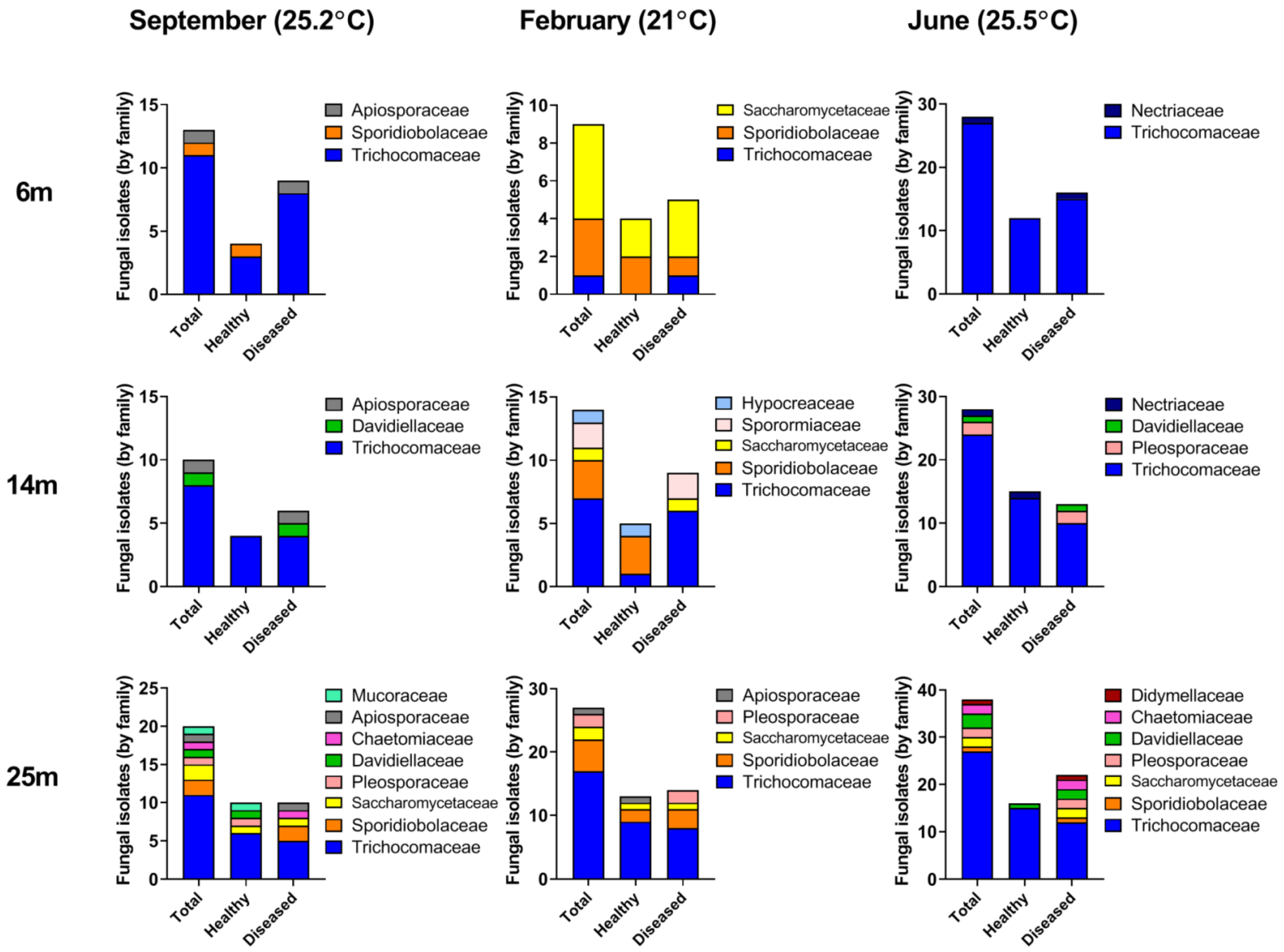
3.1. Fungal Diversity in A. loripes
3.2. Fungal Diversity in A. loripes along the Reef Slope
3.3. Seasonal Fluctuations in Fungal Diversity
3.4. Changes in Fungal Abundance and Diversity in Apparently-Healthy Corals vs. Corals Exhibiting Lesions
3.5. Presence of Fungi in Water and Sediment
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Jones, E.B.G. Fifty years of marine mycology. Fungal Divers. 2011, 50, 73–112. [Google Scholar] [CrossRef]
- Yarden, O. Fungal association with sessile marine invertebrates. Front. Microbiol. 2014, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, T.D.; Fordyce, A.J.; Camp, E.F. The other microeukaryotes of the coral reef microbiome. Trends Microbiol. 2017, 25, 980–991. [Google Scholar] [CrossRef]
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Mera, H.; Bourne, D.G. Disentangling causation: Complex roles of coral-associated microorganisms in disease. Environ. Microbiol. 2018, 20, 431–449. [Google Scholar] [CrossRef]
- Raghukumar, C.; Ravindran, J. Fungi and their role in corals and coral reef ecosystems. Adv. Biochem. Eng. Biotechnol. 2012, 53, 89–113. [Google Scholar] [CrossRef]
- Morrison-Gardiner, S. Dominant fungi from Australian coral reefs. Water Resour. Manag. 2000, 14, 105–121. [Google Scholar]
- Wang, G.; Li, Q.; Zhu, P. Phylogenetic diversity of culturable fungi associated with the Hawaiian Sponges Suberites zeteki and Gelliodes fibrosa. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2008, 93, 163–174. [Google Scholar] [CrossRef]
- Góes-Neto, A.; Marcelino, V.R.; Verbruggen, H.; da Silva, F.F.; Badotti, F. Biodiversity of endolithic fungi in coral skeletons and other reef substrates revealed with 18S rDNA metabarcoding. Coral Reefs 2020, 39, 229–238. [Google Scholar] [CrossRef]
- Paulino, G.V.B.; Félix, C.R.; Landell, M.F. Diversity of filamentous fungi associated with coral and sponges in coastal reefs of northeast Brazil. J. Basic Microbiol. 2019, 60, 103–111. [Google Scholar] [CrossRef]
- Amend, A.S.; Barshis, D.J.; Oliver, T.A. Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J. 2012, 6, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Littman, R.; Willis, B.L.; Bourne, D.G. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 2011, 3, 651–660. [Google Scholar] [CrossRef]
- Thurber, R.V.; Willner-Hall, D.; Rodriguez-Mueller, B.; Desnues, C.; Edwards, R.A.; Angly, F.; Dinsdale, E.; Kelly, L.; Rohwer, F. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 2009, 11, 2148–2163. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.W.; Groenewald, J.Z.; Pfenning, L.H.; Yarden, O.; Crous, P.W.; Cai, L. The phoma-like dilemma. Stud. Mycol. 2020. [Google Scholar] [CrossRef]
- Yarden, O.; Ainsworth, T.D.; Roff, G.; Leggat, W.; Fine, M.; Hoegh-Guldberg, O. Increased prevalence of ubiquitous ascomycetes in an acropoid coral (Acropora formosa) exhibiting symptoms of brown band syndrome and skeletal eroding band disease. Appl. Environ. Microbiol. 2007, 73, 2755–2757. [Google Scholar] [CrossRef]
- Sutherland, K.P.; Porter, J.W.; Torres, C. Disease and immunity in caribbean and indo-pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 2004, 266, 273–302. [Google Scholar] [CrossRef]
- Smith, G.W.; Ives, L.D.; Nagelkerken, I.A.; Ritchie, K.B. Caribbean sea-fan mortalities. Nature 1996, 383, 487. [Google Scholar] [CrossRef]
- Geiser, D.M.; Taylor, J.W.; Ritchie, K.B.; Smith, G.W. Cause of sea fan death in the West Indies. Nature 1998, 394, 137–138. [Google Scholar] [CrossRef]
- Toledo-Hernández, C.; Zuluaga-Montero, A.; Bones-González, A.; Rodríguez, J.A.; Sabat, A.M.; Bayman, P. Fungi in healthy and diseased sea fans (Gorgonia ventalina): Is Aspergillus sydowii always the pathogen? Coral Reefs 2008, 27, 707–714. [Google Scholar] [CrossRef]
- Ein-Gil, N.; Ilan, M.; Carmeli, S.; Smith, G.W.; Pawlik, J.R.; Yarden, O. Presence of Aspergillus sydowii, a pathogen of gorgonian sea fans in the marine sponge Spongia obscura. ISME J. 2009, 3, 752–755. [Google Scholar] [CrossRef]
- Alker, A.P.; Smith, G.W.; Kim, K. Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen of Caribbean sea fan corals. In Hydrobiologia; Springer: New York, NY, USA, 2001; Volume 460, pp. 105–111. [Google Scholar]
- Ward, J.R.; Kim, K.; Harvell, C.D. Temperature affects coral disease resistance and pathogen growth. Mar. Ecol. Prog. Ser. 2007, 329, 115–121. [Google Scholar] [CrossRef]
- Zhou, G.; Whong, W.Z.; Ong, T.; Chen, B. Development of a fungus-specific PCR assay for detecting low-level fungi in an indoor environment. Mol. Cell. Probes 2000, 14, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Genin, A.; Lazar, B.; Brenner, S. Vertical mixing and coral death in the red sea following the eruption of Mount Pinatubo. Nature 1995, 377, 507–510. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Volkmann-kohlmeyer, B. Lulworthiales, a New Order of marine Ascomycota. Mycologia 2000, 92, 453–458. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10. [Google Scholar] [CrossRef]
- Gleason, F.H.; Gadd, G.M.; Pitt, J.I.; Larkum, A.W.D. The roles of endolithic fungi in bioerosion and disease in marine ecosystems. II. Potential facultatively parasitic anamorphic ascomycetes can cause disease in corals and molluscs. Mycology 2017, 8, 216–227. [Google Scholar] [CrossRef]
- Paz, Z.; Komon-Zelazowska, M.; Druzhinina, I.S.; Aveskamp, M.M.; Shnaiderman, A.; Aluma, Y.; Carmeli, S.; Ilan, M.; Yarden, O. Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 2010, 42, 17–26. [Google Scholar] [CrossRef]
- Hyde, K.D.; Farrant, C.A.; Jones, E.B.G. Isolation and Culture of Marine Fungi. Bot. Mar. 1987, 30, 291–304. [Google Scholar] [CrossRef]
- López-Legentil, S.; Erwin, P.M.; Turon, M.; Yarden, O. Diversity of fungi isolated from three temperate ascidians. Symbiosis 2015, 66, 99–106. [Google Scholar] [CrossRef]
- Overy, D.P.; Rämä, T.; Oosterhuis, R.; Walker, A.K.; Pang, K.L. The neglected marine fungi, sensu stricto, and their isolation for natural products’ discovery. Mar. Drugs 2019, 17, 42. [Google Scholar] [CrossRef]
- Overy, D.P.; Bayman, P.; Kerr, R.G.; Bills, G.F. An assessment of natural product discovery from marine (sensu strictu) and marine-derived fungi. Mycology 2014, 5, 145–167. [Google Scholar] [CrossRef]
- Morales, S.E.; Biswas, A.; Herndl, G.J.; Baltar, F. Global structuring of phylogenetic and functional diversity of pelagic fungi by depth and temperature. Front. Mar. Sci. 2019, 6, 131. [Google Scholar] [CrossRef]
- Gektidis, M.; Dubinsky, Z.; Goffredo, S. Microendoliths of the shallow euphotic zone in open and shaded habitats at 30°N—Eilat, Israel—Paleoecological implications. Facies 2007, 53, 43–55. [Google Scholar] [CrossRef]
- Dishon, G.; Dubinsky, Z.; Fine, M.; Iluz, D. Underwater light field patterns in subtropical coastal waters: A case study from the Gulf of Eilat (Aqaba). Isr. J. Plant Sci. 2012, 60, 265–275. [Google Scholar] [CrossRef]
- Cohen, I.; Dishon, G.; Iluz, D.; Dubinsky, Z. UV-B as a photoacclimatory enhancer of the hermatypic Coral Stylophora pistillata. Open J. Mar. Sci. 2013, 3, 15–27. [Google Scholar] [CrossRef]
- Wolf-Vecht, A.; Paldor, N.; Brenner, S. Hydrographic indications of advection/convection effects in the Gulf of Elat. Deep Sea Res. Part A Oceanogr. Res. Pap. 1992, 39, 1393–1401. [Google Scholar] [CrossRef]
- Arvas, M.; Kivioja, T.; Mitchell, A.; Saloheimo, M.; Ussery, D.; Penttila, M.; Oliver, S. Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina. BMC Genom. 2007, 8, 325. [Google Scholar] [CrossRef]
- Wegley, L.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719. [Google Scholar] [CrossRef]
- Marubini, F.; Davies, P.S. Nitrate increases zooxanthellae population density and reduces skeletogenesis in corals. Mar. Biol. 1996, 127, 319–328. [Google Scholar] [CrossRef]
- Tanaka, Y.; Miyajima, T.; Koike, I.; Hayashibara, T.; Ogawa, H. Translocation and conservation of organic nitrogen within the coral-zooxanthella symbiotic system of Acropora pulchra, as demonstrated by dual isotope-labeling techniques. J. Exp. Mar. Biol. Ecol. 2006, 336, 110–119. [Google Scholar] [CrossRef]
- Le Campion Alsumard, T.; Golubic, S.; Priess, K. Fungi in corals: Symbiosis or disease? Interaction between polyps and fungi causes pearl-like skeleton biomineralization. Mar. Ecol. Prog. Ser. 1995, 117, 137–148. [Google Scholar] [CrossRef]
- Roitman, S.; Joseph Pollock, F.; Medina, M. Coral microbiomes as bioindicators of reef health. In Population Genomics; Springer: Cham, Switzerland, 2018; pp. 39–57. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lifshitz, N.; Hazanov, L.; Fine, M.; Yarden, O. Seasonal Variations in the Culturable Mycobiome of Acropora loripes along a Depth Gradient. Microorganisms 2020, 8, 1139. https://doi.org/10.3390/microorganisms8081139
Lifshitz N, Hazanov L, Fine M, Yarden O. Seasonal Variations in the Culturable Mycobiome of Acropora loripes along a Depth Gradient. Microorganisms. 2020; 8(8):1139. https://doi.org/10.3390/microorganisms8081139
Chicago/Turabian StyleLifshitz, Nofar, Lena Hazanov, Maoz Fine, and Oded Yarden. 2020. "Seasonal Variations in the Culturable Mycobiome of Acropora loripes along a Depth Gradient" Microorganisms 8, no. 8: 1139. https://doi.org/10.3390/microorganisms8081139
APA StyleLifshitz, N., Hazanov, L., Fine, M., & Yarden, O. (2020). Seasonal Variations in the Culturable Mycobiome of Acropora loripes along a Depth Gradient. Microorganisms, 8(8), 1139. https://doi.org/10.3390/microorganisms8081139





