Evaluation of Product Distribution in Chemostat and Batch Fermentation in Lactic Acid-Producing Komagataella phaffii Strains Utilizing Glycerol as Substrate
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
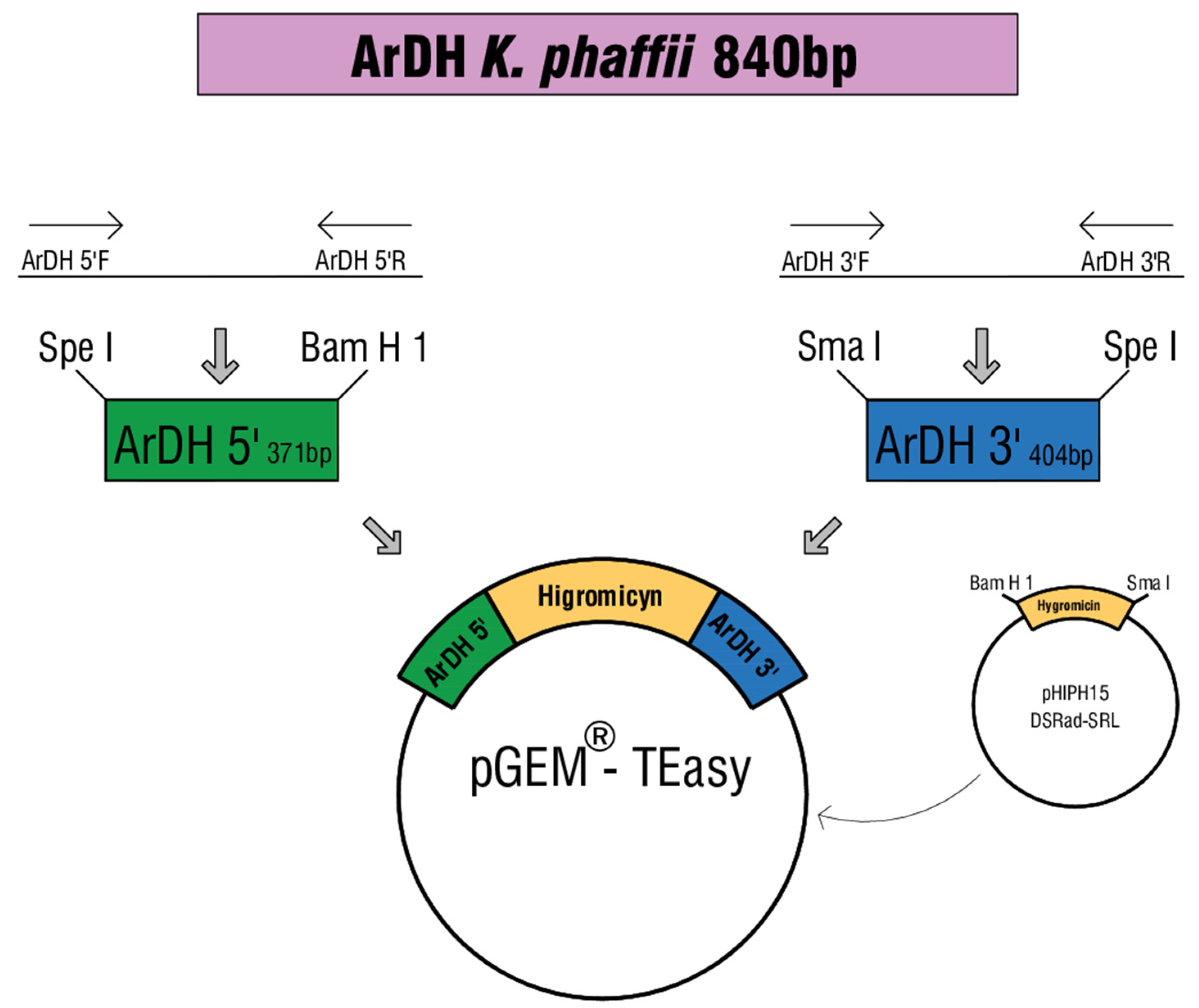
2.2. Construction of the ArDH Knockout Cassette
2.3. ArDH Knockout in K. phaffii
2.4. Cultivation in Bioreactor
2.4.1. Medium
2.4.2. Chemostat Cultivations
2.4.3. Batch Fermentation
2.4.4. Fed-Batch Fermentations
2.5. Biomass Determination
2.6. Substrate Consumption and Cellular Products Quantification
2.7. Statistical Analysis
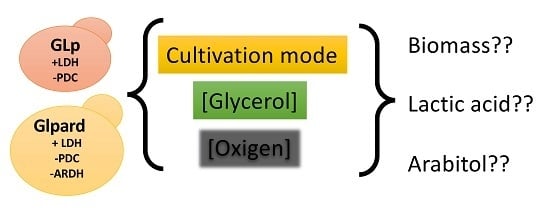
3. Results and discussion
3.1. Effect of the Dilution Rate and Oxygen Concentration for the Production of Lactic Acid in Glycerol-Limited Chemostat Cultures
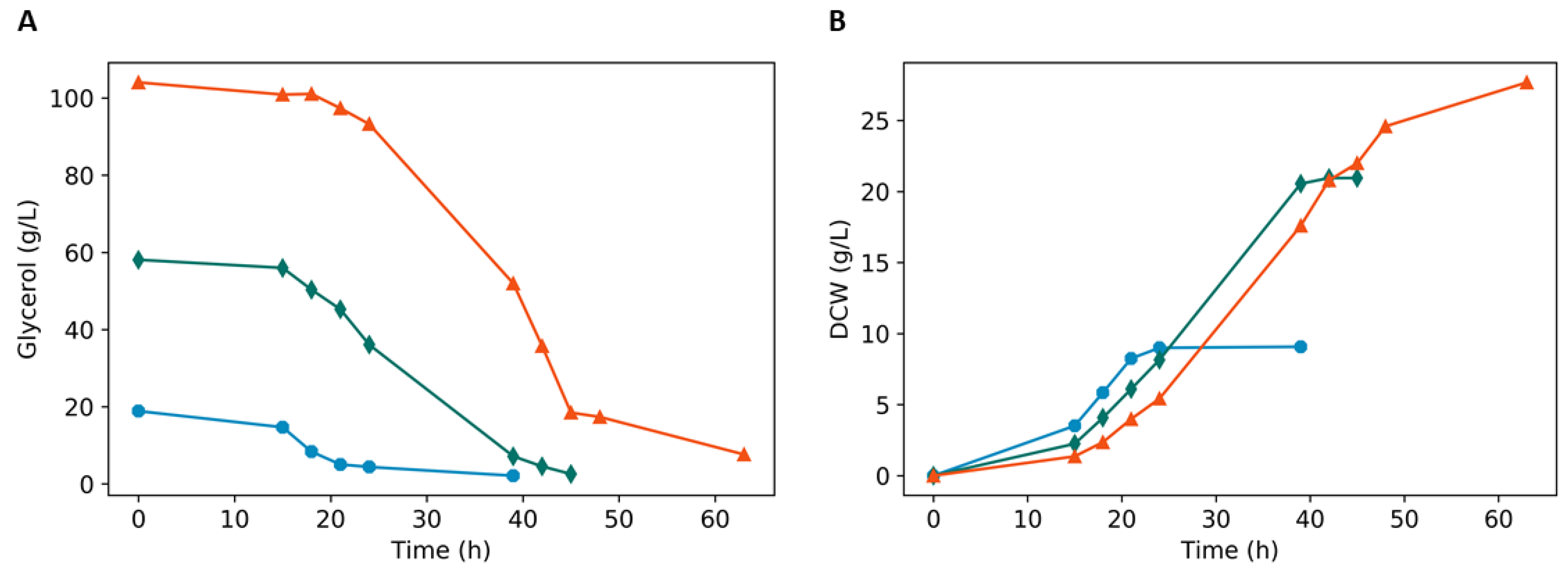
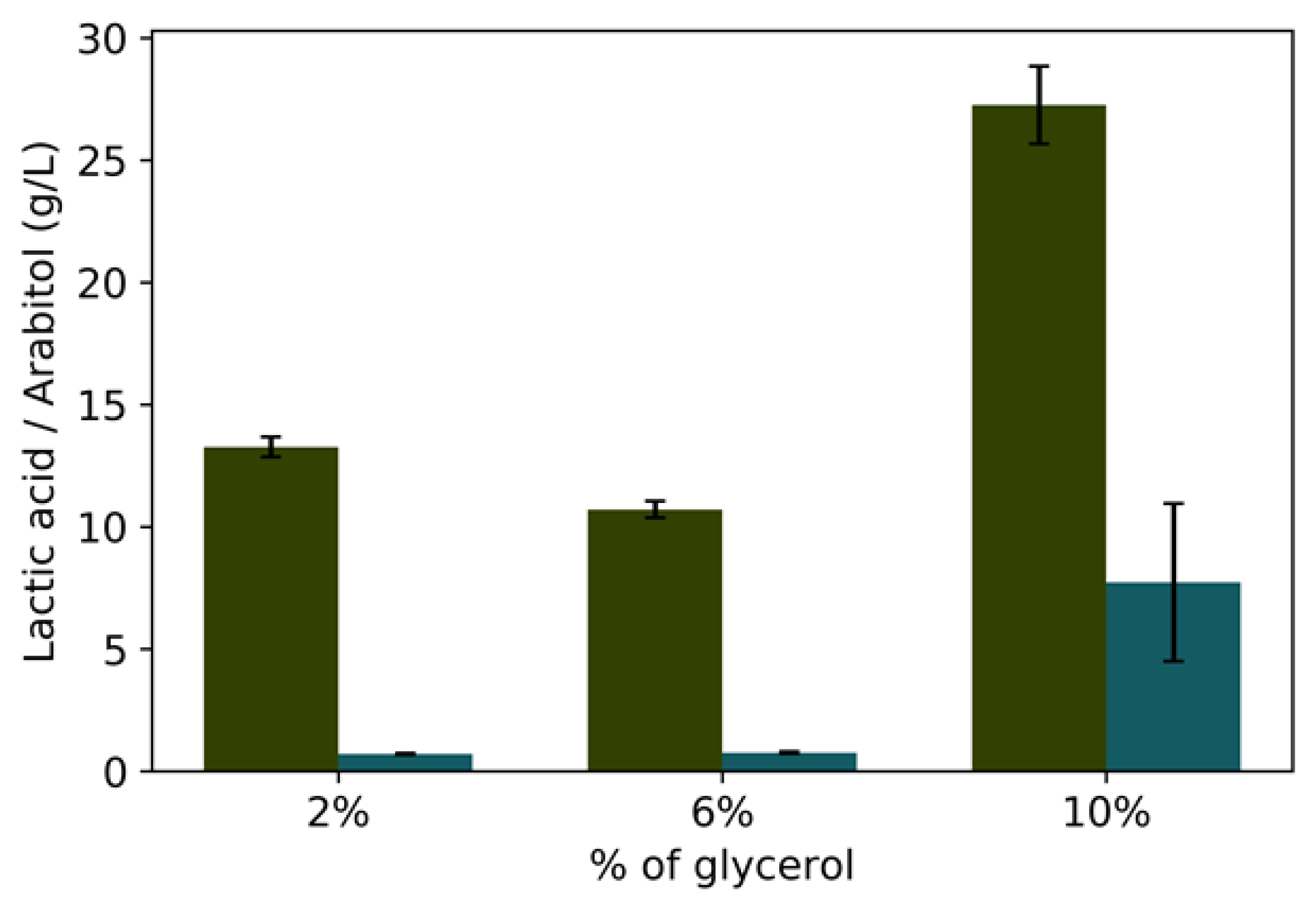
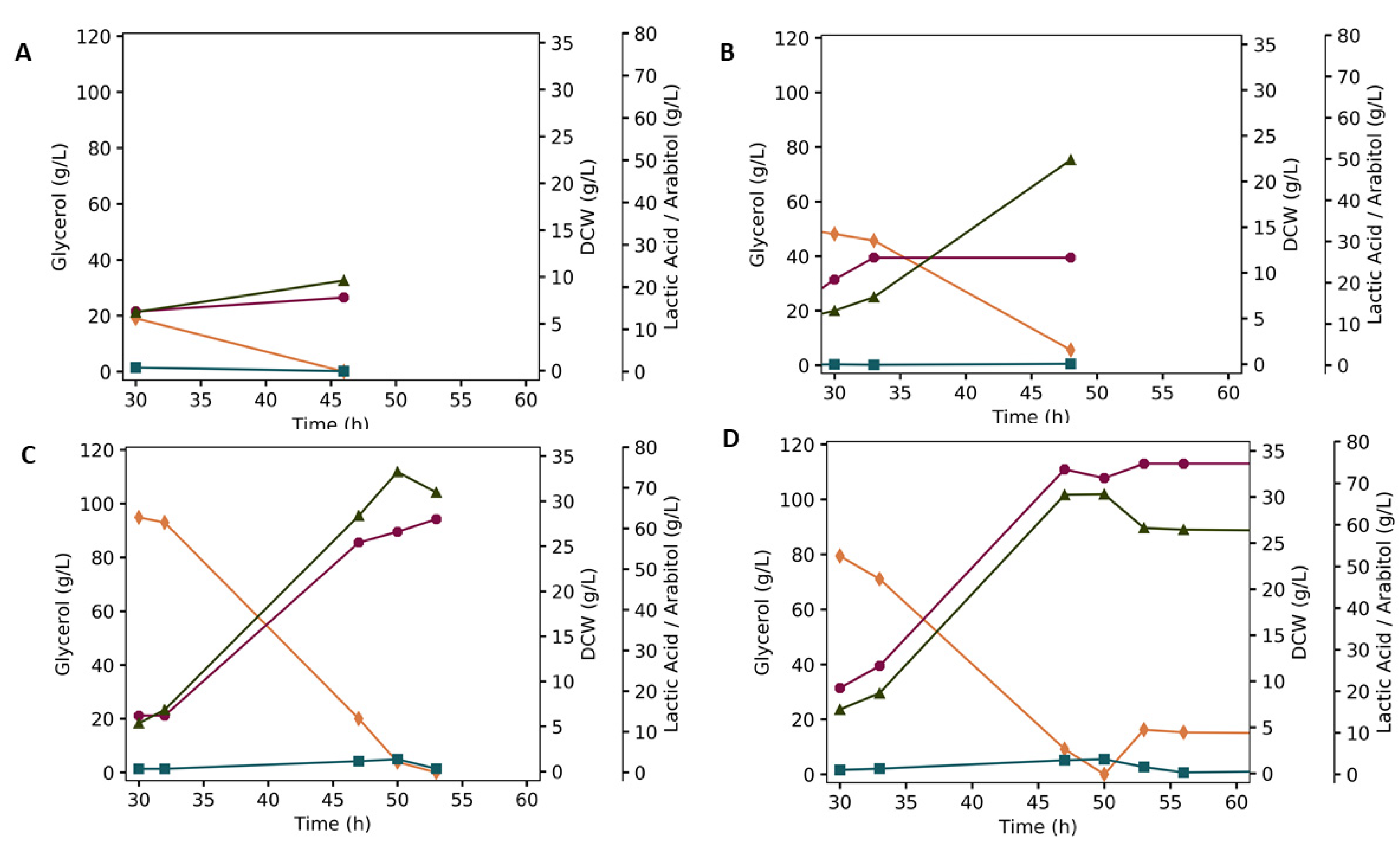
3.2. Batch Fermentation
4. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Alves de Oliveira, R.; Komesu, A.; Vaz Rossell, C.E.; Maciel Filho, R. Challenges and opportunities in lactic acid bioprocess design—From economic to production aspects. Biochem. Eng. J. 2018, 133, 219–239. [Google Scholar] [CrossRef]
- Castillo Martinez, F.A.; Balciunas, E.M.; Salgado, J.M.; Domínguez González, J.M.; Converti, A.; de Souza Oliveira, R.P. Lactic acid properties, applications and production: A review. Trends Food Sci. Technol. 2013, 30, 70–83. [Google Scholar] [CrossRef]
- Wee, Y.J.; Kim, J.N.; Ryu, H.W. Biotechnological production of lactic acid and its recent applications. Food Technol. Biotechnol. 2006, 44, 163–172. [Google Scholar]
- Willem, A.; Thibault, J.; Lacroix, C. Lactobacillus helveticus growth and lactic acid production during pH- controlled batch cultures in whey permeate/yeast extract medium. Part II: Kinetic modeling and model validation. Enzyme Microbial Technol. 2002, 30, 187–194. [Google Scholar]
- Demirci, A.; Iii, A.L.P.; Lee, B.; Hinz, P.N. Media Evaluation of Lactic Acid Repeated-Batch Fermentation with Lactobacillus plantarum and Lactobacillus casei Subsp. rhamnosus †. J. Agric. Food Chem. East. 1998, 46, 4771–4774. [Google Scholar] [CrossRef]
- Wang, Y.D.; Liao, J.Y.; Chiang, C.J.; Chao, Y.P. A simple strategy to effectively produce d-lactate in crude glycerol-utilizing Escherichia coli. Biotechnol. Biofuels 2019, 12, 1–9. [Google Scholar] [CrossRef]
- López-Gómez, J.P.; Alexandri, M.; Schneider, R.; Venus, J. A review on the current developments in continuous lactic acid fermentations and case studies utilising inexpensive raw materials. Process. Biochem. 2019, 79, 1–10. [Google Scholar] [CrossRef]
- Miller, C.; Fosmer, A.; Rush, B.; McMullin, T.; Beacom, D.; Suominen, P. Industrial Production of Lactic Acid. Ref. Modul. Life Sci. 2017, 1–10. [Google Scholar]
- Kong, X.; Zhang, B.; Hua, Y.; Zhu, Y.; Li, W.; Wang, D.; Hong, J. Bioresource Technology E ffi cient L -lactic acid production from corncob residue using metabolically engineered thermo-tolerant yeast. Bioresour. Technol. 2019, 273, 220–230. [Google Scholar] [CrossRef]
- Anastácio, G.S.; Santos, K.O.; Suarez, P.A.Z.; Torres, F.A.G.; De Marco, J.L.; Parachin, N.S. Utilization of glycerin byproduct derived from soybean oil biodiesel as a carbon source for heterologous protein production in Pichia pastoris. Bioresour. Technol. 2014, 152, 505–510. [Google Scholar] [CrossRef]
- Koganti, S.; Kuo, T.M.; Kurtzman, C.P.; Smith, N.; Ju, L.K. Production of arabitol from glycerol: Strain screening and study of factors affecting production yield. Appl. Microbiol. Biotechnol. 2011, 90, 257–267. [Google Scholar] [CrossRef] [PubMed]
- da Silva, G.P.; Mack, M.; Contiero, J. Glycerol: A promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 2009, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Tomàs-gamisans, M.; Sebastian, A.; Ødum, R.; Workman, M.; Ferrer, P.; Albiol, J. Glycerol metabolism of Pichia pastoris ( Komagataella spp.) characterised by C-based metabolic fl ux analysis. N. Biotechnol. 2019, 50, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Lima, P.B.A.; Mulder, K.C.L.; Melo, N.T.M.; Carvalho, L.S.; Menino, G.S.; Mulinari, E.; Castro, V.H.; Reis, T.F.; Goldman, G.H.; Magalhães, B.S.; et al. Novel homologous lactate transporter improves l-lactic acid production from glycerol in recombinant strains of Pichia pastoris. Microb. Cell Fact. 2016, 15, 1–9. [Google Scholar]
- Peña, D.A.; Gasser, B.; Zanghellini, J.; Steiger, M.G.; Mattanovich, D. Metabolic engineering of Pichia pastoris. Metab. Eng. 2018, 50, 2–15. [Google Scholar] [CrossRef]
- Siripong, W.; Wolf, P.; Kusumoputri, T.P.; Downes, J.J.; Kocharin, K.; Tanapongpipat, S.; Runguphan, W. Metabolic engineering of Pichia pastoris for production of isobutanol and isobutyl acetate. Biotechnol. Biofuels 2018, 11, 1–16. [Google Scholar] [CrossRef]
- Tokuhiro, K.; Ishida, N.; Nagamori, E.; Saitoh, S.; Onishi, T.; Kondo, A.; Takahashi, H. Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl. Microbiol. Biotechnol. 2009, 82, 883–890. [Google Scholar] [CrossRef]
- Ishida, N.; Saitoh, S.; Tokuhiro, K.; Nagamori, E.; Matsuyama, T.; Kitamoto, K.; Takahashi, H. Efficient production of L-lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Appl. Environ. Microbiol. 2005, 71, 1964–1970. [Google Scholar] [CrossRef]
- Ishida, N.; Saitoh, S.; Onishi, T.; Tokuhiro, K.; Nagamori, E.; Kitamoto, K.; Takahashi, H. The Effect of Pyruvate Decarboxylase Gene Knockout in Saccharomyces cerevisiae on L -Lactic Acid Production. Biosci. Biotechnol. Biochem. 2006, 70, 1148–1153. [Google Scholar] [CrossRef]
- Skory, C.D. Induction of Rhizopus oryzae pyruvate decarboxylase genes. Curr. Microbiol. 2003, 47, 59–64. [Google Scholar] [CrossRef]
- Melo, N.T.M.; Mulder, K.C.L.; Nicola, A.M.; Carvalho, L.S.; Menino, G.S.; Mulinari, E.; Parachin, N.S. Effect of pyruvate decarboxylase knockout on product distribution using Pichia pastoris (Komagataella phaffii) engineered for lactic acid production. Bioengineering 2018, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Carnicer, M.; Baumann, K.; Töplitz, I.; Sánchez-, F.; Mattanovich, D.; Ferrer, P.; Albiol, J. Macromolecular and elemental composition analysis and extracellular metabolite balances of Pichia pastoris growing at different oxygen levels. Microb. Cell Fact. 2009, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Saraya, R.; Krikken, A.M.; Kiel, J.A.K.W.; Baerends, R.J.S.; Veenhuis, M.; van der Klei, I.J. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 2012, 12, 271–278. [Google Scholar] [CrossRef]
- Olsson, L.; Nielsen, J. On-line and in situ monitoring of biomass in submerged cultivations. Elsevier Sci. Ltd. 1997, 15, 517–522. [Google Scholar] [CrossRef]
- Tomàs-Gamisans, M.; Ferrer, P.; Albiol, J. Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources. Microb. Biotechnol. 2018, 11, 224–237. [Google Scholar] [CrossRef]
- de Jesus, S.S.; Peltier, S.; Ferrer, P.; Albiol, J.; Jorda, J. Metabolic flux analysis of recombinant Pichia pastoris growing on different glycerol/methanol mixtures by iterative fitting of NMR-derived 13 C- labelling data from proteinogenic amino acids. N. Biotechnol. 2014, 31, 120–131. [Google Scholar]
- Baumann, K.; Carnicer, M.; Dragosits, M.; Graf, A.B.; Stadlmann, J.; Jouhten, P.; Maaheimo, H.; Gasser, B.; Albiol, J.; Mattanovich, D.; et al. A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC Syst. Biol. 2010, 4, 141. [Google Scholar] [CrossRef]
- Cheng, H.; Lv, J.; Wang, H.; Wang, B.; Li, Z.; Deng, Z. Genetically engineered Pichia pastoris yeast for conversion of glucose to xylitol by a single-fermentation process. Appl. Microbiol. Biotechnol. 2014, 98, 3539–3552. [Google Scholar] [CrossRef]
- Zhang, G.; Lin, Y.; He, P.; Li, L.; Wang, Q.; Ma, Y. Characterization of the sugar alcohol-producing yeast Pichia anomala. J. Ind. Microbiol. Biotechnol. 2014, 41, 41–48. [Google Scholar] [CrossRef]
- Baumann, K.; Maurer, M.; Dragosits, M.; Cos, O.; Ferrer, P.; Mattanovich, D. Hypoxic fed-batch cultivation of Pichia pastoris increases specific and volumetric productivity of recombinant proteins. Biotechnol. Bioeng. 2008, 100, 177–183. [Google Scholar] [CrossRef]
- Nocon, J.; Steiger, M.; Mairinger, T.; Hohlweg, J.; Rußmayer, H.; Hann, S.; Gasser, B.; Mattanovich, D. Increasing pentose phosphate pathway flux enhances recombinant protein production in Pichia pastoris. Appl. Microbiol. Biotechnol. 2016, 100, 5955–5963. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kang, C.D.; Lee, S.H.; Park, Y.K.; Cho, K.M. Engineering cellular redox balance in Saccharomyces cerevisiae for improved production of L-lactic acid. Biotechnol. Bioeng. 2015, 112, 751–758. [Google Scholar] [CrossRef] [PubMed]
| Plasmids and Strains | Relevant Genotype | Ref. |
|---|---|---|
| pGEM®-T Easy | The pre-linearized Vector contains 3’-T protrusions at the insertion site to provide a compatible protrusion for PCR products. | Promega Corporation |
| pGEMHY ardh | ArDH 5′+Hygromyicin+ArDH3′ | This study |
| pHIPH15 DsRed-SKL | Ogataea polymorpha vector with Hygromycin mark | [23] |
| GLp | GS115: Δpdc + pGAP-LDH Bos taurus | [21] |
| GLpard | GS115: Δpdc + pGAP-LDH Bos taurus +Δardh | This study |
| Primers | Sequence (5′ 3′) | Restriction Site |
|---|---|---|
| Primer ArDH 5′ F | CGGGCATGCCCGATGTCGTTATCGTTAGACAACATAGTTCCA | SphI |
| Primer ArDH 5′ R | CGCGGATCCAGTATCCGGCGCAATGTACTAAG | BamHI |
| Primer ArDH 3′ F | TCCCCCGGGGAAATCCGGCCACAGAAACGG | SmaI |
| Primer ArDH 3′ R | CGGACTAGTCCGTTACCAGCATTCGTAACCACCATCG | SpeI |
| Primer HIGRO 5′ F | CCGGGATCCCGCCCACACACCATAGCTTCAAAATGTT | BamHI |
| Primer HIGRO 3′ R | ATATCCCCCCGGGGGCGTTTTCGACACTGGATGGCG | SmaI |
| 21% Oxygen | 4% Oxygen | |||
|---|---|---|---|---|
| Dilution rate (h−1) | 0.052 | 0.090 | 0.140 | 0.050 |
| YBiomass | 0.736 ± 0.003 | 0.663 ± 0.023 | 0.689 ± 0.047 | 0.534 ± 0.023 |
| YCO2 | 0.298 ± 0.003 | 0.269 ± 0.006 | 0.242 ± 0.027 | 0.190 ± 0.005 |
| YLactate | 0.007 ± 0.006 | 0.030 ± 0.007 | 0.049 ± 0.001 | 0.237 ± 0.016 |
| YArabitol | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 |
| Glycerol Consumption rate (g.L−1.h−1) | 0.51 ± 0.0162 | 0.890 ± 0.0480 | 1.39 ± 0.0268 | 0.50± 0.0989 |
| CRecovery | 1.014 ± 0.006 | 0.984 ± 0.023 | 0.983 ± 0.020 | 0.964 ± 0.016 |
| Glycerol Concentration | Residual Glycerol (g/L) | Yx/g | Ylac/g | Yara/g | |
|---|---|---|---|---|---|
| Oxygen 20% | GLp 2% | 0.982 ± 0.009 | 0.60 ± 0.130 | 0.370 ± 0.02 | 0.01 ± 0.008 |
| GLp 6% | 1.81 ± 0.121 | 0.46 ± 0.018 | 0.200± 0.003 | 0.02 ± 0.010 | |
| GLp10% | 3.307 ± 0.231 | 0.37 ± 0.086 | 0.300 ± 0.055 | 0.12 ± 0.073 | |
| Oxygen-limited | GLp2% | 0.608 ± 0.023 | 0.143 ± 0.037 | 0.559 ± 0.369 | 0.028 ± 0.000 |
| GLp 6% | 1.16 ±0.008 | 0.087 ± 0.014 | 0.855 ± 0.077 | 0.012 ± 0.014 | |
| GLp 8% | 2.77 ± 0.044 | 0.239 ± 0.081 | 0.665 ± 0.025 | 0.072 ± 0.064 | |
| GLpard 6% | 1.975 ± 0.035 | 0.304 ± 0.067 | 0.8555 ± 0.66 | 0.004 ± 0.023 | |
| Glpard 8% | 2.134 ± 0.098 | 0.448 ± 0.013 | 0.763 ± 0.034 | 0.034 ± 0.015 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamires Moreira Melo, N.; Pontes, G.C.; Procópio, D.P.; de Gois e Cunha, G.C.; Eliodório, K.P.; Costa Paes, H.; Basso, T.O.; Parachin, N.S. Evaluation of Product Distribution in Chemostat and Batch Fermentation in Lactic Acid-Producing Komagataella phaffii Strains Utilizing Glycerol as Substrate. Microorganisms 2020, 8, 781. https://doi.org/10.3390/microorganisms8050781
Tamires Moreira Melo N, Pontes GC, Procópio DP, de Gois e Cunha GC, Eliodório KP, Costa Paes H, Basso TO, Parachin NS. Evaluation of Product Distribution in Chemostat and Batch Fermentation in Lactic Acid-Producing Komagataella phaffii Strains Utilizing Glycerol as Substrate. Microorganisms. 2020; 8(5):781. https://doi.org/10.3390/microorganisms8050781
Chicago/Turabian StyleTamires Moreira Melo, Nadielle, Gabriela Coimbra Pontes, Dielle Pierotti Procópio, Gabriel Caetano de Gois e Cunha, Kevy Pontes Eliodório, Hugo Costa Paes, Thiago Olitta Basso, and Nádia Skorupa Parachin. 2020. "Evaluation of Product Distribution in Chemostat and Batch Fermentation in Lactic Acid-Producing Komagataella phaffii Strains Utilizing Glycerol as Substrate" Microorganisms 8, no. 5: 781. https://doi.org/10.3390/microorganisms8050781
APA StyleTamires Moreira Melo, N., Pontes, G. C., Procópio, D. P., de Gois e Cunha, G. C., Eliodório, K. P., Costa Paes, H., Basso, T. O., & Parachin, N. S. (2020). Evaluation of Product Distribution in Chemostat and Batch Fermentation in Lactic Acid-Producing Komagataella phaffii Strains Utilizing Glycerol as Substrate. Microorganisms, 8(5), 781. https://doi.org/10.3390/microorganisms8050781