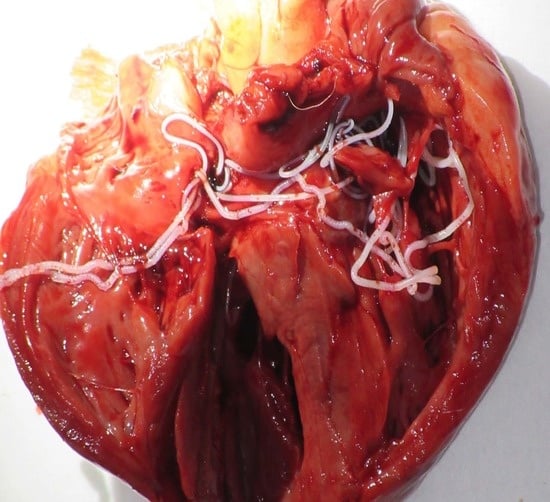
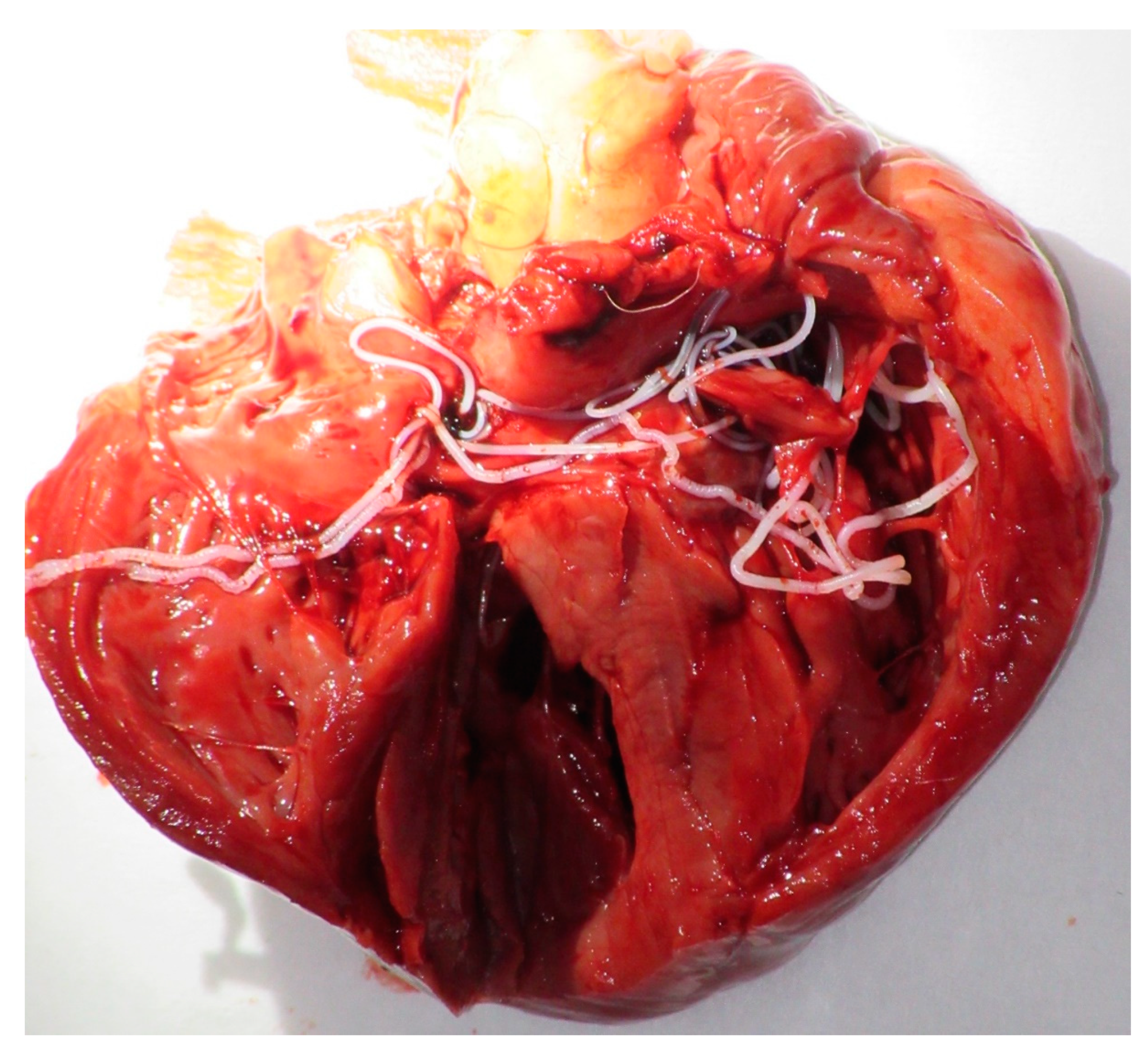
Detection of Canine Vector-Borne Filariasis and Their Wolbachia Endosymbionts in French Guiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Study Area
2.2. Ethics Approval
2.3. Microfilaria and Heartworm Antigen Tests
2.4. DNA Extraction
2.5. Molecular Screening for Filarioid and Wolbachia DNAs
2.6. Molecular and Phylogenetic Characterization of Filarioid and Wolbachia
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dantas-Torres, F. Canine vector-borne diseases in Brazil. Parasit. Vectors 2008, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Otranto, D.; Dantas-Torres, F.; Breitschwerdt, E.B. Managing canine vector-borne diseases of zoonotic concern: Part one. Trends Parasitol. 2009, 25, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Medkour, H.; Davoust, B.; Dulieu, F.; Maurizi, L.; Lamour, T.; Marié, J.-L.; Mediannikov, O. Potential animal reservoirs (dogs and bats) of human visceral leishmaniasis due to Leishmania infantum in French Guiana. PLoS Negl. Trop. Dis. 2019, 13, e0007456. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, M.; Marié, J.-L.; Mediannikov, O.; Raoult, D.; Davoust, B. First Identification of Anaplasma platys in the blood of dogs from French Guiana. Vector Borne Zoonotic Dis. 2015, 15, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Tahir, D.; Davoust, B.; Heu, K.; Lamour, T.; Demar, M.; Marié, J.-L.; Blanchet, D. Molecular and serological investigation of Trypanosoma cruzi infection in dogs in French Guiana. Vet. Parasitol. Reg. Stud. Rep. 2017, 12, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Davoust, B.; Keundjian, A.; Rous, V.; Maurizi, L.; Parzy, D. Validation of chemoprevention of canine monocytic ehrlichiosis with doxycycline. Vet. Microbiol. 2005, 107, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Llagonne-Barets, M.; Icard, V.; Leparc-Goffart, I.; Prat, C.; Perpoint, T.; Andre, P.; Ramière, C. A case of Mayaro virus infection imported from French Guiana. J. Clin. Virol. 2016, 77, 66–68. [Google Scholar] [CrossRef]
- Simón, F.; Siles-Lucas, M.; Morchón, R.; González-Miguel, J.; Mellado, I.; Carretón, E.; Montoya-Alonso, J.A. Human and animal dirofilariasis: The emergence of a zoonotic mosaic. Clin. Microbiol. Rev. 2012, 25, 507–544. [Google Scholar] [CrossRef]
- Laidoudi, Y.; Ringot, D.; Watier-Grillot, S.; Davoust, B.; Mediannikov, O. A cardiac and subcutaneous canine dirofilariosis outbreak in a kennel in central France. Parasite 2019, 26, 72. [Google Scholar] [CrossRef]
- Satjawongvanit, H.; Phumee, A.; Tiawsirisup, S.; Sungpradit, S.; Brownell, N.; Siriyasatien, P.; Preativatanyou, K. Molecular analysis of canine filaria and its Wolbachia endosymbionts in domestic dogs collected from two animal university hospitals in Bangkok Metropolitan Region, Thailand. Pathogens 2019, 8, 114. [Google Scholar] [CrossRef]
- Ambily, V.; Pillai, U.N.; Arun, R.; Pramod, S.; Jayakumar, K. Detection of human filarial parasite Brugia malayi in dogs by histochemical staining and molecular techniques. Vet. Parasitol. 2011, 181, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, R.; Varghese, S.; Nair, S.N.; Balan, V.M.; Lakshmanan, B.; Ashruf, R.M.; Kumar, S.S.; Gopalan, A.K.K.; Nair, A.S.; Malayil, A.; et al. Canine filarial infections in a human Brugia malayi endemic area of India. BioMed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.; Wiseman, S. Increasing incidence of Dirofilaria immitis in dogs in USA with focus on the southeast region 2013–2016. Parasit. Vectors 2018, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Lymphatic Filariasis. Available online: https://www.who.int/news-room/fact-sheets/detail/lymphatic-filariasis (accessed on 12 May 2020).
- Brianti, E.; Gaglio, G.; Napoli, E.; Giannetto, S.; Dantas-Torres, F.; Bain, O.; Otranto, D. New insights into the ecology and biology of Acanthocheilonema reconditum (Grassi, 1889) causing canine subcutaneous filariosis. Parasitology 2012, 139, 530–536. [Google Scholar] [CrossRef]
- Otranto, D.; Brianti, E.; Latrofa, M.S.; Annoscia, G.; Weigl, S.; Lia, R.P.; Gaglio, G.; Napoli, E.; Giannetto, S.; Papadopoulos, E.; et al. On a Cercopithifilaria sp. transmitted by Rhipicephalus sanguineus: A neglected, but widespread filarioid of dogs. Parasit. Vectors 2012, 5, 1. [Google Scholar] [CrossRef]
- Cortes, H.; Cardoso, L.; Giannelli, A.; Latrofa, M.S.; Dantas-Torres, F.; Otranto, D. Diversity of Cercopithifilaria species in dogs from Portugal. Parasit. Vectors 2014, 7, 261. [Google Scholar] [CrossRef]
- Nelson, G.S. Dipetalonema reconditum (Grassi, 1889) from the dog with a note on its development in the flea, Ctenocephalides felis and the louse, Heterodoxus spiniger. J. Helminthol. 1962, 36, 297. [Google Scholar] [CrossRef]
- Muñoz, C.; Gonzálvez, M.; Rojas, A.; Martínez-Carrasco, C.; Baneth, G.; Berriatua, E.; Ortiz, J. Massive microfilaremia in a dog subclinically infected with Acanthocheilonema dracunculoides. Parasitol. Int. 2020, 76, 102070. [Google Scholar] [CrossRef]
- Franchini, D.; Giannelli, A.; Di Paola, G.; Cortes, H.; Cardoso, L.; Lia, R.P.; Campbell, B.; Dantas-Torres, F.; Lenoci, D.; Abu Assad, E.; et al. Image diagnosis of zoonotic onchocercosis by Onchocerca lupi. Vet. Parasitol. 2014, 203, 91–95. [Google Scholar] [CrossRef]
- Martin, C.; Gavotte, L. The bacteria Wolbachia in filariae, a biological Russian dolls’ system: New trends in antifilarial treatments. Parasite 2010, 17, 79–89. [Google Scholar] [CrossRef][Green Version]
- Sabūnas, V.; Radzijevskaja, J.; Sakalauskas, P.; Petkevicius, S.; Karvelienė, B.; Žiliukienė, J.; Lipatova, I.; Paulauskas, A. Dirofilaria repens in dogs and humans in Lithuania. Parasit. Vectors 2019, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Bandi, C.; Anderson, T.J.C.; Genchi, C.; Blaxter, M. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. B Boil. Sci. 1998, 265, 2407–2413. [Google Scholar] [CrossRef] [PubMed]
- Turba, M.E.; Zambon, E.; Zannoni, A.; Russo, S.; Gentilini, F. Detection of Wolbachia DNA in blood for diagnosing filaria-associated syndromes in cats. J. Clin. Microbiol. 2012, 50, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Dingman, P.; Levy, J.K.; Kramer, L.H.; Johnson, C.M.; Lappin, M.R.; Greiner, E.C.; Courtney, C.H.; Tucker, S.J.; Morchón, R. Association of Wolbachia with heartworm disease in cats and dogs. Vet. Parasitol. 2010, 170, 50–60. [Google Scholar] [CrossRef]
- Maia, C.; Altet, L.; Serrano, L.; Cristóvão, J.; Tabar, M.-D.; Francino, O.; Cardoso, L.; Campino, L.; Roura, X. Molecular detection of Leishmania infantum, filariae and Wolbachia spp. in dogs from southern Portugal. Parasit. Vectors 2016, 9, 170. [Google Scholar] [CrossRef]
- Laidoudi, Y.; Davoust, B.; Varloud, M.; El Hadj, A.N.; Fenollar, F.; Mediannikov, O. Development of a multiplexed qPCRs-based approach for the diagnosis of Dirofilaria immitis, D. repens, Acanthocheilonema reconditum and the others filariosis. BioRxiv 2019, 24, 842575. [Google Scholar] [CrossRef]
- De Argôlo, E.G.G.; Reis, T.; Fontes, D.A.T.; Gonçalves, E.C.; Giese, E.G.; Melo, F.T.V.; Dos Santos, J.N.; Furtado, A.P. Canine filariasis in the Amazon: Species diversity and epidemiology of these emergent and neglected zoonoses. PLoS ONE 2018, 13, e0200419. [Google Scholar] [CrossRef]
- Schneider, M.C.; Aguilera, X.; Junior, J.B.D.S.; Ault, S.K.; Najera, P.; Martínez, J.; Requejo, R.; Nicholls, R.S.; Yadon, Z.E.; Silva, J.C.; et al. Elimination of neglected diseases in Latin America and the Caribbean: A mapping of selected diseases. PLoS Negl. Trop. Dis. 2011, 5, e964. [Google Scholar] [CrossRef]
- Werren, J.H.; Windsor, D.M. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc. R. Soc. B Boil. Sci. 2000, 267, 1277–1285. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Howe, K.L.; Bolt, B.J.; Shafie, M.; Kersey, P.; Berriman, M. WormBase ParaSite—A comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017, 215, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Boil. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; p. 333. [Google Scholar]
- Hasegawa, M.; Kishino, H.; Yano, T. aki Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Dantas-Torres, F.; Otranto, D. Dirofilariosis in the Americas: A more virulent Dirofilaria immitis? Parasit. Vectors 2013, 6, 288. [Google Scholar] [CrossRef]
- Otranto, D.; Dantas-Torres, F.; Breitschwerdt, E. Managing canine vector-borne diseases of zoonotic concern: Part two. Trends Parasitol. 2009, 25, 228–235. [Google Scholar] [CrossRef]
- Aroch, I.; Rojas, A.; Slon, P.; Lavy, E.; Segev, G.; Baneth, G. Serological cross-reactivity of three commercial in-house immunoassays for detection of Dirofilaria immitis antigens with Spirocerca lupi in dogs with benign esophageal spirocercosis. Vet. Parasitol. 2015, 211, 303–305. [Google Scholar] [CrossRef]
- Mallawarachchi, C.H.; Chandrasena, N.T.G.A.; Wickramasinghe, S.; Premaratna, R.; Gunawardane, N.Y.I.S.; Mallawarachchi, N.S.M.S.M.; De Silva, N.R. A preliminary survey of filarial parasites in dogs and cats in Sri Lanka. PLoS ONE 2018, 13, e0206633. [Google Scholar] [CrossRef]
- Bowman, D.D.; Mannella, C. Macrocyclic lactones and Dirofilaria immitis microfilariae. Top. Companion Anim. Med. 2011, 26, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.I.D.; Paiva, J.; Bendas, A.; Mendes-De-Almeida, F.; Knackfuss, F.; Miranda, M.; Guerrero, J.; Fernandes, O.; Labarthe, N.V. Effects of doxycycline on the endosymbiont Wolbachia in Dirofilaria immitis (Leidy, 1856)—Naturally infected dogs. Vet. Parasitol. 2010, 174, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.C.; Bradner, J.L.; Savadelis, M.D.; Nelson, C.T.; Moorhead, A.R. Acetic acid as an alternative reagent in the modified Knott test. Vet. Parasitol. 2019, 276, 108975. [Google Scholar] [CrossRef]
- Orihal, T.C. Canine filariasis in British Guiana. J. Parasitol. 1964, 50, 33. [Google Scholar]
- Guilarte, D.V.; Martínez, E.G.; El Hen, F.; Guzmán, R.; Blondell, D.; Díaz, M.T.; Santiago, J. Diagnóstico de Dirofilaria immitis en el municipio Sucre, estado Sucre, Venezuela. Bol. Mal. Salud Amb. 2011, 51, 51–58. [Google Scholar]
- Duran-Struuck, R.; Jost, C.; Hernandez, A.H. Dirofilaria immitis prevalence in a canine population in the Samana Peninsula (Dominican Republic)—June 2001. Vet. Parasitol. 2005, 133, 323–327. [Google Scholar] [CrossRef]
- Gillis, J.M.; Smith, R.D.; Todd, K.S. Diagnostic criteria for an enzyme-linked immunosorbent assay for occult heartworm disease: Standardization of the test system in naturally exposed dogs. Am. J. Veter Res. 1984, 45, 2289–2292. [Google Scholar]
- Bartolo, A. La spirocercose canine à Spirocerca lupi. Point Vét. 2006, 269, 20–26. [Google Scholar]
- Bain, O.; Otranto, D.; Diniz, D.; Dos Santos, J.N.; De Oliveira, N.P.; Almeida, I.; De Almeida, R.N.F.; De Almeida, L.N.F.; Dantas-Torres, F.; Sobrinho, E.F.D.A. Human intraocular filariasis caused by Pelecitus sp. Nematode, Brazil. Emerg. Infect. Dis. 2011, 17, 867–869. [Google Scholar] [CrossRef]
- Mar, P.-H.; Yang, I.-C.; Chang, G.-N.; Fei, C.-Y. Specific polymerase chain reaction for differential diagnosis of Dirofilaria immitis and Dipetalonema reconditum using primers derived from internal transcribed spacer region 2 (ITS2). Vet. Parasitol. 2002, 106, 243–252. [Google Scholar] [CrossRef]
- López, J.; Valiente-Echeverría, F.; Carrasco, M.; Mercado, R.; Abarca Villaseca, K. Morphological and molecular identification of canine filariae in a semi-rural district of the Metropolitan Region in Chile. Rev. Chil. Infectol. 2012, 29, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, S.; León-Galván, M.F.; Salas-Alatorre, M.; Lechuga-Arana, A.A.; Valencia-Posadas, M.; Gutiérrez-Chávez, A.J. First molecular identification of Dirofilaria repens in a dog blood sample from Guanajuato, Mexico. Vector Borne Zoonotic Dis. 2016, 16, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Lent, H.; De Freitas, J.F.T. Dirofilariose sub-cutanea dos cães no Brasil. Memórias Inst. Oswaldo Cruz 1937, 32, 443–448. [Google Scholar] [CrossRef][Green Version]
- Huynh, T.; Thean, J.; Maini, R. Dipetalonema reconditum in the human eye. Br. J. Ophthalmol. 2001, 85, 1391–1392. [Google Scholar] [CrossRef]
- Rojas, A.; Rojas, D.; Montenegro, V.M.; Baneth, G. Detection of Dirofilaria immitis and other arthropod-borne filarioids by an HRM real-time qPCR, blood-concentrating techniques and a serological assay in dogs from Costa Rica. Parasit. Vectors 2015, 8, 170. [Google Scholar] [CrossRef]
- Otranto, D.; Brianti, E.; Abramo, F.; Gaglio, G.; Napoli, E.; Latrofa, M.S.; Ramos, R.A.; Dantas-Torres, F.; Bain, O. Cutaneous distribution and localization of Cercopithifilaria sp. microfilariae in dogs. Vet. Parasitol. 2012, 190, 143–150. [Google Scholar] [CrossRef]
- Ramos, R.A.N.; Rêgo, A.G.D.O.D.; Firmino, E.D.D.F.; Ramos, C.A.D.N.; De Carvalho, G.A.; Dantas-Torres, F.; Otranto, D.; Alves, L.C. Filarioids infecting dogs in northeastern Brazil. Vet. Parasitol. 2016, 226, 26–29. [Google Scholar] [CrossRef]
- Paniz-Mondolfi, A.E.; Gárate, T.; Stavropoulos, C.; Fan, W.; González, L.M.; Eberhard, M.; Kimmelstiel, F.; Sordillo, E.M. Zoonotic filariasis caused by novel Brugia sp. nematode, United States, 2011. Emerg. Infect. Dis. 2014, 20, 1248–1250. [Google Scholar] [CrossRef]
- Moraes, M.F.D.; Da Silva, M.X.; Magalhães-Matos, P.C.; Albuquerque, A.C.A.; Tebaldi, J.H.; Mathias, L.A.; Hoppe, E.G.L. Filarial nematodes with zoonotic potential in ring-tailed coatis (Nasua nasua Linnaeus, 1766, Carnivora: Procyonidae) and domestic dogs from Iguaçu National Park, Brazil. Vet. Parasitol. Reg. Stud. Rep. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Orihel, T.C. Brugia guyanensis sp. n. (Nematoda: Filarioidea) from the Coatimundi (Nasua nasua vittata) in British Guiana. J. Parasitol. 1964, 50, 115. [Google Scholar] [CrossRef]




| Application | System Name’s | Target Gene | Primer & Probes Name’s | Sequences 5′–3′ | Amplicon Size (pb) | Tm/Elongation Time | Specificity | References |
|---|---|---|---|---|---|---|---|---|
| qPCR | Pan-Fil 28S | 28S LSU rRNA | qFil-28S-F | TTGTTTGAGATTGCAGCCCA | 151 | 60 °C/30″ | Filarial species | [27] |
| qFil-28S-R | GTTTCCATCTCAGCGGTTTC | |||||||
| qFil-28S-P | 6FAM-5′-CAAGTACCGTGAGGGAAAGT-3′-TAMRA | |||||||
| All-Wol-16S | 16S rRNA gene | all.Wol.16S.301-F | TGGAACTGAGATACGGTCCAG | 177 | 60 °C/30″ | Wolbachia sp. | ||
| all.Wol.16S.478-R | GCACGGAGTTAGCCAGGACT | |||||||
| all.Wol.16S.347-P | 6FAM-5′-AATATTGGACAATGGGCGAA-3′-TAMRA | |||||||
| Triplex TaqMan COI | cox1 | Fil.COI.749-F | CATCCTGAGGTTTATGTTATTATTTT | 166 | 60 °C/30″ | FAM: D. immitis VIC: D. repens Cy5: A. reconditum | ||
| Fil.COI.914-R | CWGTATACATATGATGRCCYCA | |||||||
| D.imm.COI.777-P | 6FAM-CGGTGTTTGGGATTGTTAGTG-TAMRA | |||||||
| D.rep.COI.871-P | 6VIC-TGCTGTTTTAGGTACTTCTGTTTGAG-TAMRA | |||||||
| A.rec.COI.866-P | Cy5-TGAATTGCTGTACTGGGAACT-BHQ-3 | |||||||
| Wol-Diro ftsZ | Prokaryotic homolog of the eukaryotic protein tubulin | WDiro.ftsZ.490-F | AAGCCATTTRGCTTYGAAGGTG | 111 | 60 °C/30″ | FAM: Wolbachia of D. immitis VIC: Wolbachia of D. repens | ||
| WDiro.ftsZ.600-R | AAACAAGTTTTGRTTTGGAATAACAAT | |||||||
| WDimm.ftsZ.523-P | 6FAM-CGTATTGCAGAGCTCGGATTA-TAMRA | |||||||
| WDrep.ftsZ.525-P | 6VIC-CATTGCAGAACTGGGACTGG-TAMRA | |||||||
| PCR | 16S W-Spec | 16S rRNA | W-Specf | CATACC TATTCGAAGGGATAG | 438 | 60 °C/1′ | Wolbachia sp. | [30] |
| W-Specr | AGCTTCGAGTGAA ACCAATTC | |||||||
| Pan-Nem 18S | 18S SSU rRNA | Fwd.18S.631 | TCGTCATTGCTGCGGTTAAA | 1127–1155 | 54 °C/1′30″ | Nematodes | [9] | |
| Rwd.18S.1825r | GGTTCAAGCCACTGCGATTAA | |||||||
| Pan-Fil cox1 | cox1 | Fwd.957 | ATRGTTTATCAGTCTTTTTTTATTGG | 509 | 52 °C/45″ | Filarial species | [27] | |
| Rwd.1465 | GCAATYCAAATAGAAGCAAAAGT | |||||||
| Pan-Fil 12S | 12S rRNA | Fwd.12S.110 | TCCAGAATAATCGGCTATACATTTT | 497 to 570 | 56 °C/45″ | Filarial species | The present study | |
| Rwd.12S.681 | CCATTGACGGATGGTTTGTA |
| Sample Description | Parasitology | Serology | Filarioid | Wolbachia | Decision | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qPCR-Based Detection | Multi-Locus Genotyping | qPCR-Based Detection | 16S Genotyping | ||||||||||||
| Dog Code | Location | Thin Smear | Witness Dirofilaria | Filarial DNA | D. imm | A. rec | Accession Number (AN) | Species | Wolbachia | WDim | 16S (AN) | Wolbachia Clade/Strains | |||
| 18S | Cox1 | 12S | |||||||||||||
| CMT 01 | Cayenne | Neg. | Neg. | Pos. | N/A | Pos. | MN795082 MN795087 | MT193075 MT193074 | MT252011 | A. reconditum Brugia sp. | Pos. | N/A | MT231951 | D/WBr. | A. reconditum Brugia sp. |
| CMT 12 | Cayenne | Neg. | Neg. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | Pos. | N/A | MT231952 | D/WBr. | Brugia sp. |
| CMT 13 | Cayenne | Neg. | Neg. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | Pos. | N/A | MT231953 | D/WBr. | Brugia sp. |
| CMT 14 | Cayenne | Neg. | Pos. | Pos. | N/A | Pos. | MN795083 | MT193076 | MT252012 | A. reconditum | N/A | N/A | ..-.. | ..-.. | A. reconditum/ O-Heartworm |
| CMT 18 | Cayenne | Pos. | Neg. | Pos. | N/A | Pos. | MN795084 | MT193077 | MT252013 | A. reconditum | N/A | N/A | ..-.. | ..-.. | A. reconditum |
| CMT 19 | Cayenne | Pos. | Pos. | Pos. | Pos. | N/A | MN795071 | MT193078 | MT252014 | D. immitis | Pos. | Pos. | MT231954 | C/WDim. | D. immitis |
| CMT 32 | Kourou | Neg. | Pos. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | N/A | N/A | ..-.. | ..-.. | O-Heartworm |
| CMT 34 | Kourou | Neg. | Neg. | Pos. | Pos. | N/A | MN795072 | MT193079 | MT252015 | D. immitis | N/A | N/A | ..-.. | ..-.. | D. immitis |
| CMT 36 | Kourou | Neg. | Pos. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | N/A | N/A | ..-.. | ..-.. | O-Heartworm |
| CMT 38 | Kourou | Neg. | Pos. | Pos. | Pos. | N/A | MN795073 | MT193080 | MT252016 | D. immitis | N/A | N/A | ..-.. | ..-.. | D. immitis |
| CMT 40 | Kourou | Pos. | Pos. | Pos. | Pos. | N/A | MN795074 | MT193081 | MT252017 | D. immitis | N/A | N/A | ..-.. | ..-.. | D. immitis |
| CMT 41 | Kourou | Neg. | Pos. | Pos. | Pos. | N/A | MN795075 | MT193082 | MT252018 | D. immitis | Pos. | Pos. | MT231955 | C/WDim. | D. immitis |
| CMT 43 | Kourou | Neg. | Neg. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | Pos. | N/A | MT231956 | D/WBr. | Brugia sp. |
| CMT 52 | Kourou | Neg. | Pos. | Pos. | Pos. | N/A | MN795076 | MT193083 | MT252019 | D. immitis | N/A | N/A | ..-.. | ..-.. | D. immitis |
| CMT 53 | Kourou | Neg. | Pos. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | N/A | N/A | ..-.. | ..-.. | O-Heartworm |
| CMT 54 | Kourou | Neg. | Pos. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | N/A | N/A | ..-.. | ..-.. | O-Heartworm |
| CMT 61 | Cayenne | Neg. | Pos. | N/A | N/A | N/A | ..-.. | ..-.. | ..-.. | ..-.. | N/A | N/A | ..-.. | ..-.. | O-Heartworm |
| CMT 71 | Cayenne | Pos. | Pos. | Pos. | Pos. | N/A | MN795077 | MT193084 | MT252020 | D. immitis | Pos. | Pos. | MT231957 | C/WDim. | D. immitis |
| CMT 75 | Cayenne | Pos. | Pos. | Pos. | Pos. | N/A | MN795078 | MT193085 | MT252021 | D. immitis | Pos. | Pos. | MT231958 | C/WDim. | D. immitis |
| CMT 76 | Cayenne | Neg. | Neg. | Pos. | Pos. | N/A | MN795079 | MT193086 | MT252022 | D. immitis | N/A | N/A | ..-.. | ..-.. | D. immitis |
| CMT 89 | Cayenne | Neg. | Neg. | Pos. | N/A | N/A | MN795085 | ..-.. | MN795631 | C. bainae | N/A | N/A | ..-.. | ..-.. | C. bainae |
| CMT 90 | Cayenne | Neg. | Neg. | Pos. | N/A | N/A | MN795086 | ..-.. | MN795632 | C. bainae | N/A | N/A | ..-.. | ..-.. | C. bainae |
| CMT 91 | Cayenne | Pos. | Pos. | Pos. | Pos. | N/A | MN795080 | MT193087 | MT252023 | D. immitis | Pos. | Pos. | MT231959 | C/WDim. | D. immitis |
| CMT 97 | Cayenne | Neg. | Pos. | Pos. | Pos. | N/A | MN795081 | MT193088 | MT252024 | D. immitis | Pos. | Pos. | MT231960 | C/WDim. | D. immitis |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laidoudi, Y.; Marié, J.-L.; Tahir, D.; Watier-Grillot, S.; Mediannikov, O.; Davoust, B. Detection of Canine Vector-Borne Filariasis and Their Wolbachia Endosymbionts in French Guiana. Microorganisms 2020, 8, 770. https://doi.org/10.3390/microorganisms8050770
Laidoudi Y, Marié J-L, Tahir D, Watier-Grillot S, Mediannikov O, Davoust B. Detection of Canine Vector-Borne Filariasis and Their Wolbachia Endosymbionts in French Guiana. Microorganisms. 2020; 8(5):770. https://doi.org/10.3390/microorganisms8050770
Chicago/Turabian StyleLaidoudi, Younes, Jean-Lou Marié, Djamel Tahir, Stéphanie Watier-Grillot, Oleg Mediannikov, and Bernard Davoust. 2020. "Detection of Canine Vector-Borne Filariasis and Their Wolbachia Endosymbionts in French Guiana" Microorganisms 8, no. 5: 770. https://doi.org/10.3390/microorganisms8050770
APA StyleLaidoudi, Y., Marié, J.-L., Tahir, D., Watier-Grillot, S., Mediannikov, O., & Davoust, B. (2020). Detection of Canine Vector-Borne Filariasis and Their Wolbachia Endosymbionts in French Guiana. Microorganisms, 8(5), 770. https://doi.org/10.3390/microorganisms8050770