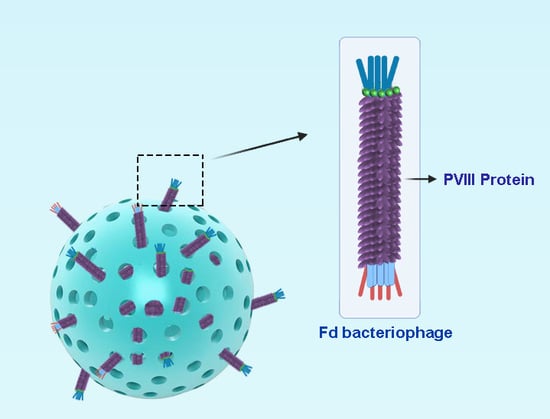
Recombinant Filamentous Bacteriophages Encapsulated in Biodegradable Polymeric Microparticles for Stimulation of Innate and Adaptive Immune Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteriophage Purification
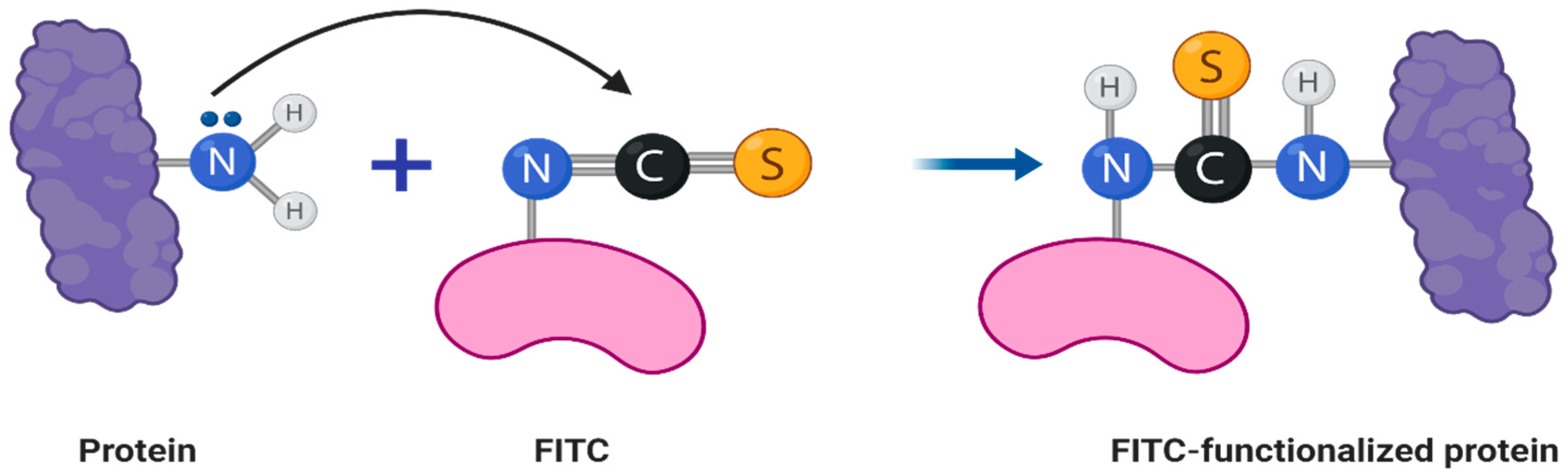
2.2. Fluorescent Labeling of Bacteriophage Virions
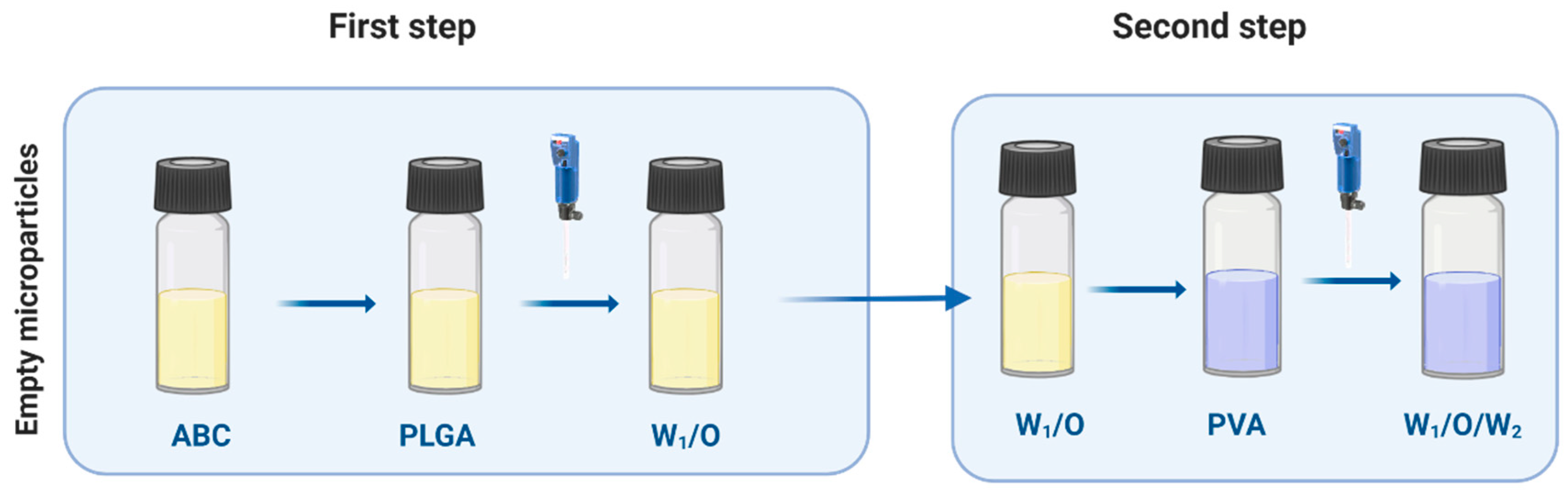
2.3. PLGA Empty MPs
2.4. Bacteriophage Loading on Empty MPs
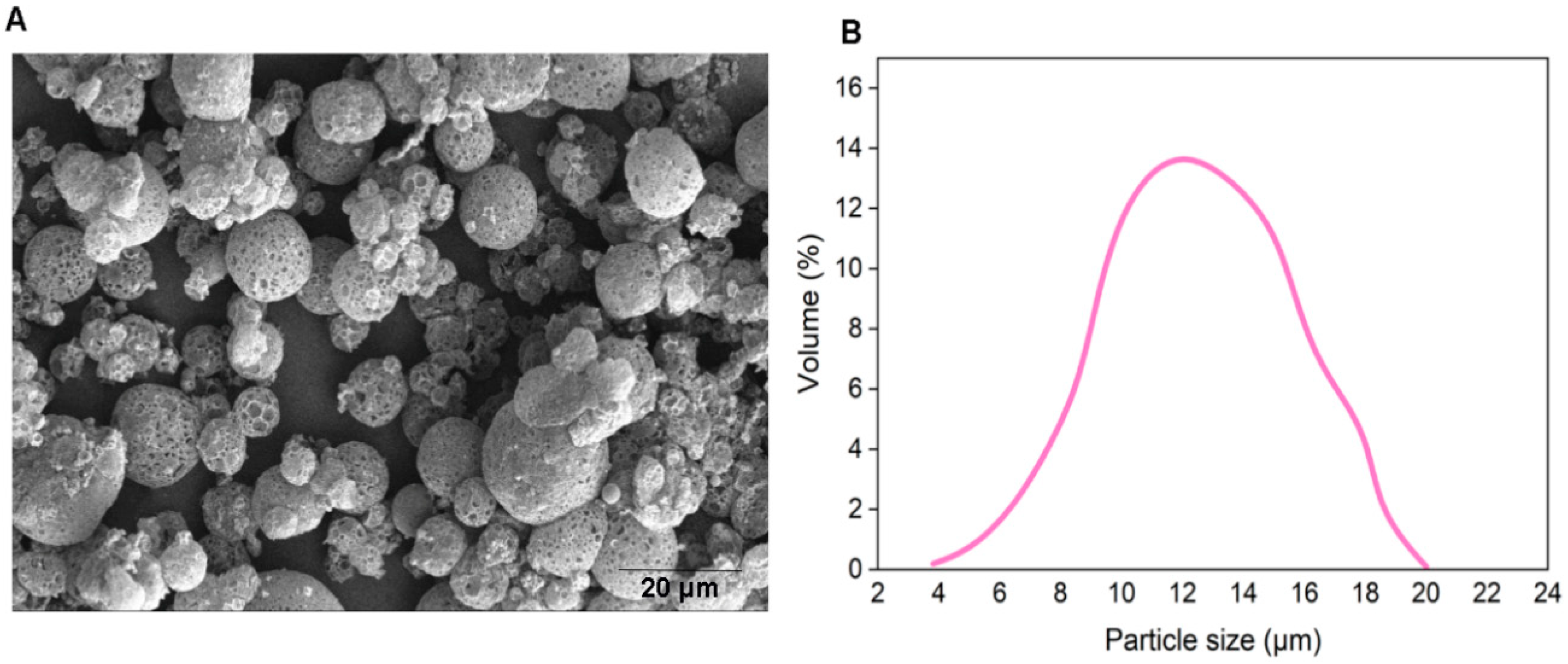
2.5. Particle Size
2.6. Scanning Electron Microscopy of MPs
2.7. Confocal Microscopy
2.8. Bacteriophage Release from Particles
2.9. Encapsulation Efficacy
2.10. Circular Dichroism
2.11. Colony-Forming Unit Determination
2.12. Bone Marrow Derived-Dendritic Cells Generation
2.13. DC Presentation Assay
3. Results
3.1. Bacteriophage-Encapsulated MPs
3.2. Fluorescent Labeling of Bacteriophage Virions
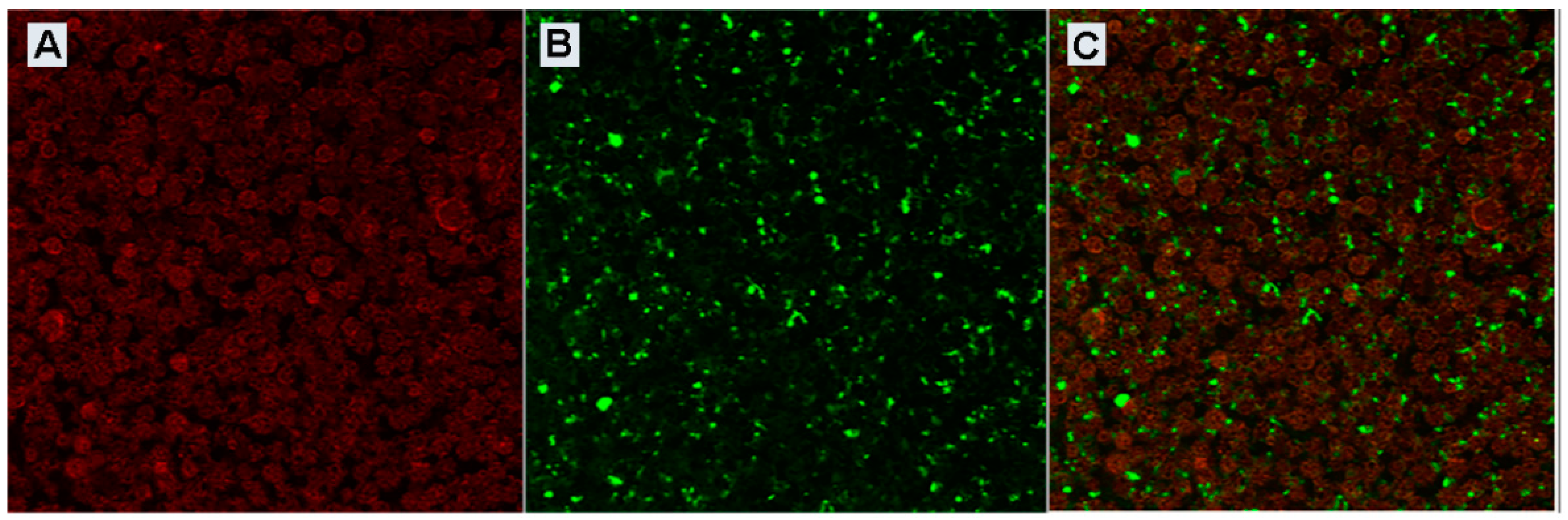
3.3. Confocal Characterization of Bacteriophage-Loaded MPs
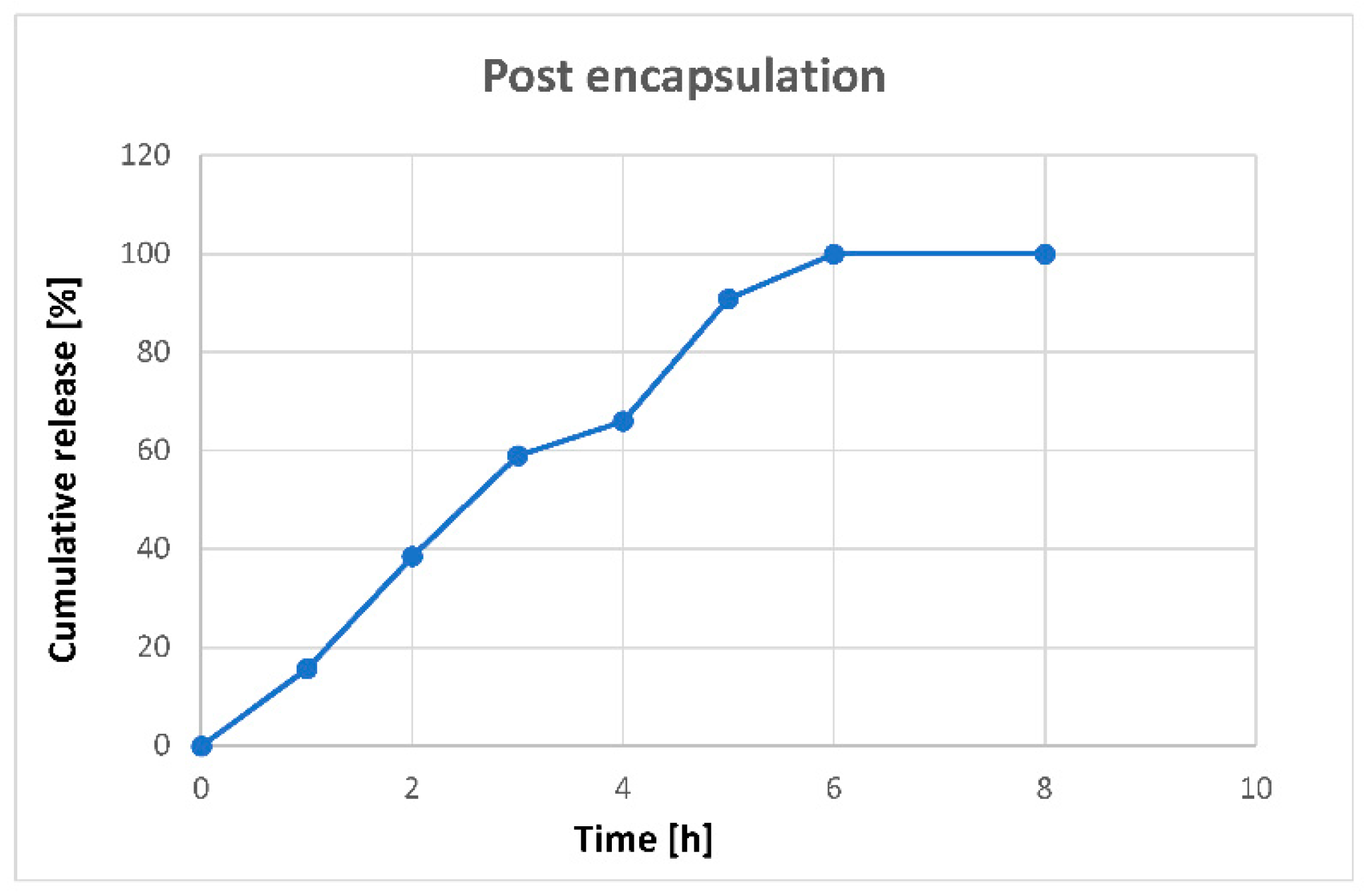
3.4. In Vitro Controlled Release
3.5. Encapsulation Efficiency
3.6. Circular Dichroism
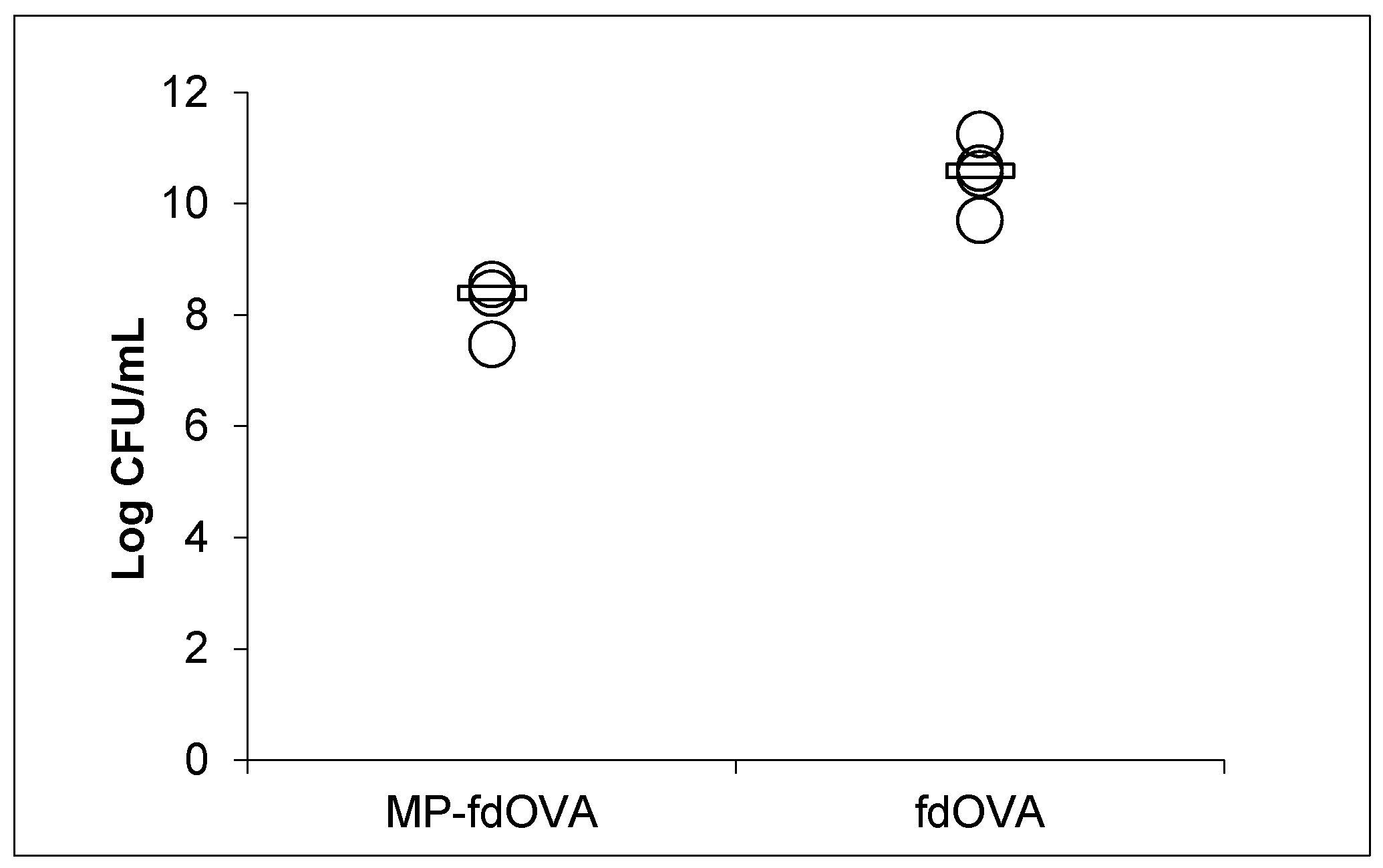
3.7. Biological Activity of Filamentous Bacteriophage after Encapsulation
3.8. Antigen-Specific Immune Response to Encapsulated fdOVA Bacteriophage
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ghosh, D.; Kohli, A.G.; Moser, F.; Endy, D.; Belcher, A.M. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth. Biol. 2012, 1, 576–582. [Google Scholar] [CrossRef]
- Lee, Y.J.; Yi, H.; Kim, W.-J.; Kang, K.; Yun, D.S.; Strano, M.S.; Ceder, G.; Belcher, A.M. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 2009, 324, 1051–1055. [Google Scholar] [CrossRef]
- Cahoon, L. Bugs Build Batteries. Science 2009, 312, 885. [Google Scholar]
- Karimi, M.; Mirshekari, H.; Basri, S.M.M.; Bahrami, S.; Moghoofei, M.; Hamblin, M.R. Bacteriophages and phage-inspired nanocarriers for targeted delivery of therapeutic cargos. Adv. Drug Deliv. Rev. 2016, 106, 45–62. [Google Scholar] [CrossRef]
- Prisco, A.; De Berardinis, P. Filamentous bacteriophage fd as an antigen delivery system in vaccination. Int. J. Mol. Sci. 2012, 13, 5179–5194. [Google Scholar] [CrossRef] [PubMed]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51. [Google Scholar] [PubMed]
- Sartorius, R.; Pisu, P.; D’Apice, L.; Pizzella, L.; Romano, C.; Cortese, G.; Giorgini, A.; Santoni, A.; Velotti, F.; De Berardinis, P. The use of filamentous bacteriophage fd to deliver MAGE-A10 or MAGE-A3 HLA-A2-restricted peptides and to induce strong antitumor CTL responses. J. Immunol. Res. 2008, 180, 3719–3728. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Ahmed, R. Immunological mechanisms of vaccination. Nat. Immunol. 2011, 12, 509. [Google Scholar] [CrossRef] [PubMed]
- Hunter, G.J.; Rowitch, D.H.; Perham, R.N. Interactions between DNA and coat protein in the structure and assembly of filamentous bacteriophage fd. Nature 1987, 327, 252. [Google Scholar] [CrossRef]
- Banner, D.; Nave, C.; Marvin, D. Structure of the protein and DNA in fd filamentous bacterial virus. Nature 1981, 289, 814. [Google Scholar] [CrossRef]
- Oakes, R.S.; Froimchuk, E.; Jewell, C.M. Engineering Biomaterials to Direct Innate Immunity. Adv. Ther. 2019, 2, 1800157. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.Z.; Diambou, I.; Kim, J.R.; Wang, B.; Champion, J.A. Host-and pathogen-derived adjuvant coatings on protein nanoparticle vaccines. Bioeng. Trans. Med. 2017, 2, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Hess, K.L.; Andorko, J.I.; Tostanoski, L.H.; Jewell, C.M. Polyplexes assembled from self-peptides and regulatory nucleic acids blunt toll-like receptor signaling to combat autoimmunity. Biomaterials 2017, 118, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Hess, K.L.; Jewell, C.M. Phage display as a tool for vaccine and immunotherapy development. Bioeng. Trans. Med. 2020, 5, e10142. [Google Scholar] [CrossRef]
- Allahyari, M.; Mohit, E. Peptide/protein vaccine delivery system based on PLGA particles. Hum. Vacc. Immunother. 2016, 12, 806–828. [Google Scholar] [CrossRef]
- Cruz, L.J.; Tacken, P.J.; Fokkink, R.; Joosten, B.; Stuart, M.C.; Albericio, F.; Torensma, R.; Figdor, C.G. Targeted PLGA nano-but not microparticles specifically deliver antigen to human dendritic cells via DC-SIGN in vitro. J. Control. Release 2010, 144, 118–126. [Google Scholar] [CrossRef]
- Jamaledin, R.; Di Natale, C.; Onesto, V.; Taraghdari, Z.B.; Zare, E.N.; Makvandi, P.; Vecchione, R.; Netti, P.A. Progress in Microneedle-Mediated Protein Delivery. J. Clin. Med. 2020, 9, 542. [Google Scholar] [CrossRef]
- Sartorius, R.; D’apice, L.; Trovato, M.; Cuccaro, F.; Costa, V.; De Leo, M.G.; Marzullo, V.M.; Biondo, C.; D’auria, S.; De Matteis, M.A. Antigen delivery by filamentous bacteriophage fd displaying an anti-DEC-205 single-chain variable fragment confers adjuvanticity by triggering a TLR9-mediated immune response. EMBO Mol. Med. 2015, 7, 973–988. [Google Scholar] [CrossRef]
- D’Apice, L.; Costa, V.; Sartorius, R.; Trovato, M.; Aprile, M.; De Berardinis, P. Stimulation of innate and adaptive immunity by using filamentous bacteriophage fd targeted to DEC-205. J. Immunol. Res. 2015, 2015, 585078. [Google Scholar] [CrossRef]
- D’Apice, L.; Sartorius, R.; Caivano, A.; Mascolo, D.; Del Pozzo, G.; Di Mase, D.S.; Ricca, E.; Pira, G.L.; Manca, F.; Malanga, D. Comparative analysis of new innovative vaccine formulations based on the use of procaryotic display systems. Vaccine 2007, 25, 1993–2000. [Google Scholar] [CrossRef]
- Sartorius, R.; Bettua, C.; D’Apice, L.; Caivano, A.; Trovato, M.; Russo, D.; Zanoni, I.; Granucci, F.; Mascolo, D.; Barba, P. Vaccination with filamentous bacteriophages targeting DEC-205 induces DC maturation and potent anti-tumor T-cell responses in the absence of adjuvants. Eur. J. Immunol. 2011, 41, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, K.; Switała-Jelen, K.; Opolski, A.; Weber-Dabrowska, B.; Gorski, A. Bacteriophage penetration in vertebrates. J. Appl. Microbiol. 2005, 98, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Costa, A.R.; Santos, J.F.; Siliakus, M.F.; Van Lent, J.W.; Kengen, S.W.; Azeredo, J.; Kluskens, L.D. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci. Rep. 2016, 6, 39235. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pacan, J.C.; Wang, Q.; Xu, Y.; Huang, X.; Korenevsky, A.; Sabour, P.M. Microencapsulation of bacteriophage Felix O1 into chitosan-alginate microspheres for oral delivery. Appl. Environ. Microbiol. 2008, 74, 4799–4805. [Google Scholar] [CrossRef]
- Vinner, G.K.; Vladisavljević, G.T.; Clokie, M.R.; Malik, D.J. Microencapsulation of Clostridium difficile specific bacteriophages using microfluidic glass capillary devices for colon delivery using pH triggered release. PLoS ONE 2017, 12, e0186239. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.; Garton, N.J.; Stapley, A.G.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.; Mönkäre, J.; Bouwstra, J.; Kersten, G. Dissolving microneedle patches for dermal vaccination. Pharm. Res. 2017, 34, 2223–2240. [Google Scholar] [CrossRef]
- Nazarzadeh Zare, E.; Jamaledin, R.; Naserzadeh, P.; Afjeh-Dana, E.; Ashtari, B.; Hosseinzadeh, M.; Vecchione, R.; Wu, A.; R Tay, F.; Borzacchiello, A. Metal-based Nanostructures/PLGA Nanocomposites: Antimicrobial Activity, Cytotoxicity and Their Biomedical Applications. ACS Appl. Mater. Interfaces 2020, 12, 3279–3300. [Google Scholar] [CrossRef]
- Mazzara, J.M.; Ochyl, L.J.; Hong, J.K.; Moon, J.J.; Prausnitz, M.R.; Schwendeman, S.P. Self-healing encapsulation and controlled release of vaccine antigens from PLGA microparticles delivered by microneedle patches. Bioeng. Transl. Med. 2019, 4, 116–128. [Google Scholar] [CrossRef]
- Del Pozzo, G.; Mascolo, D.; Sartorius, R.; Citro, A.; Barba, P.; D’Apice, L.; De Berardinis, P. Triggering DTH and CTL Activity by fd Filamentous Bacteriophages: Role of CD4. BioMed Res. Int. 2010, 2010, 894971. [Google Scholar]
- Malik, P.; Perham, R.N. Simultaneous display of different peptides on the surface of filamentous bacteriophage. Nucleic Acids Res. 1997, 25, 915–916. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Terry, T.D.; Perham, R.N. Multiple display of foreign peptide epitopes on filamentous bacteriophage virions. In Phage Display of Peptides and Proteins; Elsevier: Amsterdam, The Netherlands, 1996; pp. 127–139. [Google Scholar]
- Di Natale, C.; Celetti, G.; Scognamiglio, P.L.; Cosenza, C.; Battista, E.; Causa, F.; Netti, P.A. Molecularly endowed hydrogel with an in silico-assisted screened peptide for highly sensitive small molecule harvesting. Chem. Commun. 2018, 54, 10088–10091. [Google Scholar] [CrossRef] [PubMed]
- De Alteriis, R.; Vecchione, R.; Attanasio, C.; De Gregorio, M.; Porzio, M.; Battista, E.; Netti, P.A. A method to tune the shape of protein-encapsulated polymeric microspheres. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Battisti, M.; Vecchione, R.; Casale, C.; Pennacchio, F.A.; Lettera, V.; Jamaledin, R.; Profeta, M.; Di Natale, C.; Imparato, G.; Urciuolo, F. Non-invasive production of multi-compartmental biodegradable polymer microneedles for controlled intradermal drug release of labile molecules. Front. Bioeng. Biotechnol. 2019, 7, 296. [Google Scholar] [CrossRef]
- Di Natale, C.; Onesto, V.; Lagreca, E.; Vecchione, R.; Netti, P.A. Tunable Release of Curcumin with an In Silico-Supported Approach from Mixtures of Highly Porous PLGA Microparticles. Materials 2020, 13, 1807. [Google Scholar] [CrossRef] [PubMed]
- Celetti, G.; Di Natale, C.; Causa, F.; Battista, E.; Netti, P.A. Functionalized poly (ethylene glycol) diacrylate microgels by microfluidics: In situ peptide encapsulation for in serum selective protein detection. Colloids Surf. B 2016, 145, 21–29. [Google Scholar] [CrossRef]
- Fotticchia, T.; Vecchione, R.; Scognamiglio, P.L.; Guarnieri, D.; Calcagno, V.; Di Natale, C.; Attanasio, C.; De Gregorio, M.; Di Cicco, C.; Quagliariello, V. Enhanced drug delivery into cell cytosol via glycoprotein H-derived peptide conjugated nanoemulsions. ACS Nano 2017, 11, 9802–9813. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Liposome-encapsulated bacteriophages for enhanced oral phage therapy against Salmonella spp. Appl. Environ. Microbiol. 2015, 81, 4841–4849. [Google Scholar] [CrossRef]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Aríñez-Soriano, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Microencapsulation with alginate/CaCO3: A strategy for improved phage therapy. Sci. Rep. 2017, 7, 41441. [Google Scholar] [CrossRef]
- Puapermpoonsiri, U.; Spencer, J.; van der Walle, C.F. A freeze-dried formulation of bacteriophage encapsulated in biodegradable microspheres. Eur. J. Pharm. Biopharm. 2009, 72, 26–33. [Google Scholar] [CrossRef]
- Han, F.Y.; Thurecht, K.J.; Whittaker, A.K.; Smith, M.T. Bioerodable PLGA-based microparticles for producing sustained-release drug formulations and strategies for improving drug loading. Front. Pharmacol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Essa, D.; Kondiah, P.P.; Choonara, Y.E.; Pillay, V. The design of poly (lactide-co-glycolide) nanocarriers for medical applications. Front. Bioeng. Biotechnol. 2020, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Gupta, R.K.; Deshpande, M.C.; Schwendeman, S.P. Biodegradable poly (lactic-co-glycolic acid) microparticles for injectable delivery of vaccine antigens. Adv. Drug Deliv. Rev. 2005, 57, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Tel, J.; Sittig, S.P.; Blom, R.A.; Cruz, L.J.; Schreibelt, G.; Figdor, C.G.; de Vries, I.J.M. Targeting uptake receptors on human plasmacytoid dendritic cells triggers antigen cross-presentation and robust type I IFN secretion. J. Immunol. 2013, 191, 5005–5012. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.; Kempf, M.; Merkle, H.P.; Walter, E. Immobilisation of GM-CSF onto particulate vaccine carrier systems. Int. J. Pharm. 2004, 269, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Kempf, M.; Mandal, B.; Jilek, S.; Thiele, L.; Vörös, J.; Textor, M.; Merkle, H.P.; Walter, E. Improved stimulation of human dendritic cells by receptor engagement with surface-modified microparticles. J. Drug Target. 2003, 11, 11–18. [Google Scholar] [CrossRef]
- Yoo, J.; Park, C.; Yi, G.; Lee, D.; Koo, H. Active targeting strategies using biological ligands for nanoparticle drug delivery systems. Cancers 2019, 11, 640. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamaledin, R.; Sartorius, R.; Di Natale, C.; Vecchione, R.; De Berardinis, P.; Netti, P.A. Recombinant Filamentous Bacteriophages Encapsulated in Biodegradable Polymeric Microparticles for Stimulation of Innate and Adaptive Immune Responses. Microorganisms 2020, 8, 650. https://doi.org/10.3390/microorganisms8050650
Jamaledin R, Sartorius R, Di Natale C, Vecchione R, De Berardinis P, Netti PA. Recombinant Filamentous Bacteriophages Encapsulated in Biodegradable Polymeric Microparticles for Stimulation of Innate and Adaptive Immune Responses. Microorganisms. 2020; 8(5):650. https://doi.org/10.3390/microorganisms8050650
Chicago/Turabian StyleJamaledin, Rezvan, Rossella Sartorius, Concetta Di Natale, Raffaele Vecchione, Piergiuseppe De Berardinis, and Paolo Antonio Netti. 2020. "Recombinant Filamentous Bacteriophages Encapsulated in Biodegradable Polymeric Microparticles for Stimulation of Innate and Adaptive Immune Responses" Microorganisms 8, no. 5: 650. https://doi.org/10.3390/microorganisms8050650
APA StyleJamaledin, R., Sartorius, R., Di Natale, C., Vecchione, R., De Berardinis, P., & Netti, P. A. (2020). Recombinant Filamentous Bacteriophages Encapsulated in Biodegradable Polymeric Microparticles for Stimulation of Innate and Adaptive Immune Responses. Microorganisms, 8(5), 650. https://doi.org/10.3390/microorganisms8050650