Abstract
In this investigation, we explored the oenological significance of Candida zemplinina (syn. Starmerella bacillaris) isolates from Apulian grape musts. Moreover, we provide the first evidence of the impact of different C. zemplinina strains on the wine aromatic properties tested as monocultures. We described the diversity of C. zemplinina strains isolated from grapes and the variability of ‘volatile’ phenotypes associated with this intraspecific variability. Thirty-three isolates were characterized at strain level by PCR-based approach and, among these, 16 strains were identified and then tested by microfermentation tests carried out in grape must. Analyzed strains were low producers of acetic acid and hydrogen sulphide, not able to decarboxylate a panel of representative amino acids, whereas they showed fructophilic character and significant glycerol production. Volatile profiles of produced wines were investigated by gas chromatography–mass spectrometry. The Odor Activity Values of all molecules were calculated and 12 compounds showed values above their odor thresholds. Two selected strains (35NC1 and 15PR1) could be considered as possible starter cultures since they were able to positively affect the sensory properties of obtained wine. This report firstly supplies evidence on the strain-specific impact of different C. zemplinina strains on the final aroma of produced wines.
1. Introduction
The transformation of grape juice into wine is a complex microbial reaction characterized by the sequential development of various species and strains of oenological yeasts. Traditionally, the spontaneous fermentation process is driven by indigenous microbes associated with grapes and the winery environment [1,2,3]. Among these yeasts, the early stages of fermentation are characterized by progressive development of non-Saccharomyces yeasts, such as Candida spp., Hanseniaspora spp., Kluyveromyces spp., Pichia spp., and Rhodotorula spp., and subsequently, S. cerevisiae proliferates, dominating and completing the wine fermentation [4]. Generally, non-Saccharomyces yeasts were considered of secondary significance or undesirable to the process, but, in the last years, their important role in the fermentation process has been considered [4,5,6]. Recent studies have shown that non-Saccharomyces yeasts have different oenological properties compared to those of S. cerevisiae, and can be used to modulate and improve the aroma and complexity of wines [7,8,9,10]. In fact, when used in combination with Saccharomyces strains, these species are able to improve wine organoleptic quality and sensory notes [4,5,11]. Among non-Saccharomyces yeasts, Torulaspora delbrueckii, Metschnikowia pulcherrima, Pichia kluyveri, Lachancea thermotolerans are already commercialized as oenological starter cultures [12], while the oenological properties of other species, such as Hanseniaspora uvarum and Candida zemplinina (syn. Starmerella bacillaris), have been the subject of several studies [13,14,15]. Recent studies delved into the oenological significance of H. uvarum strains isolated from Apulian musts [16,17] but no information is available about the fermentative properties of C. zemplinina strains identified in the above conditions [18,19,20].
It was already shown that C. zemplinina strains may play relevant activities during winemaking, due to their remarkably fructophilic nature and low ethanol production rate [14,21]. Strains belonging to this species, when inoculated at first, were able to alleviate the osmotic stress of S. cerevisiae cells by selectively consuming sugars [22]. Several studies assessed the oenological significance of C. zemplinina strains employed in combination with S. cerevisiae [22,23,24,25]. Recently, Barbera wines produced with C. zemplinina and S. cerevisiae were characterized by higher amounts of glycerol and esters responsible for fruity notes [26]. Englezos and coworkers [26] demonstrated that the wines produced with mixed cultures of C. zemplinina and S. cerevisiae contained higher values of esters compared to wines fermented with S. cerevisiae alone. To the best of our knowledge, only two studies [24,27] evaluated the impact of C. zemplinina monoculture inoculation on wine volatiles and each of these studies tested only one strain. The novelty of the present investigation is mainly to increase the limited knowledge available about the specific impact of different C. zemplinina strains during the winemaking process.
The present study describes the genetic characterization of a collection of Apulian C. zemplinina isolates at the strain level. The identified strains were tested as monocultures in microfermentation trials monitoring the chemical and volatile profiles during the fermentative process, allowing an investigation of the strain-specific influence of the selected strains on produced volatile compounds.
2. Materials and Methods
2.1. Yeast Strains
Yeast strains used in the present study were deposited in Agro-Food Microbial Culture Collection of ISPA (http://www.ispacnr.it/collezioni-microbiche). Yeasts were cultured in YPD broth (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose, 20 g/L agar) at 28 °C for 24 h, and maintained at −80 °C in glycerol 50% [28]
2.2. Molecular Characterization
Total genomic DNA was extracted as previously described by Tufariello et al. [29]. The genetic analysis was carried out by using the following primers: R5 (5′-AACGCGCAAC-3′; Tm 38 °C), RF2 (5′-CGGCCCCTGT-3′; Tm 42 °C) [30], 1283 (5′-GCGATCCCCA-3′; Tm 42 °C), M13 (5′-GAGGGTGGCGGTTCT -3′; Tm 53 °C), (GTG)5 (5′-GTGGTGGTGGTGGTG -3′; Tm 53 °C) [31]. The reactions were carried out using Taq DNA polymerase (Thermo Scientific, Waltham, MA, USA) and the following parameters: 40 cycles of 1 min at 94 °C, 1 min at the primer-specific Tm, 2 min at 72 °C, with a final cycle of 10 min at 72 °C. The amplicon profiles obtained in the above described PCR reactions were analyzed with the Gel Compar 3.1 software (Applied Math, Kortrijk, Belgium).
2.3. Microfermentations
The strain-specific fermentative properties of the identified strains were assessed by microfermentation in must from Negroamaro grapes (sugars 201 g/L, 21.1 °Brix, pH 3.4, assimilable nitrogen concentration 141.14 g/L). The must was firstly centrifuged (10 min at 8000× g) and then sterilized by membrane filtration (0.45 mm Ø membrane). Five hundred milliliters of treated must were aliquoted in sterile Erlenmeyer flasks and then inoculated with 106 CFU/mL of C. zemplinina precultured in the same must, for 48 h at 25 °C. The kinetics of the fermentations were monitored daily by gravimetric determinations, recording the weight decrease caused by the release of CO2. When the sample reached a constant weight, they were directly processed or stored at –20 °C for further analysis. Each fermentation experiment was achieved by carrying out three simultaneous independent repetitions.
2.4. Technological Characterization
The qualitative hydrogen sulphide production was determined by the blackening of the PbAcO paper inserted between the plug and inner wall of the Erlenmeyer, above the level of the liquid. Based on the results obtained, the isolates were classified as high (+++), medium (++), low (+), and no (-) sulphide producers [32]. The identification of yeast strains keeping an amino acid (acids histidine, tyrosine, phenylalanine, tryptophan, lysine, leucine, and arginine) decarboxylation activity was assessed by a plate assay method [20].
2.5. Chemical Analysis
The chemical analysis of wines and musts was carried out by Fourier transform infrared spectroscopy (FTIR) using the WineScan Flex (FOSS Analytical, Hillerød, Denmark). Samples were firstly centrifuged at 8000× g for 10 min and then subjected to the analysis [33]. Extraction of volatile compounds in wines was carried out by means of solid-phase extraction (SPE), according to Fragasso et al. [34]. The chromatographic analysis was conducted as described by Tufariello et al. [35]. To 50 mL of each wine sample and each standard solution were added 300 µL of 2-octanol solution (100 mg/mL). The extraction of volatile compounds was carried out by adsorption/desorption on strata polymeric SPE sorbents (styrene–divinylbenzene) prepacked in 500 mg/6 mL cartridges (Phenomenex, Torrance, CA, USA). The analyses were performed in triplicate. The concentration of each volatile compound was expressed as μg internal standard equivalents/L wine, obtained by normalizing the compound peak area to that of the internal standard and multiplying by the concentration of the internal standard (2-octanol). The analysis of volatile wine compounds previously extracted by SPE method was performed by gas chromatography coupled to quadrupolar mass spectrometry, using an Agilent 5973 Network detector (Agilent Technologies, Palo Alto, CA, USA). Analytes were thermally desorbed at 250 °C, and then separated on an HP-INNOWAX capillary column (60 m × 0.25 mm, 0.25 μm, J&W Scientific Inc., Folsom, CA, USA). The elution program started at a temperature of 40 °C, which was held for 5 min, then increased to 220 °C at a rate of 3 °C/min; the final temperature was held for 15 min before returning to the initial value. The carrier gas (He) flow rate was 1 mL/min. The separated analytes were transferred into the ion source of the mass spectrometer, kept at 240 °C, through a transfer line, kept at 230 °C. Their detection was carried out by electron ionization mass spectrometry, operated in total ion current (TIC) mode, using ionization energy of 70 eV. The m/z acquisition range was 35–400. Data were collected with the HP Chemstation software (Agilent Technologies). Thirteen compounds were identified by comparing their retention times and mass spectra with those of pure compounds analyzed under the same conditions. For other extracted compounds, the comparison of MS fragmentation patterns with those included in the National Institute for Standards and Technology database (NIST 02, p > 80) was employed to achieve a tentative identification.
2.6. Statistical Analysis
Significant differences among samples were determined for each chemical compound by analysis of variance (post hoc Tukey, α = 0.05). Statistical data processing was performed using the free software package STATISTICA 7.0 software (StatSoft software package, Tulsa, OK, USA). Cluster analysis and microbial diversity indices calculation were performed using the free software package PAST [36]. Aroma profile data pretreatment (calculation of standardized concentrations), statistical analysis (Principal Component Analysis), and presentation of results were carried out using routines written in the MATLAB programming environment.
3. Results
3.1. Genetic, Molecular, and Technological Characterization of the C. zemplinina Strains
The analyzed population was composed of 33 C. zemplinina isolates (Table S1) selected from spontaneous fermentations of Negroamaro grape must and formerly characterized at the species level [20]. To assess the diversity within this population, the discerning capacity of three PCR-based methods were evaluated (i.e., Random Amplification of Polymorphic DNA (RAPD), minisatellites and microsatellites analysis) by separately using means of the following primers: (i) R5, R12, and 1283 (RAPD-PCR; Figures S1–S3); (ii) M13 (minisatellite sequences evaluation; Figure S4); (iii) (GTG)5 (macrosatellite sequences analysis; Figure S5). The dendrogram analysis of the data set was based on the presence or absence of major bands produced by the above five primers. The statistical analysis of the obtained data allowed the identification in each population of three distinct strain clusters with a similarity value within 90%. The obtained clustering indicated that the analyzed population was composed of 16 different strains, thus showing a biodiversity value equal to 80%, within the population itself (Figure 1). The technological and oenological properties of the above-selected strains (7NC1, 35NC1, 19NC1, 31NC1, 3NC1, 23PR2, 20NT1, 5PR1, 15PR1, 19PR1, 18PR1, 3T16, 3KUT7, FG6, FG21, FG27) were then assessed by microfermentative assays in order to evaluate properties of oenological interest.
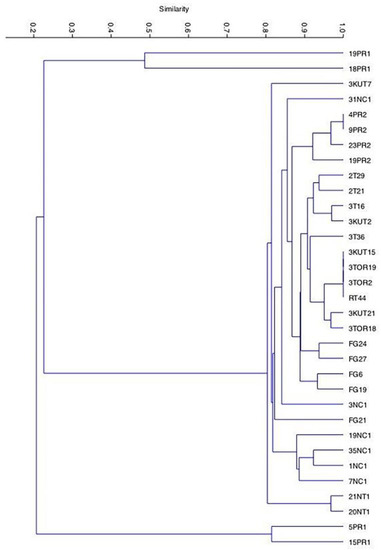
Figure 1.
The Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendrogram generated by cluster analysis of interdelta region patterns obtained from the 33 Apulian C. zemplinina strains. The percentage of similarity is indicated.
All the analyzed strains showed the same fermentation kinetics characterized by the start of the alcoholic fermentation process, 48 h after inoculation (Figure S6). The qualitative analysis of the microfermentations showed a reduced production of H2S for all strains. Hydrogen sulphide, a secondary metabolite in the synthesis of sulphur amino acids and produced in fermentation by yeasts, is an undesirable compound because it causes a rotten egg odor. The ability of yeasts to produce biogenic amines by decarboxylation of the corresponding amino acid (His, Tyr, Phe, Trp, Lys, Leu, and Arg) has been determined (Table S2). According to data obtained, the 19NC1 and 18PR1 strains were able to decarboxylate arginine, whereas the FG6 and FG27 strains showed decarboxylation of tyrosine. All the other strains showed no activity on the amino acids tested. The results of the chemical analysis of the produced wines are shown in Table 1.

Table 1.
Mean concentrations (g/L) and standard deviations (SD) of main chemical parameters in wines fermented by C. zemplinina strains.
We found ethanol content varying between 8.24% and 10.44%. All the strains produced a significant amount of glycerol, whose concentration ranged from 8.80 to 9.98 g/L in the obtained wines. Concerning acetic acid production expressed as volatile acidity, no values higher than 0.6 g/L were detected and, in particular, the 35NC1 showed the lowest production (0.35 g/L). All 16 C. zemplinina strains showed preferential consumption of fructose.
3.2. Analysis of Volatile Compounds
The strain-specific aptitude to produce volatile compounds involved in the wine flavor was evaluated and the results are shown in Table 2 and Table 3. Twenty-four volatile molecules were identified and quantified in wines, including alcohols, esters, terpenes, volatile phenols and acids, lactones, and norisoprenoids.

Table 2.
Volatile compounds identified in wines fermented by the indicated C. zemplinina strains.

Table 3.
Volatile compounds identified in wines fermented by the indicated C. zemplinina strains.
Alcohols represented the major group for all the wines with concentrations ranging from 55.83 mg/L (7NC1) to 131.06 mg/L (35NC1) followed by esters and terpenes. In particular, 3-methyl-1-butanol and phenylethanol were detected at higher concentrations in all samples ranging from 24 mg/L (3T16) to 65.83 mg/L (35NC1) and from 11.97 mg/L (7NC1) to 46.69 mg/L (35NC1), respectively. The Odor Activity Values (OAVs), a marker of the influence on wine aroma of individual volatile molecules, were calculated by dividing the mean concentration of each compound by its odor threshold value (OTH) as previously described [35,36]. Table 4 and Table 5 show the molecules with OAV > 1 considered as odor-active compounds.

Table 4.
Odor threshold (Cs) and odor activity values (OAVs) of wines fermented by the C. zemplinina strains.

Table 5.
Odor threshold (Cs) and odor activity values (OAVs) of wines fermented by the C. zemplinina strains.
The 3-methyl-1-butanol and phenylethanol are the molecules with OAV > 1 contributing fine fruity and rose odor to the wines [35]. 1-Hexanol, 3-hexen-1-ol cis (Z) and trans (E), and 2-methyl-1-propanol were also detected, but at amounts singularly low for their impact on the wine aroma (OAVs < 1). Regarding esters, the concentrations revealed ranged from 0.34 mg/L in wine fermented by FG27 strain to 7.99 mg/L in wine added with 35NC1 strain. The ethyl esters of fatty acids (ethyl octanoate and decanoate) showed important variations in their concentrations (Table 2 and Table 3) which were wider for ethyl decanoate (0.76 mg/L–6.70 mg/L). As shown in Table 4, ethyl decanoate exhibits an OAV > 8 in all samples except FG27, FG6, and 15PR1, susceptible to imparting notes of fruitiness and sweetness to these wines. Regarding odor activity, the second most representative ester was ethyl octanoate revealed in concentrations above its odor threshold in wines fermented by 35NC1, 3KUT7, 31NC1, and FG6. Fatty acids have been described as giving a general rise to fruity, cheesy, fatty, and rancid notes [35]. In our case, we detected two acids, methyl butanoic and methyl propanoic acids, in concentrations below their odor threshold, except in wines produced by 31NC1 and FG21 characterized by 3-methylbutanoic acid at values above its odor threshold (Table 4 and Table 5). The norisoprenoid β-damascenone was found in amounts ranging from 0.08 to 0.75mg/L in the wines produced with 35NC1, 3KUT7, FG21, FG27B, 5PR1, and 19NC1. This compound is related to flowery, sweet, and fruity notes, and its concentration in all samples was above its odor threshold. Finally, six terpenes were detected in wines. They were linalool, α-terpineol, citronellol, geraniol, ho-trienol (42 μg/L), and [E,E]-farnesol. Their concentrations were low, but linalool, geraniol, and citronellal had OAVs higher than one.
In order to understand the variability among the strains and the impact on the aroma profile, the volatile compounds present at concentrations above their odor threshold (Table 4 and Table 5) were submitted to the principal component analysis (PCA; Figure 2). For each molecule, standardized concentrations were obtained by scaling absolute concentrations to the same sample mean (equal to zero) and sample standard deviation (equal to one).
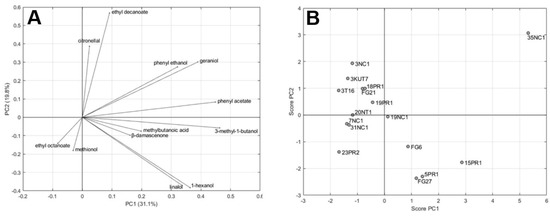
Figure 2.
Two-dimensional principal component analysis (PCA). Loading plot (A) for volatiles having OAV > 1 and scores plot (B) for selected strains as variables.
Figure 2A shows the loadings plot of the first two PCs, which jointly accounted for 50.9% of the total variance (31.1% and 19.8% by PC1 and PC2, respectively), used to establish the relative importance of each volatile component in order to relate volatile compounds with positive odor impact to one another and with samples. Figure 2B shows the scores scatter plot of the 16 samples.
The loading plot shows that almost all the volatiles are located in the first and fourth plane (positive PC1 values). This allows discriminating in the corresponding score plot the samples with an overall volatile concentration below the average (3NC1, 3KUT7, 3T16, FG21, 18PR1, 19PR1, 20NT1, 7NC1, 31NC1, and 23PR2) and above the average (19NC1, FG6, FG27, 5PR1, 15PR1, and 35NC1). This statistical representation confirmed strong oenological similarities shown by strains 3KUT7 and FG21. The wines made inoculating these two biotypes presented very close levels of ethanol and sugars (Table 1), and behaved similarly when aroma compounds were considered (Table 2; Figure 2).
The loading plot also suggests that PC2 can be used to highlight a different aromatic profile of the samples. Positive values of PC2 correspond to relatively high concentrations of certain compounds such as citronellol, etyl decanoate, phenyl ethanol, and gerianol. Negative values correspond to relatively higher concentrations of compounds such as linalol, 1-hexanol, ethyl octanate, and methionol.
On the whole, the analysis of the scatter plot allows highlighting two samples (35NC1 and 15PR1) that differ from the others in their relatively higher overall concentration of the compounds and from each other in their aromatic profile.
A significant presence of geraniol, phenylethanol, phenyl acetate, ethyl decanoate, and citronellol positively characterized the 35NC1 sample, while higher values of 3-methyl-1-butanol, methylbutanoic acid, β-damascenone, 1-hexanol, and linalool distinguished wines fermented with the 15PR1 strain. The statistical validation allowed identifying the above two strains as significant producers of important aroma-contributing compounds (OAV > 1). The standardized concentrations of these two samples are compared in Figure 3. For each standardized concentration, the error bars are representative of the standard deviation obtained by considering the six replicates.
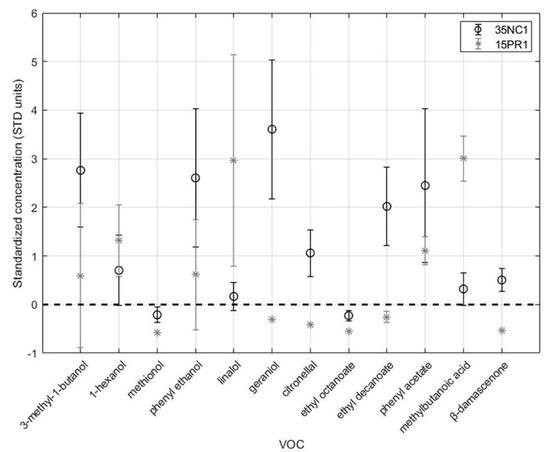
Figure 3.
Comparison of standardized concentrations of volatile compounds having OAV > 1 in wines produced with 35NC1 and 15PR1B strains.
4. Discussion
The interest in the use of autochthonous non-Saccharomyces strains is growing thanks to their valuable contribution to the fermentation process, the possibility to valorize specific grape musts, and the preservation of biodiversity in particular geographical areas, thus strengthening the terroir concept [37,38,39]. The main aim of this work was to broaden knowledge about the fermentation performance of C. zemplinina strains. In particular, for the first time, the production of volatile compounds in wines produced by monocultures of different strains belonging to this species was described.
Concerning the chemical composition, the absence of residual fructose confirmed the fructophilic character of C. zemplinina [21,40]. The production of relevant quantities of glycerol and low amounts of acetic acid was also observed, in agreement with previous studies [21,30]. Within the Candida genus, C. stellata and C. versatilis have been reported to be able to produce biogenic amines in wine and in soy sauce, respectively [41]. However, most of the strains considered did not show amino acid decarboxylation. Regarding the ‘volatile’ phenotype, alcohols and esters were the most representative compounds in all the wines’ volatile profiles and are produced by yeasts during the fermentative process. Among alcohols, 3-methyl-1-butanol and phenylethanol represented more than 80% of the alcoholic fraction. Overall, concentrations below 300 mg/L of these alcohols can have a positive impact on the wine, by conferring fruity and floral notes [42]. Under the conditions of this study, the wines produced by all the C. zemplinina monocultures did not exceed this concentration. We found a strain-specific release for several aroma compounds belonging to eight different families such as alcohols, esters, terpenes, acids, phenols, lactones, and furanic compounds. In particular, monoculture trial confirmed the strain-specific variability in higher alcohols, esters, fatty acids, terpenes, and carbonyl compounds reported by Binati et al. [23]. In addition, results produced the first evidence of a strain-specific production of norisoprenoids and lactones. Aware of the pros and the cons of Odor Activity Values (OAVs) as an indicator of the potential aroma contribution of individual compounds [36], we found that 12 compounds showed values above their odor thresholds. Among these, for the first time, we demonstrated (i) moderate, (ii) consistent, and (iii) high strain-dependent variability for (i) 3-methyl-1-butanol, (ii) 1-hexanol, phenylethanol, phenyl acetate, methyl butanoic acid, and (iii) methyonol, linalool, geraniol, citronellol, ethyl octanoate, ethyl decanoate, β-damascenone, respectively. It is important to highlight that these 12 compounds are only a part of the complex variability reported for volatiles. The reported findings provide phenotypical confirmation of the high genetic diversity depicted in winemaking environments for C. zemplinina [14,31] and, more in general, of the intraspecific variability of non-Saccharomyces in terms of oenological significance [4,5]. It is conceivable that phenotypic variability depends on both genetic properties of different strains and the influence of environmental factors [43]. In this regard, it is interesting to highlight that the similarity found in terms of oenological properties between strains genotypically distant, as it was the case of the two strains 3KUT7 and FG21, underlined the importance of studying the genetic basis associated with natural variation in oenological traits in C. zemplinina (as already reported in the model species S. cerevisiae [44,45]).
Considering geographical interest, it is important to underline that Apulia (Southern Italy) is the second region in Italy for wine production, particularly relevant for red and rosé wines [46]. Several studies delved into the study and the characterization of microbial diversity associated with grapes and wine fermentations [33,47,48]. Reporting the first study on a C. zemplinina population isolated from grapes/wines in the Apulian region, this investigation provides further insights into the Apulian microbiological diversity of oenological significance. The potential future perspectives of the present work mainly include the evaluation of the selected strains (i) at the industrial scale of wine production, (ii) in combination with S. cerevisiae strains, (iii) in terms of compatibility with malolactic starter cultures, (iv) as candidate starter cultures to produce alcoholic beverages from fruits other than grapevines [16,29,49,50,51,52,53,54].
This investigation, for the first time, highlights the strain-specific property of different C. zemplinina strains to modulate the volatile profile of the produced wine. Studies are now under way where the two selected strains, 35NC1 and 15PR1, will be tested as starter cultures for the industrial-scale production of regional typical wines.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/5/628/s1.
Author Contributions
M.T. (Maria Tufariello): Data curation, Writing—original draft, Visualization, Investigation. P.R.: Investigation, Writing—original draft, Writing—review & editing. R.R.: Data curation, Visualization, Investigation. M.T. (Mariana Tristezza): Data curation, Visualization, Investigation. M.T. (Marco Taurino): Methodology, Investigation. L.P.: Software, Statistical Validation. V.C.: Conceptualization, Supervision, Writing—review & editing. C.G.R.: Writing—review & editing. F.G.: Conceptualization, Resources, Supervision, Writing—review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Apulia Region project DOMINA APULIAE (POR Puglia FESR - FSE 2014–2020-Azione 1.6.—InnoNetwork; Project code AGBGUK2). Pasquale Russo is the beneficiary of a grant by MIUR in the framework of ‘AIM: Attraction and International Mobility’ (PON R&I2014–2020) (practice code D74I18000190001). Published with a contribution from 5 × 1000 IRPEF funds in favor of the University of Foggia, in memory of Gianluca Montel.
Acknowledgments
We would like to thank the two anonymous reviewers for their suggestions and comments. The authors wish to thank Giovanni Colella for his valuable technical assistance and Henry Smith for proofreading and providing appreciated linguistic advice.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abdo, H.; Catacchio, C.R.; Ventura, M.; D’Addabbo, P.; Calabrese, F.M.; Laurent, J.; David-Vaizant, V.; Alexandre, H.; Guilloux-Bénatier, M.; Rousseaux, S. Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery. Beverages 2020, 6, 9. [Google Scholar] [CrossRef]
- Grangeteau, C.; Roullier-Gall, C.; Rousseaux, S.; Gougeon, R.D.; Schmitt-Kopplin, P.; Alexandre, H.; Guilloux-Benatier, M. Wine microbiology is driven by vineyard and winery anthropogenic factors. Microb. Biotechnol. 2017, 10, 354–370. [Google Scholar] [CrossRef] [PubMed]
- Grangeteau, C.; Gerhards, D.; von Wallbrunn, C.; Alexandre, H.; Rousseaux, S.; Guilloux-Benatier, M. Persistence of Two Non-Saccharomyces Yeasts (Hanseniaspora and Starmerella) in the Cellar. Front. Microbiol. 2016, 7, 268. [Google Scholar] [CrossRef] [PubMed]
- Tempère, S.; Marchal, A.; Barbe, J.-C.; Bely, M.; Masneuf-Pomarede, I.; Marullo, P.; Albertin, W. The complexity of wine: Clarifying the role of microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 3995–4007. [Google Scholar] [CrossRef] [PubMed]
- Berbegal, C.; Spano, G.; Tristezza, M.; Grieco, F.; Capozzi, V. Microbial Resources and Innovation in the Wine Production Sector. South Afr. J. Enol. Vitic. 2017, 38, 156–166. [Google Scholar] [CrossRef]
- Roca-Mesa, H.; Sendra, S.; Mas, A.; Beltran, G.; Torija, M.-J. Nitrogen Preferences during Alcoholic Fermentation of Different Non-Saccharomyces Yeasts of Oenological Interest. Microorganisms 2020, 8, 157. [Google Scholar] [CrossRef]
- Canonico, L.; Solomon, M.; Comitini, F.; Ciani, M.; Varela, C. Volatile profile of reduced alcohol wines fermented with selected non-Saccharomyces yeasts under different aeration conditions. Food Microbiol. 2019, 84, 103247. [Google Scholar] [CrossRef]
- Du Plessis, H.; Du Toit, M.; Nieuwoudt, H.; Van der Rijst, M.; Hoff, J.; Jolly, N. Modulation of Wine Flavor using Hanseniaspora uvarum in Combination with Different Saccharomyces cerevisiae, Lactic Acid Bacteria Strains and Malolactic Fermentation Strategies. Fermentation 2019, 5, 64. [Google Scholar] [CrossRef]
- Berbegal, C.; Fragasso, M.; Russo, P.; Bimbo, F.; Grieco, F.; Spano, G.; Capozzi, V. Climate Changes and Food Quality: The Potential of Microbial Activities as Mitigating Strategies in the Wine Sector. Fermentation 2019, 5, 85. [Google Scholar] [CrossRef]
- Morata, A.; Escott, C.; Bañuelos, M.A.; Loira, I.; del Fresno, J.M.; González, C.; Suárez-Lepe, J.A. Contribution of Non-Saccharomyces Yeasts to Wine Freshness. A Review. Biomolecules 2020, 10, 34. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Benito, S. The Influence of Non-Saccharomyces Species on Wine Fermentation Quality Parameters. Fermentation 2019, 5, 54. [Google Scholar] [CrossRef]
- Roudil, L.; Russo, P.; Berbegal, C.; Albertin, W.; Spano, G.; Capozzi, V. Non-Saccharomyces Commercial Starter Cultures: Scientific Trends, Recent Patents and Innovation in the Wine Sector. Recent Pat. Food Nutr. Agric. 2019, 10, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Rantsiou, K.; Cravero, F.; Torchio, F.; Ortiz-Julien, A.; Gerbi, V.; Rolle, L.; Cocolin, L. Starmerella bacillaris and Saccharomyces cerevisiae mixed fermentations to reduce ethanol content in wine. Appl. Microbiol. Biotechnol. 2016, 100, 5515–5526. [Google Scholar] [CrossRef]
- Masneuf-Pomarede, I.; Juquin, E.; Miot-Sertier, C.; Renault, P.; Laizet, Y.; Salin, F.; Alexandre, H.; Capozzi, V.; Cocolin, L.; Colonna-Ceccaldi, B.; et al. The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef]
- Mateus, D.; Sousa, S.; Coimbra, C.; S Rogerson, F.; Simões, J. Identification and Characterization of Non-Saccharomyces Species Isolated from Port Wine Spontaneous Fermentations. Foods 2020, 9, 120. [Google Scholar] [CrossRef]
- Capozzi, V.; Berbegal, C.; Tufariello, M.; Grieco, F.; Spano, G.; Grieco, F. Impact of co-inoculation of Saccharomyces cerevisiae, Hanseniaspora uvarum and Oenococcus oeni autochthonous strains in controlled multi starter grape must fermentations. LWT-Food Sci. Technol. 2019, 109, 241–249. [Google Scholar] [CrossRef]
- Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G.; Grieco, F. The Oenological Potential of Hanseniaspora uvarum in Simultaneous and Sequential Co-fermentation with Saccharomyces cerevisiae for Industrial Wine Production. Front. Microbiol. 2016, 7, 670. [Google Scholar] [CrossRef]
- Garofalo, C.; Russo, P.; Beneduce, L.; Massa, S.; Spano, G.; Capozzi, V. Non-Saccharomyces biodiversity in wine and the ‘microbial terroir’: A survey on Nero di Troia wine from the Apulian region, Italy. Ann. Microbiol. 2016, 66, 143–150. [Google Scholar] [CrossRef]
- Garofalo, C.; Tristezza, M.; Grieco, F.; Spano, G.; Capozzi, V. From grape berries to wine: Population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J. Microbiol. Biotechnol. 2016, 32, 59. [Google Scholar] [CrossRef]
- Tristezza, M.; Vetrano, C.; Bleve, G.; Spano, G.; Capozzi, V.; Logrieco, A.; Mita, G.; Grieco, F. Biodiversity and safety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia Region, Italy. Food Microbiol. 2013, 36, 335–342. [Google Scholar] [CrossRef]
- Magyar, I.; Tóth, T. Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Dolci, P.; Giacosa, S.; Torchio, F.; Tofalo, R.; Torriani, S.; Suzzi, G.; Rolle, L.; Cocolin, L. Candida zemplinina can reduce acetic acid produced by Saccharomyces cerevisiae in sweet wine fermentations. Appl. Environ. Microbiol. 2012, 78, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
- Binati, R.L.; Lemos Junior, W.J.F.; Luzzini, G.; Slaghenaufi, D.; Ugliano, M.; Torriani, S. Contribution of non-Saccharomyces yeasts to wine volatile and sensory diversity: A study on Lachancea thermotolerans, Metschnikowia spp. and Starmerella bacillaris strains isolated in Italy. Int. J. Food Microbiol. 2020, 318, 108470. [Google Scholar] [CrossRef]
- Tofalo, R.; Patrignani, F.; Lanciotti, R.; Perpetuini, G.; Schirone, M.; Di Gianvito, P.; Pizzoni, D.; Arfelli, G.; Suzzi, G. Aroma Profile of Montepulciano d’Abruzzo Wine Fermented by Single and Co-culture Starters of Autochthonous Saccharomyces and Non-saccharomyces Yeasts. Front. Microbiol. 2016, 7, 610. [Google Scholar] [CrossRef] [PubMed]
- Zara, G.; Mannazzu, I.; Caro, A.D.; Budroni, M.; Pinna, M.B.; Murru, M.; Farris, G.A.; Zara, S. Wine quality improvement through the combined utilisation of yeast hulls and Candida zemplinina/Saccharomyces cerevisiae mixed starter cultures. Aust. J. Grape Wine Res. 2014, 20, 199–207. [Google Scholar] [CrossRef]
- Englezos, V.; Torchio, F.; Cravero, F.; Marengo, F.; Giacosa, S.; Gerbi, V.; Rantsiou, K.; Rolle, L.; Cocolin, L. Aroma profile and composition of Barbera wines obtained by mixed fermentations of Starmerella bacillaris (synonym Candida zemplinina) and Saccharomyces cerevisiae. LWT-Food Sci. Technol. 2016, 73, 567–575. [Google Scholar] [CrossRef]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.-J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef]
- Bleve, G.; Tufariello, M.; Durante, M.; Perbellini, E.; Ramires, F.A.; Grieco, F.; Cappello, M.S.; De Domenico, S.; Mita, G.; Tasioula-Margari, M.; et al. Physico-chemical and microbiological characterization of spontaneous fermentation of Cellina di Nardò and Leccino table olives. Front. Microbiol. 2014, 5, 570. [Google Scholar] [CrossRef]
- Tufariello, M.; Maiorano, G.; Rampino, P.; Spano, G.; Grieco, F.; Perrotta, C.; Capozzi, V.; Grieco, F. Selection of an autochthonous yeast starter culture for industrial production of Primitivo “Gioia del Colle” PDO/DOC in Apulia (Southern Italy). LWT-Food Sci. Technol. 2019, 99, 188–196. [Google Scholar] [CrossRef]
- Tofalo, R.; Schirone, M.; Torriani, S.; Rantsiou, K.; Cocolin, L.; Perpetuini, G.; Suzzi, G. Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol. 2012, 29, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Pfliegler, W.P.; Horváth, E.; Kállai, Z.; Sipiczki, M. Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiol. Res. 2014, 169, 402–410. [Google Scholar] [CrossRef]
- De Benedictis, M.; Bleve, G.; Grieco, F.; Tristezza, M.; Tufariello, M.; Grieco, F. An optimized procedure for the enological selection of non-Saccharomyces starter cultures. Antonie Van Leeuwenhoek 2011, 99, 189–200. [Google Scholar] [CrossRef]
- Tristezza, M.; Vetrano, C.; Bleve, G.; Grieco, F.; Tufariello, M.; Quarta, A.; Mita, G.; Spano, G.; Grieco, F. Autochthonous fermentation starters for the industrial production of Negroamaro wines. J. Ind. Microbiol. Biotechnol. 2012, 39, 81–92. [Google Scholar] [CrossRef]
- Fragasso, M.; Antonacci, D.; Pati, S.; Tufariello, M.; Baiano, A.; Forleo, L.R.; Caputo, A.R.; Notte, E.L. Influence of Training System on Volatile and Sensory Profiles of Primitivo Grapes and Wines. Am. J. Enol. Vitic. 2012, 63, 477–486. [Google Scholar] [CrossRef]
- Tufariello, M.; Capone, S.; Siciliano, P. Volatile components of Negroamaro red wines produced in Apulian Salento area. Food Chem. 2012, 132, 2155–2164. [Google Scholar] [CrossRef]
- Capone, S.; Tufariello, M.; Siciliano, P. Analytical characterisation of Negroamaro red wines by “Aroma Wheels”. Food Chem. 2013, 141, 2906–2915. [Google Scholar] [CrossRef]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef]
- Sgouros, G.; Chalvantzi, I.; Mallouchos, A.; Paraskevopoulos, Y.; Banilas, G.; Nisiotou, A. Biodiversity and Enological Potential of Non-Saccharomyces Yeasts from Nemean Vineyards. Fermentation 2018, 4, 32. [Google Scholar] [CrossRef]
- Capozzi, V.; Fragasso, M.; Russo, P. Microbiological Safety and the Management of Microbial Resources in Artisanal Foods and Beverages: The Need for a Transdisciplinary Assessment to Conciliate Actual Trends and Risks Avoidance. Microorganisms 2020, 8, 306. [Google Scholar] [CrossRef]
- Tofalo, R.; Schirone, M.; Telera, G.C.; Manetta, A.C.; Corsetti, A.; Suzzi, G. Influence of organic viticulture on non-Saccharomyces wine yeast populations. Ann. Microbiol. 2011, 61, 57–66. [Google Scholar] [CrossRef]
- Russo, P.; Fragasso, M.; Berbegal, C.; Grieco, F.; Spano, G.; Capozzi, V. Chapter 2: Microorganisms Able to Produce Biogenic Amines and Factors Affecting Their Activity. In Biogenic Amines in Food; Royal Society of Chemistry: Cambridge, UK, 2019; pp. 18–40. [Google Scholar]
- Tufariello, M.; Chiriatti, M.A.; Grieco, F.; Perrotta, C.; Capone, S.; Rampino, P.; Tristezza, M.; Mita, G.; Grieco, F. Influence of autochthonous Saccharomyces cerevisiae strains on volatile profile of Negroamaro wines. LWT-Food Sci. Technol. 2014, 58, 35–48. [Google Scholar] [CrossRef]
- Dzialo, M.C.; Park, R.; Steensels, J.; Lievens, B.; Verstrepen, K.J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 2017, 41, S95–S128. [Google Scholar] [CrossRef]
- Marullo, P.; Aigle, M.; Bely, M.; Masneuf-Pomarède, I.; Durrens, P.; Dubourdieu, D.; Yvert, G. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 2007, 7, 941–952. [Google Scholar] [CrossRef][Green Version]
- Salinas, F.; Cubillos, F.A.; Soto, D.; Garcia, V.; Bergström, A.; Warringer, J.; Ganga, M.A.; Louis, E.J.; Liti, G.; Martinez, C. The Genetic Basis of Natural Variation in Oenological Traits in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e49640. [Google Scholar] [CrossRef]
- Grieco, F.; Tristezza, M.; Vetrano, C.; Bleve, G.; Panico, E.; Grieco, F.; Mita, G.; Logrieco, A. Exploitation of autochthonous micro-organism potential to enhance the quality of Apulian wines. Ann. Microbiol. 2011, 61, 67–73. [Google Scholar] [CrossRef]
- Berbegal, C.; Peña, N.; Russo, P.; Grieco, F.; Pardo, I.; Ferrer, S.; Spano, G.; Capozzi, V. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 2016, 57, 187–194. [Google Scholar] [CrossRef]
- Di Toro, M.R.; Capozzi, V.; Beneduce, L.; Alexandre, H.; Tristezza, M.; Durante, M.; Tufariello, M.; Grieco, F.; Spano, G. Intraspecific biodiversity and ‘spoilage potential’ of Brettanomyces bruxellensis in Apulian wines. LWT-Food Sci. Technol. 2015, 60, 102–108. [Google Scholar] [CrossRef]
- Berbegal, C.; Borruso, L.; Fragasso, M.; Tufariello, M.; Russo, P.; Brusetti, L.; Spano, G.; Capozzi, V. A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine. Int. J. Mol. Sci. 2019, 20, 3980. [Google Scholar] [CrossRef] [PubMed]
- Kirchmayr, M.R.; Segura-García, L.E.; Lappe-Oliveras, P.; Moreno-Terrazas, R.; de la Rosa, M.; Gschaedler Mathis, A. Impact of environmental conditions and process modifications on microbial diversity, fermentation efficiency and chemical profile during the fermentation of Mezcal in Oaxaca. LWT-Food Sci. Technol. 2017, 79, 160–169. [Google Scholar] [CrossRef]
- Hu, L.; Liu, R.; Wang, X.; Zhang, X. The Sensory Quality Improvement of Citrus Wine through Co-Fermentations with Selected Non-Saccharomyces Yeast Strains and Saccharomyces cerevisiae. Microorganisms 2020, 8, 323. [Google Scholar] [CrossRef]
- Jiang, X.; Lu, Y.; Liu, S.Q. Effects of Different Yeasts on Physicochemical and Oenological Properties of Red Dragon Fruit Wine Fermented with Saccharomyces cerevisiae, Torulaspora delbrueckii and Lachancea thermotolerans. Microorganisms 2020, 8, 315. [Google Scholar] [CrossRef] [PubMed]
- Nardi, T. Microbial Resources as a Tool for Enhancing Sustainability in Winemaking. Microorganisms 2020, 8, 507. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Englezos, V.; Capozzi, V.; Pollon, M.; Río Segade, S.; Rantsiou, K.; Spano, G.; Cocolin, L. Effect of mixed fermentations with Starmerella bacillaris and Saccharomyces cerevisiae on management of malolactic fermentation. Food Res. Int. 2020, 134, 109246. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).