Antimicrobial Peptides from Rat-Tailed Maggots of the Drone Fly Eristalis tenax Show Potent Activity against Multidrug-Resistant Gram-Negative Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA-Seq and de novo Transcriptome Assembly
2.2. Synthetic Peptides
2.3. Strains and Culture Conditions
2.4. Inhibition of Microbial Growth
2.5. Checkerboard Assay
2.6. Serial-passage Mutagenesis
2.7. Hemolysis of Human Erythrocytes
2.8. Cytotoxicity Assay Based on ATP Quantification
2.9. Cytotoxicity Assay Based on Neutral Red Uptake
2.10. Interaction with the Human ERG (ether-a-go-go related gene) Potassium Channel
2.11. Plasmastability
2.12. Metabolic Stability
3. Results
3.1. Transcriptome Assembly and AMP Identification
3.2. Antimicrobial Activity against Reference Strains
3.3. Activity against an Extended Panel of Gram-Negative Clinical Isolates
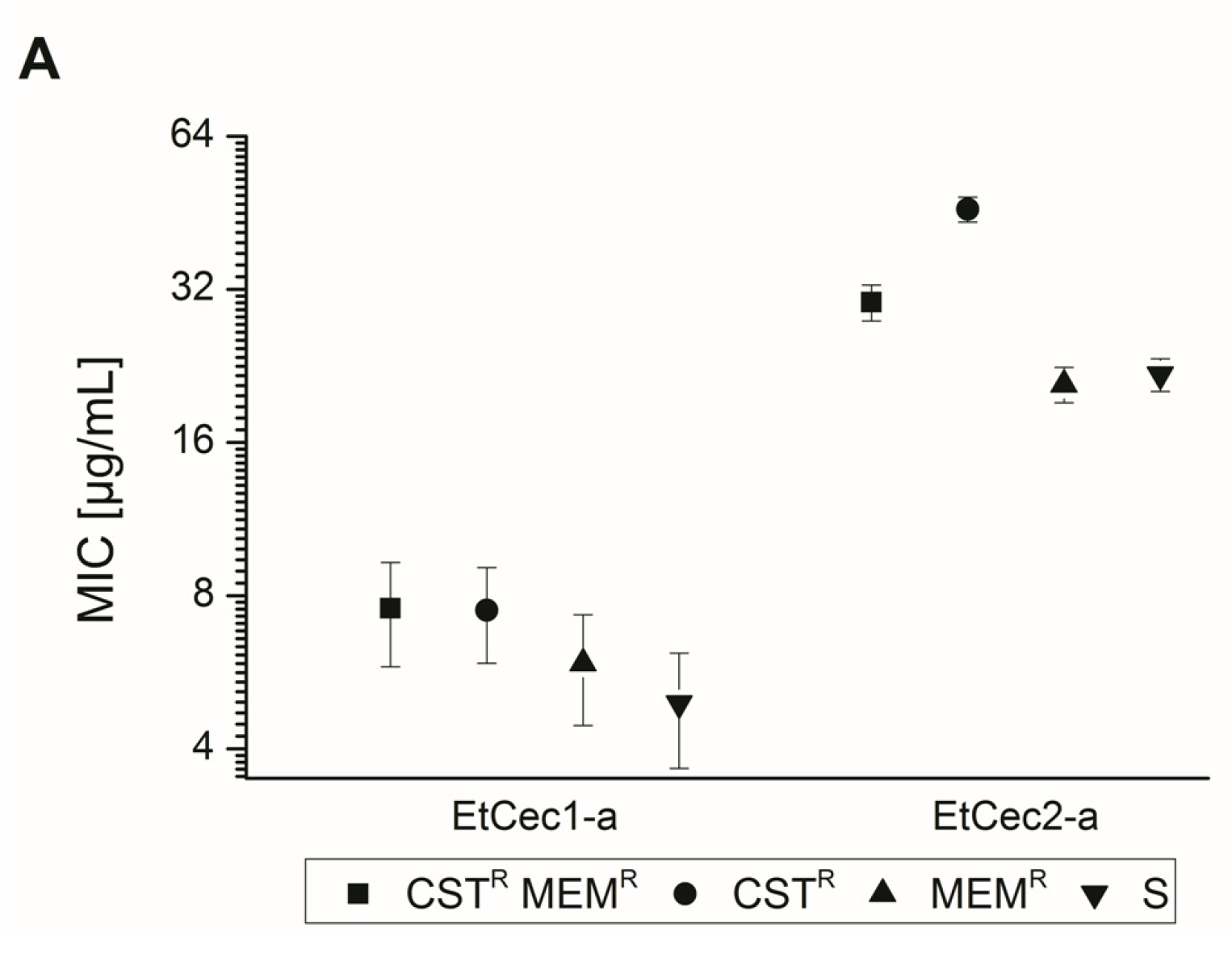
3.4. Impact of the C-terminal Amidation
3.5. Activity under Simulated Physiological Conditions
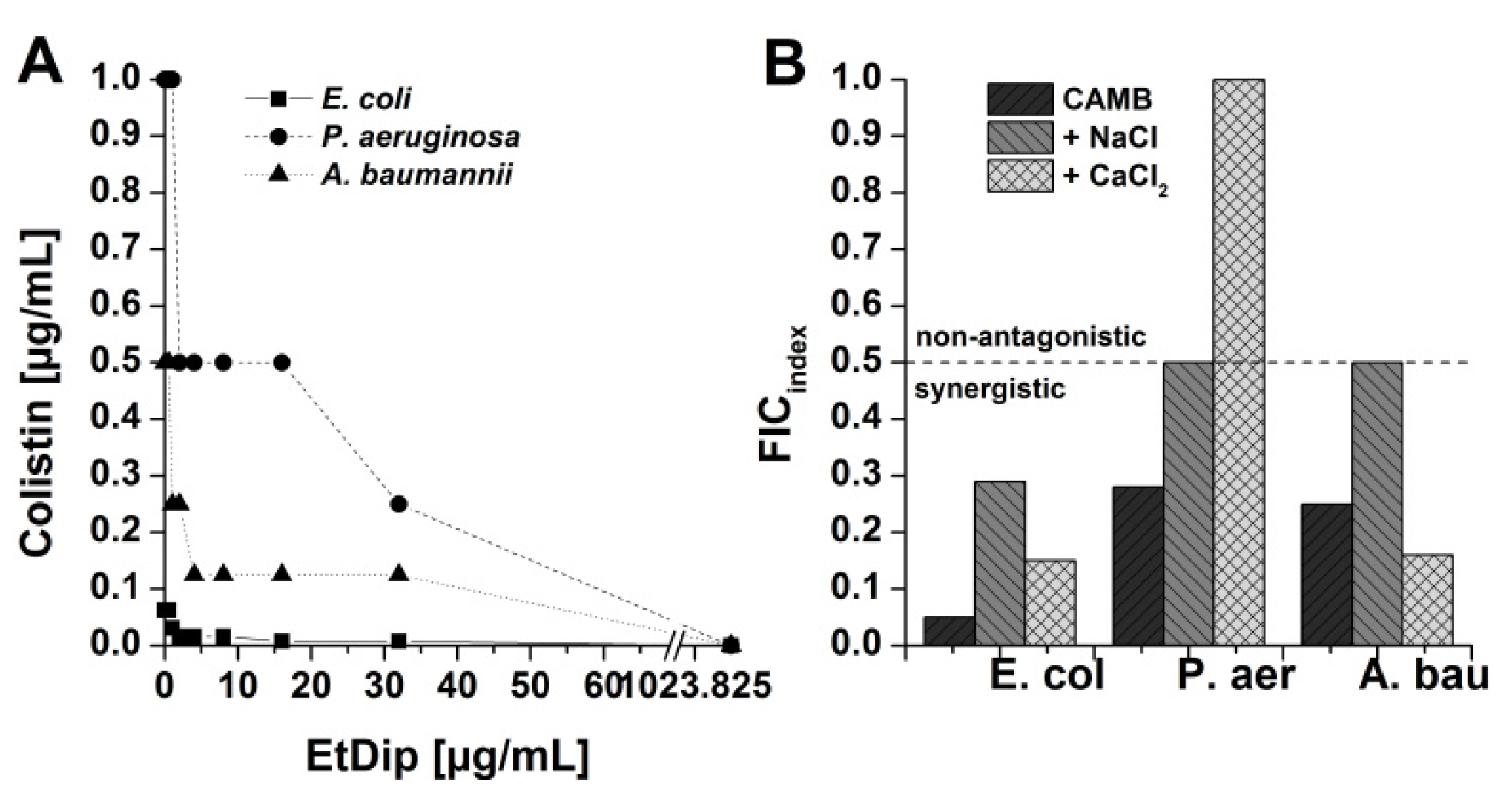
3.6. Antibacterial Activity in Combination with Approved Antibiotics
3.7. Preliminary Qualitative SAR Studies on Interaction of AMPs with Polymyxin Derivatives
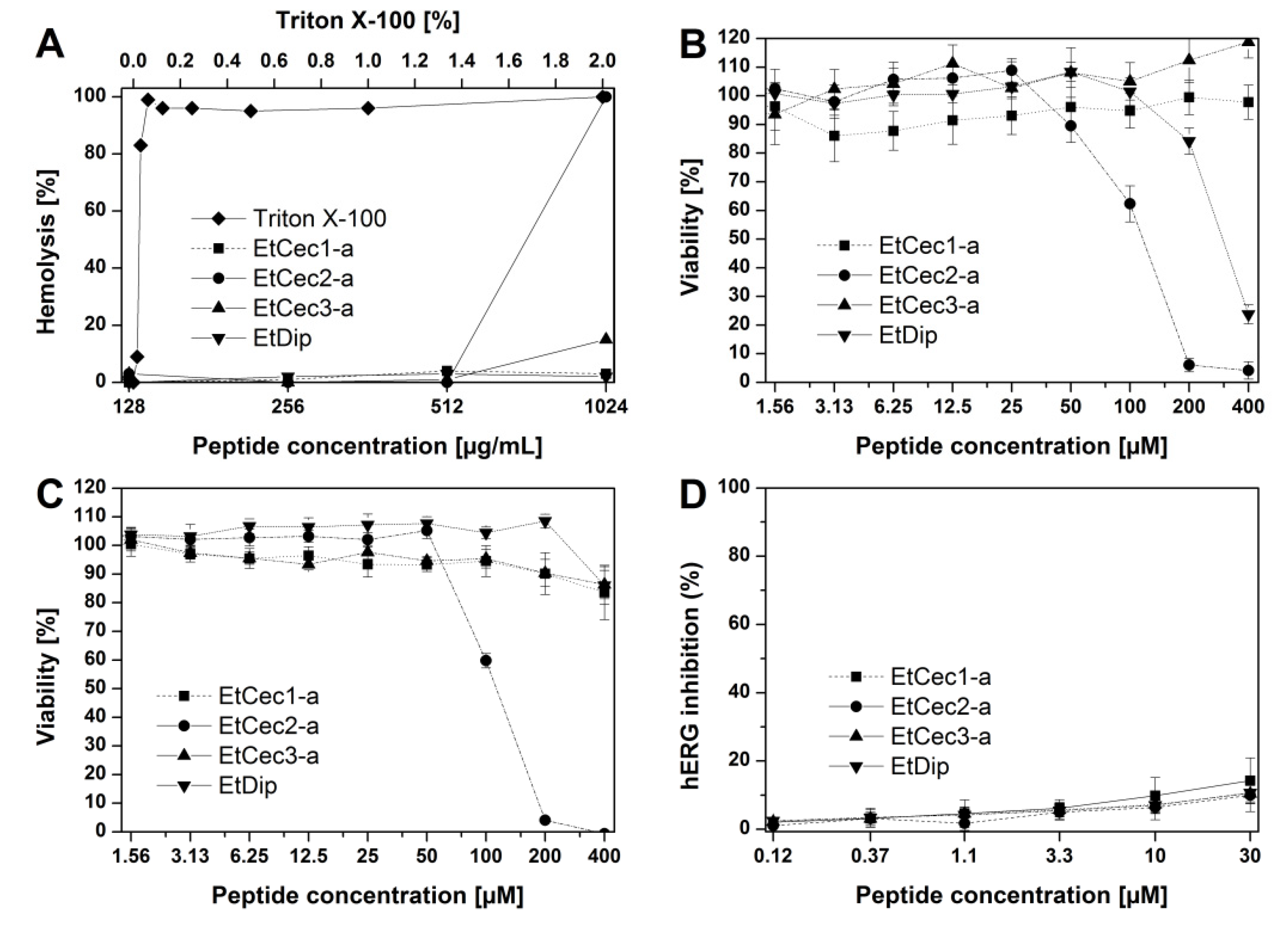
3.8. Hemolytic Activity
3.9. Toxicity Studies
3.10. In vitro Stability Studies
3.11. Development of Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Combination Effects of Antimicrobial Peptides. Antimicrob. Agents Chemother. 2016, 60, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Vilcinskas, A. Evolutionary plasticity of insect immunity. J. Insect Physiol. 2013, 59, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.G.C.; Steiger, S.; Heckel, D.G.; Wielsch, N.; Vilcinskas, A.; Vogel, H. Sex, offspring and carcass determine antimicrobial peptide expression in the burying beetle. Sci. Rep. 2016, 6, 25409. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.; Schmidtberg, H.; Vilcinskas, A. Comparative transcriptomics in three ladybird species supports a role for immunity in invasion biology. Dev. Comp. Immunol. 2017, 67, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Altincicek, B.; Vilcinskas, A. Analysis of the immune-inducible transcriptome from microbial stress resistant, rat-tailed maggots of the drone fly Eristalis tenax. BMC Genom. 2007, 8, 326. [Google Scholar] [CrossRef]
- Boulanger, N.; Munks, R.J.; Hamilton, J.V.; Vovelle, F.; Brun, R.; Lehane, M.J.; Bulet, P. Epithelial innate immunity. A novel antimicrobial peptide with antiparasitic activity in the blood-sucking insect Stomoxys calcitrans. J. Biol. Chem. 2002, 277, 49921–49926. [Google Scholar] [CrossRef]
- Matsumoto, N.; Okada, M.; Takahashi, H.; Ming, Q.X.; Nakajima, Y.; Nakanishi, Y.; Komano, H.; Natori, S. Molecular cloning of a cDNA and assignment of the C-terminal of sarcotoxin IA, a potent antibacterial protein of Sarcophaga peregrina. Biochem. J. 1986, 239, 717–722. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Shelomi, M.; Jacobs, C.; Vilcinskas, A.; Vogel, H. The unique antimicrobial peptide repertoire of stick insects. Dev. Comp. Immunol. 2020, 103, 103471. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Houtmann, S.; Schombert, B.; Sanson, C.; Partiseti, M.; Bohme, G.A. Automated Patch-Clamp Methods for the hERG Cardiac Potassium Channel. Methods Mol. Biol. 2017, 1641, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Morris, T. Physiological parameters in laboratory animals and humans. Pharm. Res. 1993, 10, 1093–1095. [Google Scholar] [CrossRef]
- Barter, Z.E.; Bayliss, M.K.; Beaune, P.H.; Boobis, A.R.; Carlile, D.J.; Edwards, R.J.; Houston, J.B.; Lake, B.G.; Lipscomb, J.C.; Pelkonen, O.R.; et al. Scaling factors for the extrapolation of in vivo metabolic drug clearance from in vitro data: Reaching a consensus on values of human microsomal protein and hepatocellularity per gram of liver. Curr. Drug Metab. 2007, 8, 33–45. [Google Scholar] [CrossRef]
- McCoy, A.J.; Liu, H.; Falla, T.J.; Gunn, J.S. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 2001, 45, 2030–2037. [Google Scholar] [CrossRef]
- Samonis, G.; Korbila, I.P.; Maraki, S.; Michailidou, I.; Vardakas, K.Z.; Kofteridis, D.; Dimopoulou, D.; Gkogkozotou, V.K.; Falagas, M.E. Trends of isolation of intrinsically resistant to colistin Enterobacteriaceae and association with colistin use in a tertiary hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1505–1510. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 2016, 26, 43–57. [Google Scholar] [CrossRef]
- Friedrich, C.; Scott, M.G.; Karunaratne, N.; Yan, H.; Hancock, R.E. Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob. Agents Chemother. 1999, 43, 1542–1548. [Google Scholar] [CrossRef]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharm. 2006, 6, 468–472. [Google Scholar] [CrossRef]
- Wang, W.; MacKinnon, R. Cryo-EM Structure of the Open Human Ether-à-go-go-Related K+ Channel hERG. Cell 2017, 169, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.; Wiesner, J.; Marker, A.; Pfeifer, Y.; Bauer, A.; Hammann, P.E.; Vilcinskas, A. Profiling antimicrobial peptides from the medical maggot Lucilia sericata as potential antibiotics for MDR Gram-negative bacteria. J. Antimicrob. Chemother. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.; Wiesner, J.; Marker, A.; Bauer, A.; Hammann, P.E.; Vilcinskas, A. Biological Profiling of Coleoptericins and Coleoptericin-Like Antimicrobial Peptides from the Invasive Harlequin Ladybird Harmonia axyridis. Adv. Exp. Med. Biol. 2019, 1214, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Ebbensgaard, A.; Mordhorst, H.; Overgaard, M.T.; Nielsen, C.G.; Aarestrup, F.M.; Hansen, E.B. Comparative Evaluation of the Antimicrobial Activity of Different Antimicrobial Peptides against a Range of Pathogenic Bacteria. PLoS ONE 2015, 10, e0144611. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. Chem. Immunol. Allergy 2005, 86, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, J.; Falciani, C.; Roscia, G.; Pollini, S.; Bindi, S.; Scali, S.; Arrieta, U.C.; Gomez-Vallejo, V.; Quercini, L.; Ibba, E.; et al. In vitro and in vivo efficacy, toxicity, bio-distribution and resistance selection of a novel antibacterial drug candidate. Sci. Rep. 2016, 6, 26077. [Google Scholar] [CrossRef]
- Lohner, K. Membrane-active Antimicrobial Peptides as Template Structures for Novel Antibiotic Agents. Curr. Top. Med. Chem. 2017, 17, 508–519. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M.N. A short D-enantiomeric antimicrobial peptide with potent immunomodulatory and antibiofilm activity against multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Sci. Rep. 2017, 7, 6953. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Löpez, C.A.; Gnanakaran, S. Permeability Barrier of Gram-Negative Cell Envelopes and Approaches To Bypass It. ACS Infect. Dis. 2015, 1, 512–522. [Google Scholar] [CrossRef]
- Cantor, S.; Vargas, L.; Rojas, A.; Yarce, C.J.; Salamanca, C.H.; Oñate-Garzón, J. Evaluation of the Antimicrobial Activity of Cationic Peptides Loaded in Surface-Modified Nanoliposomes against Foodborne Bacteria. Int. J. Mol. Sci. 2019, 20, 680. [Google Scholar] [CrossRef]
- López Cascales, J.J.; Zenak, S.; García de la Torre, J.; Lezama, O.G.; Garro, A.; Enriz, R.D. Small Cationic Peptides: Influence of Charge on Their Antimicrobial Activity. ACS Omega 2018, 3, 5390–5398. [Google Scholar] [CrossRef] [PubMed]
- Koehbach, J. Structure-Activity Relationships of Insect Defensins. Front. Chem. 2017, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A.; Hetru, C. Insect defensins: Inducible antibacterial peptides. Immunol. Today 1992, 13, 411–415. [Google Scholar] [CrossRef]
- Tonk, M.; Knorr, E.; Cabezas-Cruz, A.; Valdes, J.; Kollewe, C.; Vilcinskas, A. Tribolium castaneum defensins are primarily active against Gram-positive bacteria. J. Invertebr. Pathol. 2015, 132, 208–215. [Google Scholar] [CrossRef]
- Li, Z.; Mao, R.; Teng, D.; Hao, Y.; Chen, H.; Wang, X.; Wang, X.; Yang, N.; Wang, J. Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 12124. [Google Scholar] [CrossRef]
- Cudic, M.; Bulet, P.; Hoffmann, R.; Craik, D.J.; Otvos, L., Jr. Chemical synthesis, antibacterial activity and conformation of diptericin, an 82-mer peptide originally isolated from insects. Eur. J. Biochem. 1999, 266, 549–558. [Google Scholar] [CrossRef]
- Keppi, E.; Pugsley, A.P.; Lambert, J.; Wicker, C.; Dimarcq, J.-L.; Hoffmann, J.A.; Hoffmann, D. Mode of action of diptericin A, a bactericidal peptide induced in the hemolymph of Phormia terranovae larvae. Arch. Insect Biochem. Physiol. 1989, 10, 229–239. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Pini, A.; Giuliani, A.; Falciani, C.; Runci, Y.; Ricci, C.; Lelli, B.; Malossi, M.; Neri, P.; Rossolini, G.M.; Bracci, L. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob. Agents Chemother. 2005, 49, 2665–2672. [Google Scholar] [CrossRef] [PubMed]
- Vila-Farres, X.; Garcia de la Maria, C.; Lopez-Rojas, R.; Pachon, J.; Giralt, E.; Vila, J. In vitro activity of several antimicrobial peptides against colistin-susceptible and colistin-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 2012, 18, 383–387. [Google Scholar] [CrossRef]
- Deslouches, B.; Steckbeck, J.D.; Craigo, J.K.; Doi, Y.; Burns, J.L.; Montelaro, R.C. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob. Agents Chemother. 2015, 59, 1329–1333. [Google Scholar] [CrossRef]
- Lam, S.J.; O’Brien-Simpson, N.M.; Pantarat, N.; Sulistio, A.; Wong, E.H.; Chen, Y.Y.; Lenzo, J.C.; Holden, J.A.; Blencowe, A.; Reynolds, E.C.; et al. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 2016, 1, 16162. [Google Scholar] [CrossRef]
- Rodriguez-Hernandez, M.J.; Saugar, J.; Docobo-Perez, F.; de la Torre, B.G.; Pachon-Ibanez, M.E.; Garcia-Curiel, A.; Fernandez-Cuenca, F.; Andreu, D.; Rivas, L.; Pachon, J. Studies on the antimicrobial activity of cecropin A-melittin hybrid peptides in colistin-resistant clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2006, 58, 95–100. [Google Scholar] [CrossRef]
- Conlon, J.M.; Sonnevend, A.; Pal, T.; Vila-Farres, X. Efficacy of six frog skin-derived antimicrobial peptides against colistin-resistant strains of the Acinetobacter baumannii group. Int. J. Antimicrob. Agents 2012, 39, 317–320. [Google Scholar] [CrossRef]
- Hashemi, M.M.; Rovig, J.; Weber, S.; Hilton, B.; Forouzan, M.M.; Savage, P.B. Susceptibility of Colistin-Resistant, Gram-Negative Bacteria to Antimicrobial Peptides and Ceragenins. Antimicrob. Agents Chemother. 2017, 61, e00292-17. [Google Scholar] [CrossRef] [PubMed]
- Uppu, D.; Konai, M.M.; Sarkar, P.; Samaddar, S.; Fensterseifer, I.C.M.; Farias-Junior, C.; Krishnamoorthy, P.; Shome, B.R.; Franco, O.L.; Haldar, J. Membrane-active macromolecules kill antibiotic-tolerant bacteria and potentiate antibiotics towards Gram-negative bacteria. PLoS ONE 2017, 12, e0183263. [Google Scholar] [CrossRef] [PubMed]
- Poppel, A.K.; Vogel, H.; Wiesner, J.; Vilcinskas, A. Antimicrobial peptides expressed in medicinal maggots of the blow fly Lucilia sericata show combinatorial activity against bacteria. Antimicrob. Agents Chemother. 2015, 59, 2508–2514. [Google Scholar] [CrossRef] [PubMed]
- Rahnamaeian, M.; Cytrynska, M.; Zdybicka-Barabas, A.; Vilcinskas, A. The functional interaction between abaecin and pore-forming peptides indicates a general mechanism of antibacterial potentiation. Peptides 2016, 78, 17–23. [Google Scholar] [CrossRef]
- Beckert, A.; Wiesner, J.; Baumann, A.; Poppel, A.K.; Vogel, H.; Vilcinskas, A. Two c-type lysozymes boost the innate immune system of the invasive ladybird Harmonia axyridis. Dev. Comp. Immunol. 2015, 49, 303–312. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Tonk, M.; Schreiber, C.; Salzig, D.; Czermak, P.; Vilcinskas, A.; Rahnamaeian, M. The potential of the Galleria mellonella innate immune system is maximized by the co-presentation of diverse antimicrobial peptides. Biol. Chem. 2016, 397, 939–945. [Google Scholar] [CrossRef]
- Rahnamaeian, M.; Cytryńska, M.; Zdybicka-Barabas, A.; Dobslaff, K.; Wiesner, J.; Twyman, R.M.; Zuchner, T.; Sadd, B.M.; Regoes, R.R.; Schmid-Hempel, P.; et al. Insect antimicrobial peptides show potentiating functional interactions against Gram-negative bacteria. Proc. Biol. Sci. 2015, 282. [Google Scholar] [CrossRef]
- Dixon, R.A.; Chopra, I. Polymyxin B and polymyxin B nonapeptide alter cytoplasmic membrane permeability in Escherichia coli. J. Antimicrob. Chemother. 1986, 18, 557–563. [Google Scholar] [CrossRef]
- Vaara, M.; Viljanen, P.; Vaara, T.; Mäkelä, P.H. An outer membrane-disorganizing peptide PMBN sensitizes E. coli strains to serum bactericidal action. J. Immunol. 1984, 132, 2582–2589. [Google Scholar]
- Chu, H.L.; Yu, H.Y.; Yip, B.S.; Chih, Y.H.; Liang, C.W.; Cheng, H.T.; Cheng, J.W. Boosting salt resistance of short antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 4050–4052. [Google Scholar] [CrossRef]
- Huang, J.; Hao, D.; Chen, Y.; Xu, Y.; Tan, J.; Huang, Y.; Li, F.; Chen, Y. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an alpha-helical antibacterial peptide against bacteria. Peptides 2011, 32, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Maisetta, G.; Di Luca, M.; Esin, S.; Florio, W.; Brancatisano, F.L.; Bottai, D.; Campa, M.; Batoni, G. Evaluation of the inhibitory effects of human serum components on bactericidal activity of human beta defensin 3. Peptides 2008, 29, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-J.; Park, S.J.; Mishig-Ochir, T.; Lee, B.-J. Antimicrobial peptides: Therapeutic potentials. Expert Rev. Anti Infect. 2014, 12, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Laverty, G.; Gilmore, B. Cationic antimicrobial peptide cytotoxicity. SOJ Microbiol. Infect. Dis. 2014, 2, 1–8. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid. Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]
- Kosikowska, P.; Lesner, A. Antimicrobial peptides (AMPs) as drug candidates: A patent review (2003–2015). Expert. Opin. Pat. 2016, 26, 689–702. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef]
- Mandell, J.B.; Deslouches, B.; Montelaro, R.C.; Shanks, R.M.Q.; Doi, Y.; Urish, K.L. Elimination of Antibiotic Resistant Surgical Implant Biofilms Using an Engineered Cationic Amphipathic Peptide WLBU2. Sci. Rep. 2017, 7, 18098. [Google Scholar] [CrossRef]
- Knappe, D.; Henklein, P.; Hoffmann, R.; Hilpert, K. Easy strategy to protect antimicrobial peptides from fast degradation in serum. Antimicrob. Agents Chemother. 2010, 54, 4003–4005. [Google Scholar] [CrossRef]
- Rao, A.; Chopra, S.; Ram, G.; Gupta, A.; Ranganathan, A. Application of the “codon-shuffling” method. Synthesis and selection of de novo proteins as antibacterials. J. Biol. Chem. 2005, 280, 23605–23614. [Google Scholar] [CrossRef]


| AMP | Sequence b | Size | mol wt (g/mol) | Charge e | Gf |
|---|---|---|---|---|---|
| EtCec1 | GFLKKIGKKLEGAVQRTRDATIQTIAVAQAAANVAATAKQG | 41 | 4195.83 | +5 | −0.07 |
| EtCec1-NH2 | GFLKKIGKKLEGAVQRTRDATIQTIAVAQAAANVAATAKQ-NH2 | 40 | 4137.79 | +6 | −0.06 |
| EtCec2 | GWLRDFGKRIERTGQNIRDATIQTIGIAQEAANVAATLKG | 40 | 4340.86 | +2 | −0.38 |
| EtCec2-NH2 | GWLRDFGKRIERTGQNIRDATIQTIGIAQEAANVAATLK-NH2 | 39 | 4282.82 | +3 | −0.38 |
| EtCec3 | GFLKKVGKKLEGASDLTRDATIQTIAVAQAAANVAATAKQG | 41 | 4113.67 | +3 | 0.002 |
| EtCec3-NH2 | GFLKKVGKKLEGASDLTRDATIQTIAVAQAAANVAATAKQ-NH2 | 40 | 4055.64 | +4 | 0.01 |
| EtDip | QFNMQGGGSPRQGFDVNANARFPIWQSQNARNSVHGTASYAQHLGGPYGNSRPNFGGGLQFT | 62 | 6654.12 | +3.2 | −0.83 |
| EtDef1 | AACSLGSLLNVGCNSCACAAHCLATRGKNGACNSQRRCVCNK c | 42 | 4221.86 | +5.1 | 0.12 |
| EtDef4 | ATCDLLSFLNVKDAACAAHCLAKGYRGGYCDGRKVCNCRK d | 40 | 4289.96 | +4.1 | −0.08 |
| Cecropin A g | KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK | 37 | 4004.77 | +6 | −0.07 |
| Hymenoptaecin g | HADPQGSLVINGKKPLSGPDRRPSLDVDYHQRVYDRNGMNADAYGGLNIRPGQPAQPHLGVQIQREYKNGFIRGYSQAERGPGGRISPSFGVGGGFRF | 98 | 10676.73 | +5.3 | −0.86 |
| Defensin 1 g | VTCDLLSAEAKGVKVNHAACAAHCLLKRKRGGYCNKRRICVCRN | 44 | 4830.76 | +7.8 | −0.17 |
| Minimal Inhibitory Concentration (MIC) (µg/mL) a,b | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| EtCec1 c | EtCec2 c | EtCec3 c | EtDip | EtDef1 | EtDef4 | ||||
| Strain | -OH | -NH2 | -OH | -NH2 | -OH | -NH2 | |||
| Staphylococcus aureus ATCC 25923 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 | >128 | 16 |
| S. aureus ATCC 33592 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 | >128 | 16 |
| Staphylococcus epidermidis ATCC 35984 | >1024 | nd | >1024 | >1024 | >1024 | nd | >1024 | >128 | >128 |
| Enterococcus faecium DSM 17050 | >1024 | nd | >1024 | >1024 | >1024 | nd | >1024 | >128 | >128 |
| Listeria monocytogenes DSM 20600 | >1024 | nd | >1024 | >1024 | >1024 | nd | >1024 | >128 | 16 |
| Micrococus luteus DSM 20030 | nd | nd | nd | nd | nd | nd | nd | >128 | 16 |
| Escherichia coli D31 | 16 | 4 | 16 | 4 | >1024 | 128 | >1024 | nd | nd |
| E. coli ATCC 25922 | 16 | 8 | 64 | 32 | >1024 | >1024 | >1024 | >128 | >128 |
| Klebsiella pneumoniae DSM 30104 | 8 | 4 | 32 | 16 | >1024 | >1024 | >1024 | >128 | >128 |
| Acinetobacter baumannii ATCC 19606 | 16 | 8 | 32 | 8 | >1024 | >1024 | >1024 | >128 | >128 |
| Pseudomonas aeruginosa ATCC 27853 | 256 | 32 | >1024 | >1024 | >1024 | >1024 | >1024 | >128 | >128 |
| Proteus mirabilis DSM 4479 | >1024 | nd | >1024 | >1024 | >1024 | nd | >1024 | >128 | >128 |
| Mycobacterium smegmatis ATCC 607 | >1024 | nd | >1024 | >1024 | >1024 | nd | 64 | >128 | 32 |
| Canida albicans FH2173 | >1024 | nd | >1024 | >1024 | >1024 | nd | >1024 | >128 | >128 |
| Species (no. Isolates) | MIC50 a,b | MIC90 a,c | ||||||
|---|---|---|---|---|---|---|---|---|
| EtCec1 -OH d | EtCec1 -NH2 e | EtCec2 -OH d | EtCec2 -NH2 e | EtCec1 -OH d | EtCec1 -NH2 e | EtCec2 -OH d | EtCec2 -NH2 e | |
| Escherichia coli (26) | 8 | 4 | 32 | 16 | 32 | 8 | 128 | 32 |
| Enterobacter cloacae (23) | 8 | 4 | 128 | 32 | 128 | 16 | >128 | 128 |
| Klebsiella pneumoniae (21) | 32 | 4 | >128 | 128 | 128 | 16 | >128 | >128 |
| Salmonella enterica (10) | 32 | 16 | >128 | 128 | 32 | 16 | >128 | 128 |
| Acinetobacter baumannii (20) | 8 | 4 | 16 | 8 | 16 | 8 | 32 | 16 |
| MIC (µg/mL) a,b | ||||||||
|---|---|---|---|---|---|---|---|---|
| EtCec1 | EtCec2 | EtCec2-NH2 | ||||||
| Strain | CAMB | NaCl | CaCl2 | CAMB | NaCl | CaCl2 | CAMB | NaCl |
| Escherichia coli ATCC 25922 | 16 | 16 | 16 | 64 | 64 | 128 | 32 | 32 |
| E. coli RKI 131/08 | 16 | 16 | 32 | 32 | 32 | 64 | 16 | 16 |
| E. coli RKI 6A-6 | 32 | nd | nd | 256 | nd | nd | 64 | nd |
| Klebsiella pneumoniae DSM 30104 | 8 | 32 | 32 | 32 | 16 | 32 | 16 | 32 |
| K. pneumoniae RKI 93/10 | 16 | 32 | 32 | 128 | 256 | 256 | 64 | 256 |
| Enterobacter cloacae RKI 146/09 | 32 | 32 | 64 | 64 | 128 | 128 | 32 | 64 |
| Acinetobacter baumannii ATCC 19606 | 16 | 16 | 32 | 32 | 8 | 64 | 8 | 8 |
| A. baumannii RKI 19/09 | 16 | 16 | nd | 16 | 16 | nd | 8 | 16 |
| Stenotrophomonas maltophilia RKI 136/09 | 64 | 32 | nd | 512 | 256 | nd | 256 | 512 |
| K. pneumoniae RKI 68/16 | 64 | nd | nd | 256 | nd | nd | 128 | nd |
| K. pneumoniae RKI 268/15 | 64 | nd | nd | 256 | nd | nd | 64 | nd |
| MIC (µg/mL) a,b | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EtCec1 c | EtCec2 c | EtCec3 c | Dip | Colistin | |||||||||||
| -OH | -NH2 | -OH | -NH2 | -OH | -NH2 | ||||||||||
| Strain | CAMB | Col | CAMB | Col | CAMB | Col | CAMB | Col | CAMB | Col | CAMB | Col | CAMB | Col | CAMB |
| Escherichia coli ATCC 25922 | 16 | 4 | 8 | 2 | 64 | 4 | 32 | 4 | >1024 | 4 | >1024 | 4 | >1024 | 4 | 0.5 |
| E. coli RKI 131/08 | 16 | 2 | 4 | 2 | 32 | 2 | 16 | 2 | >1024 | 4 | >1024 | 2 | >1024 | 4 | 0.5 |
| Klebsiella pneumoniae DSM 30104 | 8 | 2 | 4 | 2 | 32 | 2 | 16 | 2 | >1024 | 4 | >1024 | 4 | >1024 | 4 | 0.5 |
| K. pneumoniae RKI 93/10 | 16 | 8 | 8 | nd | 128 | 16 | 64 | 8 | >1024 | >1024 | >1024 | nd | >1024 | >1024 | 1 |
| Klebsiella oxytocaRKI 52/07 | 16 | 2 | nd | nd | 32 | 2 | 32 | 2 | >1024 | 4 | nd | nd | >1024 | 4 | 2 |
| Enterobacter cloacae RKI 146/09 | 32 | 4 | nd | nd | 64 | 64 | 32 | 32 | >1024 | >1024 | nd | nd | >1024 | >1024 | 2 |
| Acintobacter baumannii ATCC 19606 | 16 | 8 | 8 | 4 | 32 | 16 | 8 | 8 | >1024 | >1024 | >1024 | 512 | >1024 | >1024 | 1 |
| A. baumannii RKI 19/09 | 16 | 8 | 4 | 4 | 16 | 4 | 8 | 8 | >1024 | 512 | >1024 | 256 | >1024 | 1024 | 0.5 |
| MIC [µg/mL] a for E. coli ATCC 25922 | ||||||
|---|---|---|---|---|---|---|
| Test Condition b | EtCec1-NH2 | EtCec2-NH2 | EtCec3-NH2 | EtDip | EtDef1 | EtDef4 |
| CAMB | 16 | 32 | >256 | >256 | >256 | >256 |
| + Colistin E2 [0.016 µg/mL] | 2 | 2 | 2 | 2 | 4 | 2 |
| + PMB [0.016 µg/mL] | 2 | 2 | 2 | 2 | 1 | 2 |
| + Colistin E1 [0.016 µg/mL] | 4 | 4 | >256 | >256 | 128 | 8 |
| + A000500146A [0.0625 µg/mL] | 4 | 4 | 2 | 4 | 128 | 4 |
| + A000500059A [0.0625 µg/mL] | 16 | 256 | >256 | >256 | >256 | >256 |
| + A000499933A [0.0625 µg/mL] | 16 | 128 | >256 | >256 | >256 | >256 |
| + A000173039A [0.25 µg/mL] | 16 | >256 | >256 | >256 | >256 | 64 |
| + A000160918 [4 µg/mL] | 128 | >256 | >256 | >256 | >256 | 32 |
| + A000173031A [4 µg/mL] | 32 | >256 | >256 | >256 | >256 | 16 |
| + A000501181A [4 µg/mL] | 16 | 256 | >256 | >256 | >256 | >256 |
| + A000498432A [4 µg/mL] | 16 | >256 | >256 | >256 | >256 | >256 |
| + A000173033A [4 µg/mL] | 16 | >256 | >256 | >256 | >256 | 256 |
| + A000161246 [4 µg/mL] | 128 | >256 | >256 | >256 | >256 | 128 |
| + PMBN [4 µg/mL] | 64 | >256 | >256 | >256 | >256 | >256 |
| + A000173380A [4 µg/mL] | 64 | >256 | >256 | >256 | >256 | 256 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirsch, R.; Wiesner, J.; Bauer, A.; Marker, A.; Vogel, H.; Hammann, P.E.; Vilcinskas, A. Antimicrobial Peptides from Rat-Tailed Maggots of the Drone Fly Eristalis tenax Show Potent Activity against Multidrug-Resistant Gram-Negative Bacteria. Microorganisms 2020, 8, 626. https://doi.org/10.3390/microorganisms8050626
Hirsch R, Wiesner J, Bauer A, Marker A, Vogel H, Hammann PE, Vilcinskas A. Antimicrobial Peptides from Rat-Tailed Maggots of the Drone Fly Eristalis tenax Show Potent Activity against Multidrug-Resistant Gram-Negative Bacteria. Microorganisms. 2020; 8(5):626. https://doi.org/10.3390/microorganisms8050626
Chicago/Turabian StyleHirsch, Rolf, Jochen Wiesner, Armin Bauer, Alexander Marker, Heiko Vogel, Peter Eugen Hammann, and Andreas Vilcinskas. 2020. "Antimicrobial Peptides from Rat-Tailed Maggots of the Drone Fly Eristalis tenax Show Potent Activity against Multidrug-Resistant Gram-Negative Bacteria" Microorganisms 8, no. 5: 626. https://doi.org/10.3390/microorganisms8050626
APA StyleHirsch, R., Wiesner, J., Bauer, A., Marker, A., Vogel, H., Hammann, P. E., & Vilcinskas, A. (2020). Antimicrobial Peptides from Rat-Tailed Maggots of the Drone Fly Eristalis tenax Show Potent Activity against Multidrug-Resistant Gram-Negative Bacteria. Microorganisms, 8(5), 626. https://doi.org/10.3390/microorganisms8050626





