3.1. Antifungal Activity of R9F2 and (KFF)3K Peptides Against Candida Species
Peptides R9F2 and (KFF)3K demonstrated high antifungal efficacy against all studied
Candida species (
Table S1) The MIC value for the R9F2 peptide was about 10 μM for all studied
Candida species at a concentration of fungal cells of ~1 × 10
5/mL. At the same time, the (KFF)3K peptide showed variations in the MIC value from 20 μM to 5 μM (
Table S1). The MFC value of the R9F2 peptide for all studied Candida species was 10 or >10 μM. The (KFF)3K peptide showed an MFC value of 20 or >20 μM for all studied species, except
C. guilliermondii (10 μM) (
Table S1). In total, the MIC and MFC values of both peptides were close for each species. Chlorhexidine MFC varied noticeably (12.5–50 μM) and mostly exceeded values of both peptides for each individual Candida species (
Table S1). Thus, both peptides possessed an antifungal effect in concentrations not exceeding the concentration of chlorhexidine. Earlier, we showed the absence of hemolytic and cytotoxic activity of these peptides for mammalian cells [
32].
All subsequent experiments in this study were performed using C. albicans-34 strain (in text: C. albicans) in the middle of the logarithmic growth phase.
The ability to inhibit the growth of hyphae is one of “usual” characteristics studied in new antifungals, since hyphae and pseudohyphae are the main form of existence of
C. albicans in human hosts [
36]. We also examined the influence of R9F2 and (KFF)3K peptides on hyphal growth. First, the inhibition of hyphal growth was evaluated microscopically in 96-well plates after 24 h incubation of
C. albicans with 1.25, 2.5, or 5 μM of R9F2 or (KFF)3K or chlorhexidine. Peptide R9F2 inhibited hyphal growth by 70% at a concentration of 1.25 μM, while (KFF)3K reached this level of inhibition at 2.5 μM concentration (24 h of incubation). Chlorhexidine suppressed hyphal growth by 50% at a concentration of 2.5 μM. Both peptides and chlorhexidine completely inhibited hyphal growth at a concentration of 5 μM; all wells contained only spherical cells.
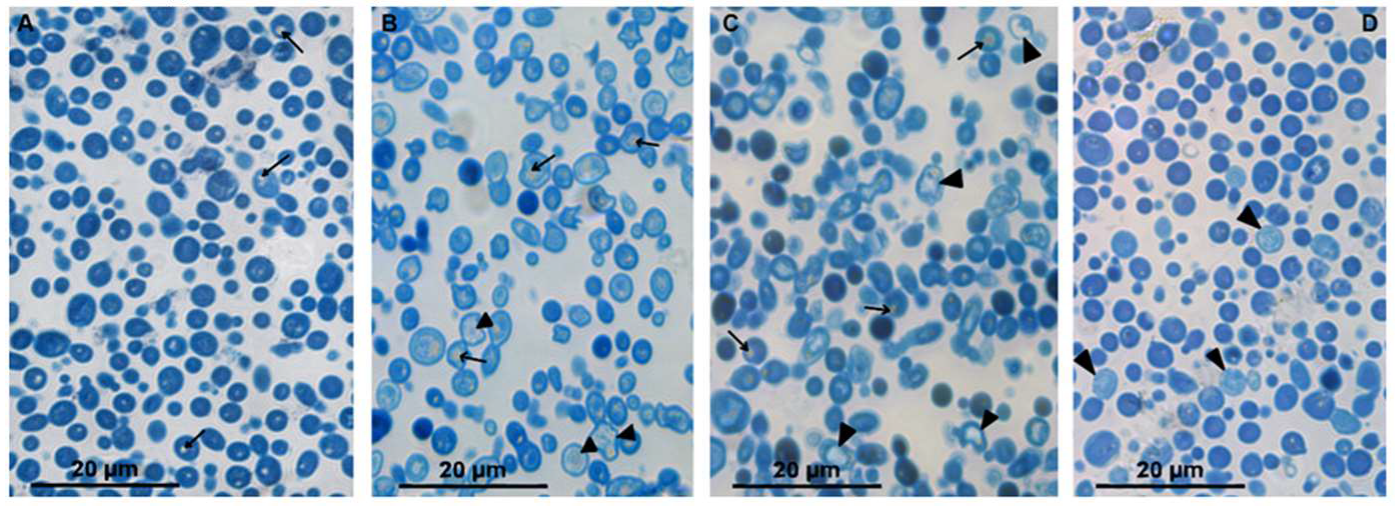
Images of the control and treated cultures of
C. albicans are presented in
Figure S1. The control culture was composed of long filamentous hyphae and rare rounded cells (
Figure S1A). Smears of
C. albicans treated with 2.5 μM (KFF)3K contained many clusters of spherical cells (
Figure S1B); similar pictures were observed in fungal smears treated with 1.25 or 2.5 μM of R9F2 or 1.25 μM (KFF)3K (images are not shown). Formation of hyphae did not occur at a concentration of both peptides of 5 μM;, the smears showed only spherical cells (
Figure S1C,D). Treatment with chlorhexidine at a concentration 2.5 μM led to the formation of clusters composed of spherical cells; many single cells were present in smears (
Figure S1E). A chlorhexydine concentration of 5 μM completely blocked hyphal formation; the smears contained only spherical cells (images are not shown).
Thus, both R9F2 and (KFF) 3K peptides showed the ability to inhibit hyphal growth in
C. albicans at concentrations similar to those of the common antiseptic chlorhexidine. Along with indicators MIC and MFC (
Table S1), these data indicate a clear antifungal action of R9F2 and (KFF)3K peptides selected for this research on the
C. albicans-34 strain.
3.2. Light Microscopy of C. albicans Cells Treated With R9F2 and (KFF) 3K Peptides
C. albicans cells in logarithmic phase actively reproduce by budding and are represented by clusters of several cells at different stages of the life cycle. Research articles are usually illustrated with images showing total views of the yeast cells in phase contrast (for example, [
17,
29,
36,
38]). Sizes, rounded shape and, especially the cell wall of
Candida spp. cells make it impossible to study them in paraffin sections. The alternative way to examine the yeast cell morphology could be the use of semithin sections made from hard blocks embedded in epoxy resin for TEM.
We conducted a study of Azure-II-stained semithin sections (about 1 μm thick) from
C. albicans samples to determine whether such sections could provide useful information for assessing the effects of peptides R9F2 and (KFF)3K. The rounded or oval cells of
C. albicans control culture (intact cells) in semithin sections (
Figure 1A) were (3.9 ± 0.1) × (3.4 ± 0.01) μm in size and were uniformly stained in bright blue, including the cell wall. Small rounded inclusions of light yellow color (lipid droplets) were occasionally observed in the cytoplasm. These morphological characteristics of intact cells did not change during 24 h of observation (data not shown).
The half-hour incubation with peptide (KFF)3K led to a noticeable decrease in the intensity of cytoplasm staining, while the cell wall remained bright blue. The cytoplasm contained “flakes” colored in different shades of blue and small lipid droplets and colorless areas. The first colorless (dead) cells bordered with bright blue cell walls were detected after 75 min of incubation with peptide (KFF)3K; their amount increased during the incubation. High cell polymorphism was observed in
C. albicans sample after 4 h of incubation, reflecting different degrees of cell damage by the peptide. Against the background of light-damaged cells, bright blue cells were distinguished, which did not visually differ from intact ones; their reproduction sometimes was observed (
Figure 1B).
Incubation of
C. albicans with the R9F2 peptide for 30 min did not cause as pronounced morphological changes as the peptide (KFF)3K. Both the cytoplasm and the cell wall remained bright blue and were indistinguishable. In some cells, colorless areas in cytoplasm and small lipid drops were noted. The first dead cells were registered after 75 min of incubation with peptide R9F2, and then their amount increased. After 4 h of incubation with the peptide, cells of various morphology were observed in the sections, as in the case of the (KFF)3K peptide. The cells showing morphology of intact cells were also present (
Figure 1C).
Incubation with chlorhexidine led to visible morphological changes in rare
C. albicans cells after 2 h; these cells were stained pale blue. After 4 h of incubation, the number of pale cells increased, and the cells looked somewhat swollen; small colorless areas were visible in the cytoplasm; and many cells had an intact morphology (
Figure 1D). Thus, semithin sections allowed us to see differences in the morphological features of the cell response to R9F2 and (KFF)3K peptides and chlorhexidine compared to intact cells.
The peptides R9F2 and (KFF)3K influenced size and shape of
C. albicans cells.
Table S2 shows values of the morphologic index (length–width ratio, [
29]), and cell length and width of intact cells and their changes during incubation with R9F2 and (KFF)3K. The changes in cell length illustrate enlargement of the fungi treated with both peptides (
Table S2). Morphological indexes varied during the time of incubation with peptides; however, they differed significantly from the control value (
Table S2). It should be noted that the cell boundaries on semithin sections are clear, which simplifies size measurements and increases their accuracy. The area of semithin sections makes it possible to analyze a large number of cells and evaluate the share of changed cells—it is almost impossible to make such assessment using ultrathin sections. Another advantage of semithin sections is that they come from specimens prepared for TEM studies and do not need special processing. We may conclude that examination of yeast cell semithin sections is informative and could give useful data.
3.3. Ultrastructure of C. albicans Intact Cells
The ability of electron microscopy to visualize cell ultrastructure and changes in cell organoids is widely used in various studies of mammalian cells, but not in studies of fungi. To a large measure, this is related to very high density of the yeast cytoplasm, which makes it difficult and sometimes impossible to recognize cell structures after routine sample processing (fixation with aldehydes, osmium postfixation, contrasting with UAc and LC [
30,
39]. Researchers studying the membrane structures of
C. albicans and other yeasts use special methods of fixation, for example, potassium permanganate, which destroys ribosomes and the cytoskeleton, making the cytoplasm “empty” [
28,
30]. The “enlightenment” of the cytoplasm could be achieved by treatment of cells with RNase [
40] and sodium metaperiodate [
41]. Obviously, such methods are poorly applicable for the analysis of experimental effects on the morphology of
C. albicans.
We suggest a simple way allowing us to visualize the structure of both the cell wall and the cell. This way (analyzing cells without contrasting of sections with UAc and LC) does not require additional processing and allows us to analyze the same sections before and after contrasting.
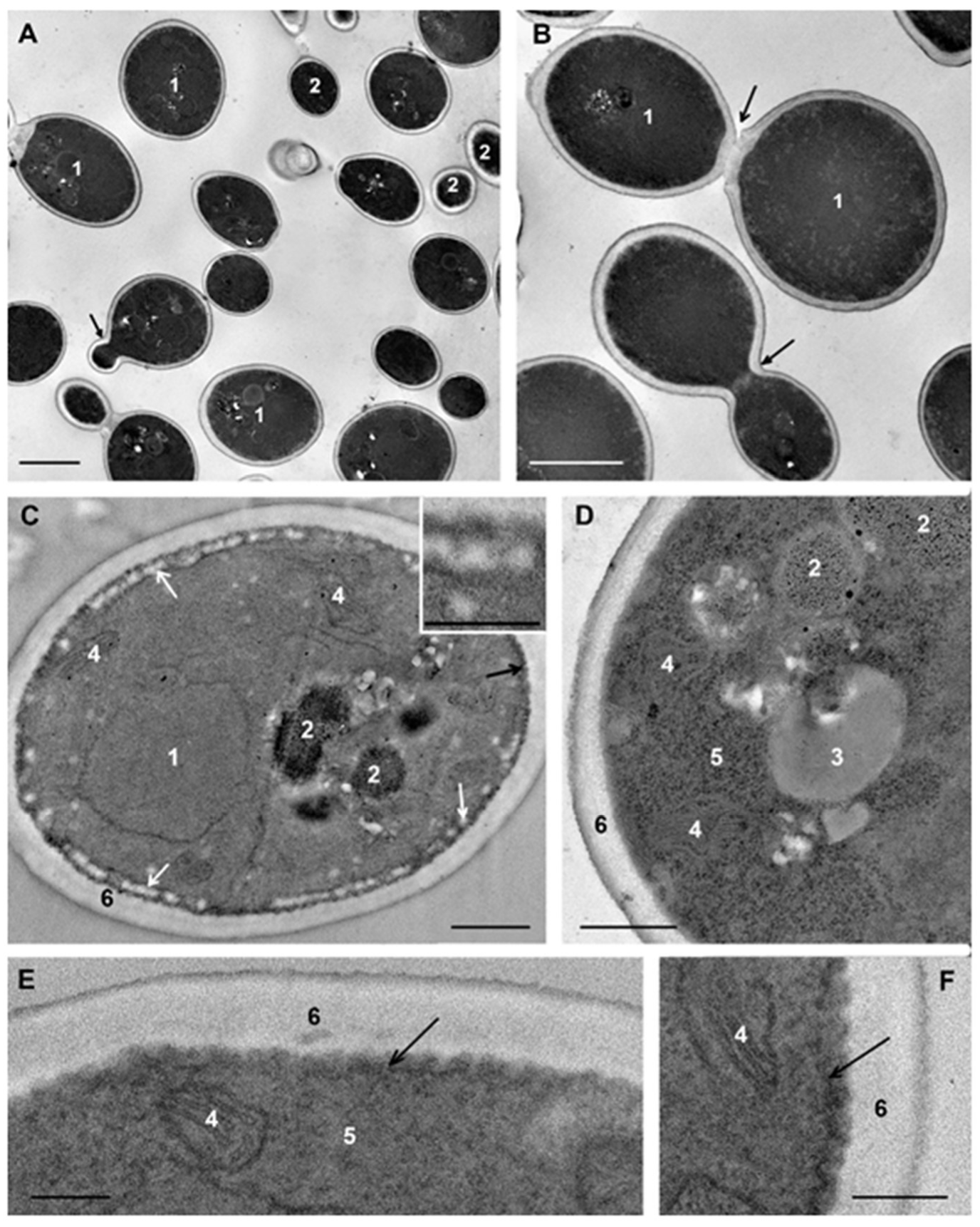
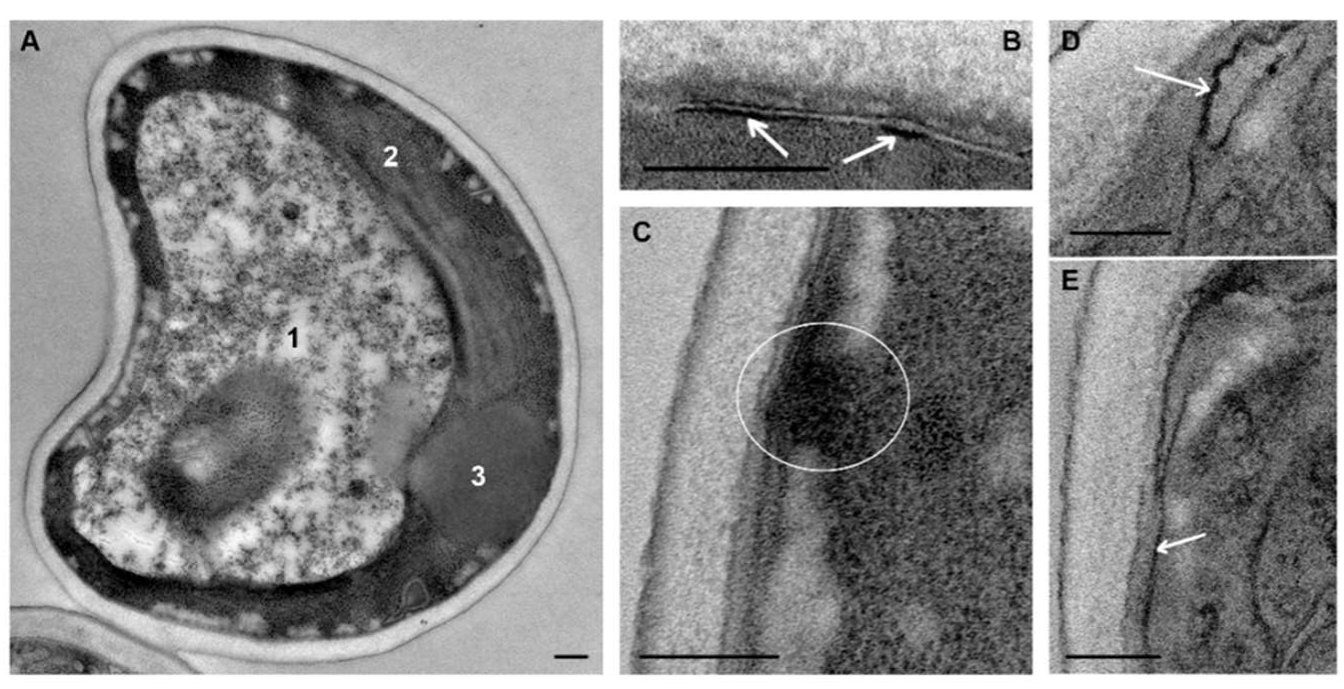
C. albicans cells that were not subjected to experimental influence (intact cells) in ultrathin sections (thickness about 70 nm) had an oval-rounded shape; pictures of budding were common. Depending on the section level, cells were represented by the central part (the cross-section size was about 4 μm), or by smaller fragments (
Figure 2A,B).
The plasma membrane adheres tightly to the cell wall and this makes difficult to distinguish membrane after contrasting. Without contrasting, the membrane is visualized better; in the sections of some cells it is smooth, while in others, numerous folds (depth up to 100 nm) increase the area of the plasma membrane giving it a wavy appearance (
Figure 2C–F). The same folds of the
C. albicans plasma membrane in the logarithmic phase were described earlier [
42].
The eukaryotic plasma membrane is a selective barrier that ensures the penetration of various compounds into the cell by different ways, including endocytosis. The process of clathrin-dependent endocytosis has been studied in detail in
S. cerevisiae cells [
43]. Similar studies of
C. albicans have not been conducted; researchers usually confine themselves to the fact that fluorescent labels enter cells and migrate into vacuoles and call this process “endocytosis” [
38,
44,
45]. This partly could be related to the active study of another very important process: endocytosis of
C. albicans by host cells.
C. albicans is able to induce this process, providing invasion of the fungus in the host tissue, and this ability is a factor of its virulence and pathogenicity [
45,
46,
47].
At the periphery of many cells, spherical structures (60–100 nm) composed of granular material of low electron density without a limiting membrane appeared as small uniform balls were observed. These “balls” often composed a “tape” encircling the cell (
Figure 2C); sometimes they formed large aggregations. The “balls” are seen in illustrations in many articles; however, the authors do not pay attention to their identification. We suppose that the “balls” could be a kind of “reserve deposit” in a cell preparing for division and growth.
The nucleus of intact
C. albicans cells (
Figure 2C,E) was mostly irregular in shape and located in the cell center, or may be displaced to the cell wall. The nucleus envelope consisted of two membranes, visible in cross sections, and the nucleoplasm was homogeneously fine-grained. The observed nucleus morphology corresponds to the published data [
48]. The cytoplasm of
C. albicans cells contains a few large mitochondria with transverse cristae (
Figure 2C–E). The Golgi apparatus could be seen after “enlightenment” of the cytoplasm [
41], so this structure did not appear in images in our study. Homogeneous lipid droplets of average and sometimes high electron density were common structures in
C. albicans cells (
Figure 2D). They had various sizes and were often in close contact with the vacuole.
In our study, vacuoles of intact
C. albicans cells were mainly represented by several non-large structures of high electron density (
Figure 2C); such structures were previously described in intact
C. albicans cells at the logarithmic growth phase [
38,
49,
50,
51]. Another variant of the vacuole was non-large structures of medium electron density containing electron-dense particles (
Figure 2E). The vacuole membrane was clearly visible only in perpendicular sections, similar to other membranes of fungal cells. Vacuoles were present but not in all sections of intact cells, and the presence was the main element determining the polymorphism of
C. albicans cells in a TEM study of ultrathin sections.
3.4. Changes of Cell Wall Ultrastructure under the Influence of R9F2 and (KFF)3K Peptides
Ultrathin sections can pass a cell at different angles to the plane of membranes and the cell wall, thereby making the membranes “invisible”, and the wall structure and its dimensions distorted. The need to select strictly perpendicular sections for studies of the structure and measurements of the cell wall was noted more than 30 years ago; the author also recommended avoiding zones of bud scars, in which the wall thickness may differ [
52].
The cell wall of
C. albicans, similar to other fungi, is a natural barrier to any substances on the way to a cell. The fungal cell wall is layered, and details of its molecular structure are described in comprehensive reviews [
11,
53]. In
Candida and
Saccharomyces species, the outer layer of the cell wall comprises highly mannosylated glycoproteins and covers the inner layer, composed mainly of β-(1,6) glucans and β-(1,3) glucans. Fungal cell walls also contain a large number of various molecules responsible for regulating the synthesis of wall components, signaling, and transport. The cell wall is considered as a complex structure that provides many critical vital processes in fungi [
53]. Cryoelectron microscopy made it possible to visualize some components of cell walls, including mannoprotein fibrils [
53,
54]. It is interesting that cell walls mainly consist of molecules that are absent in the human body, which determines interest in it as a target for drugs [
11,
52,
53].
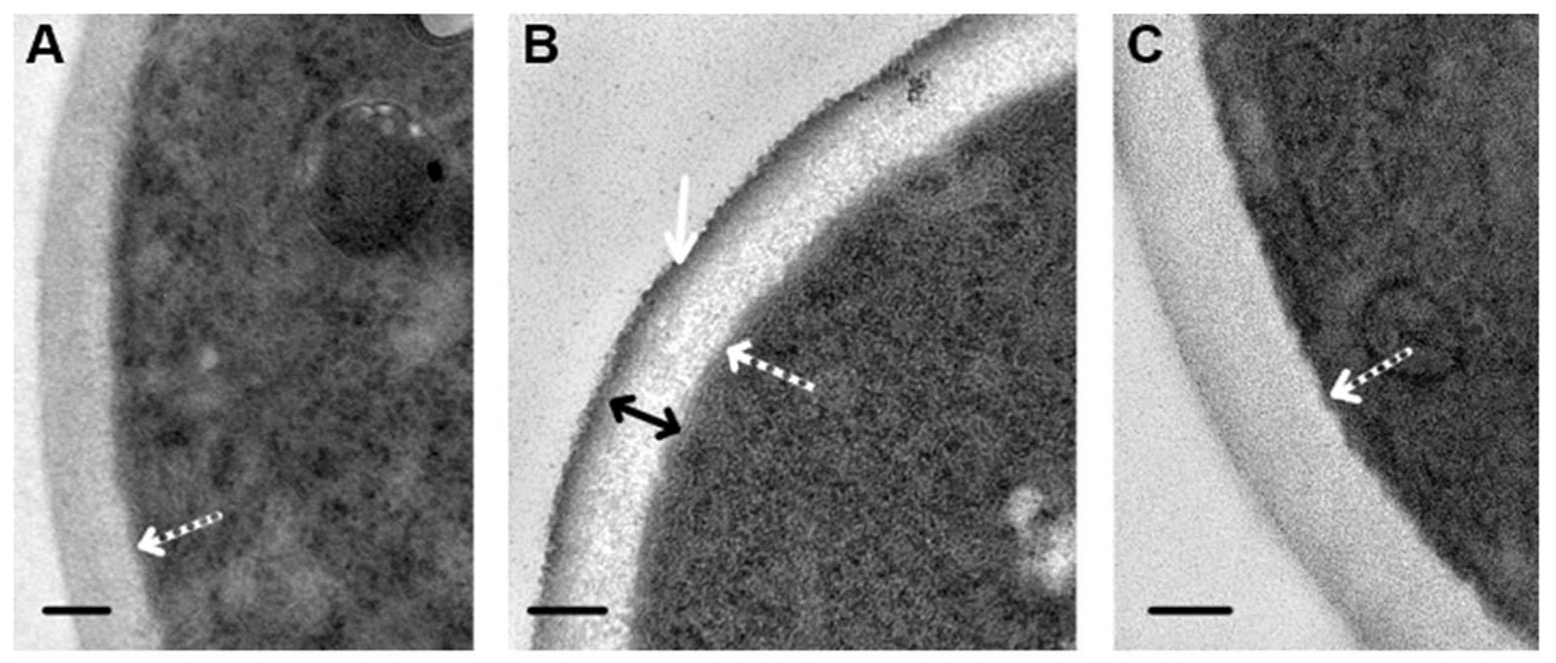
Our study revealed that visibility in the TEM of cell walls depends on the composition of the fixative solution. Fixation in GA resulted in homogeneous cell walls, despite contrasting of sections with UAc and LC (
Figure 3A). When fixed in a mixture of GA and PFA, the wall structure in contrasted sections was detailed (
Figure 3B): the outer electron-dense layer about 20 nm thick (corresponding to the mannosylated glycoproteins layer) and the inner layer (about 100 nm) formed by an electron-transparent matrix containing variously oriented filamentous structures were clearly visible. In the absence of contrasting, cell walls in the same section had a low electron density and were homogeneous, and the outer electron-dense layer was poorly distinguishable; however, it formed a clear wall boundary, and the fibers of the inner layer were not visualized (
Figure 3C). The thickness of the cell wall in perpendicular sections through the cell central part was 126.3 ± 10.53 nm. Thus, the method of fixation and contrasting influences the appearance of cell walls seen in TEM.
We observed clear differences in cell wall changes and their starting time during the incubation with peptides.
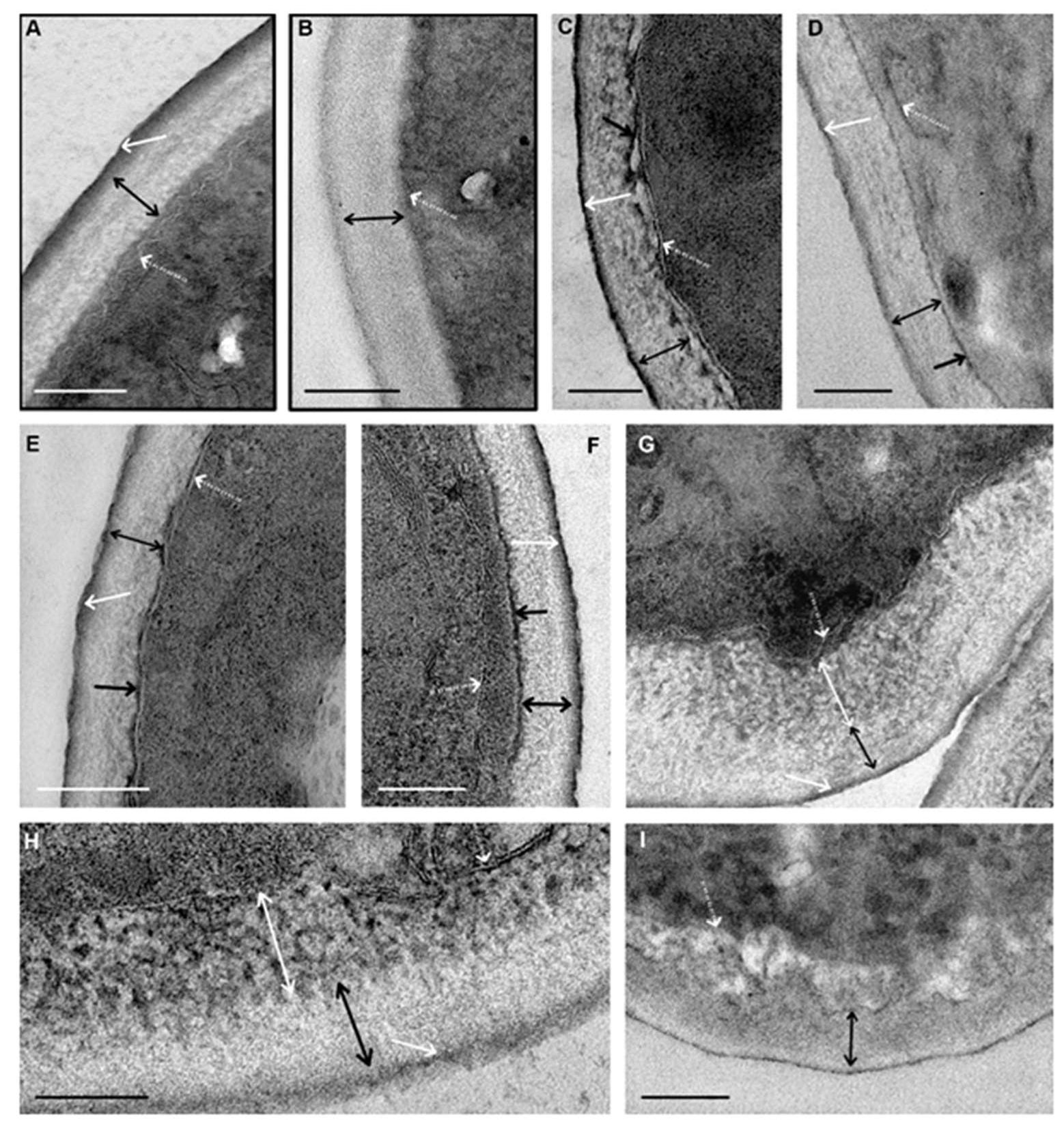
Figure 4 shows images of cell walls at the greatest severity of changes induced by each of the peptides. In these images, the reader can see the appearance of different structures mentioned in the text.
The first changes caused by peptide (KFF)3K in the cell wall ultrastructure were observed after 30 min incubation with
C. albicans suspension. Small pieces of a homogeneous electron-dense substance appeared on the inner side of the wall (
Figure 4C–F). As incubation with the peptide progressed, the thickness and length of these pieces increased, and the substance gradually covered the entire surface of the cell. The substance was visualized without contrasting; this means that it binds to osmium tetroxide (osmiophilia). It can be assumed that this substance (1) is a component of the inner layer of the wall chemically altered by the (KFF)3K peptide or (2) reflects the accumulation of the osmiophilic substance in the wall during its increased migration to or from the cell.
After 45 min of incubation with the (KFF)3K peptide, single fibers of medium electron density were detected in the wall inner layer, and their contrast and number gradually increased, and after 135 min of incubation, distinct electron-dense fibers were observed (
Figure 4C). Thus, peptide (KFF)3K caused disorganization of cell walls of
C. albicans, at the same time widening of the space between the cell wall and plasma membrane, and the wall was detached from the cell (
Figure 4C,D). Amorphous material of average electron density, electron-dense particles, and membrane fragments could present in this space. Described alterations of cell walls were observed in sections of almost all yeast cells after 135 min of incubation with peptide (KFF)3K. Incubation of
C. albicans with the peptide for 6 h resulted in deformation and loosening of cell walls, which thickened to 200 nm (
Figure S2).
Morphological changes of the cell wall (
Figure 4G–I) induced by the peptide R9F2 were registered 45 min later than in the case of peptide (KFF)3K. Thinning of the outer electron-dense layer, loosening, and thickening of the inner layer were observed in wall sections. The thinning indicates peptide R9F2 interaction with components of the outer layer (mannosylated glycoproteins); this effect was absent under incubation with peptide (KFF)3K. Wall changes caused by the R9F2 peptide progressively increased during 6 h of incubation: the outer electron-dense layer gradually disappeared; the thickness of the inner layer increased up to 200 nm. Electron-dense “branches” and “strings” become visible in the inner layer, anastomosing among themselves with the formation of a “network”; these changes were most clearly visible in cell sections after 6 h of incubation with the peptide (
Figure 4F,G).
A comparison of the nature and dynamics of ultrastructural changes in C. albicans cell walls under the influence of peptides (KFF)3K and R9F2 shows clear differences. Most significant, in our view, are the more rapid effect of peptide (KFF)3K; the occurrence of electron-dense material at the border of the cell wall under the action of peptide (KFF)3K; the disappearance of the cell wall outer layer, loosening and structuring of wall inner layer components under the action of peptide R9F2. Therefore, peptides (KFF)3K and R9F2 interact differently with cell walls of C. albicans, and morphological signs of this interaction are registered much later than the changes in the plasma membrane described below, although peptide molecules must overcome the cell wall to reach the plasma membrane.
3.5. Impact of R9F2 and (KFF)3K Peptides on C. albicans Cell Ultrastructure
The plasma membrane of
C. albicans cells serves as a selective barrier, ensuring metabolism in fungal cells and, according to the published data, differs in molecular composition from plasma membranes of bacteria and mammals; moreover, it differs even from the membrane of a close “relative”
S. cerevisiae [
11]. The results of published studies reported that the plasma membrane of
C. albicans is the first target for AFP [
16,
27].
To identify differences between effects of peptides (KFF)3K and R9F2 on C. albicans cells, it was important to study initial steps of the interaction. The need to wash cells from the culture medium determined the duration of the minimal incubation period of cells with peptides—15 min.
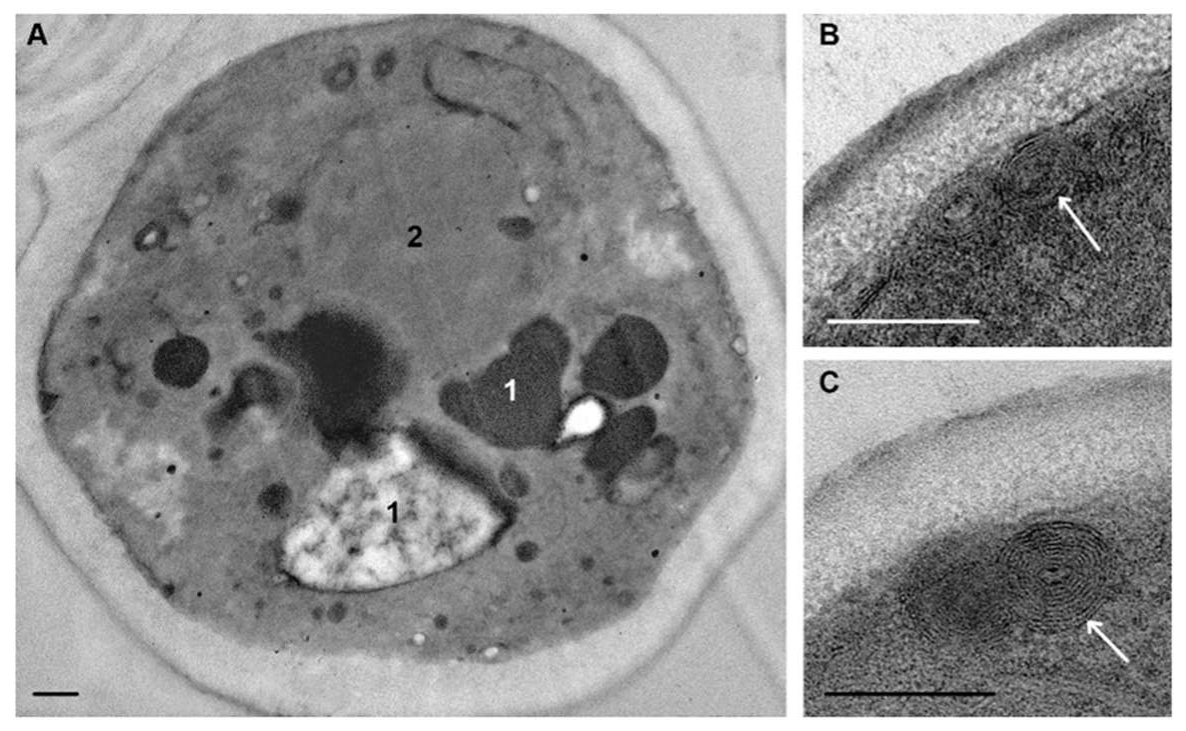
The first change in
C. albicans ultrastructure under the influence of (KFF)3K peptide was detected 15 min after peptide addition to the culture medium and involved the vacuole and plasma membrane. The vacuole increased in size, often causing cell deformation; its cavity was filled with an electron transparent material in which electron-dense particles were dispersed (
Figure 5A). It is interesting that the peptide (KFF)3K within 15 min or less caused changes in the vacuole, which is a cytoplasmic organoid. Decrease in vacuole electron density (
Figure 5A) compared to intact cells (
Figure 2) suggests a disturbance of ion transport through the vacuole membrane and the flow of water into the vacuole cavity, the membrane of which carries numerous transport channels [
49,
50,
55,
56,
57,
58].
Obviously, damage to the vacuole structure could not occur without alteration of the plasma membrane, which first came into contact with peptide (KFF)3K. Within 15 min, the membrane lost its folding, and this “alignment” was noticeable both in contrasted and non-contrasted sections. Small electron-dense areas appeared in the plasma membrane (
Figure 5B–E), the number and length of which increased during subsequent incubation with the peptide. These areas undoubtedly reflect damage to the membrane, and their independence on contrasting gives a reason to consider them as osmiophilic.
Figure 5C shows an electron-dense “bridge” between the damaged area of the plasma membrane and the cytoplasm, probably reflecting the formation of a hole in membrane through which cell contents can outflow into a space between the cell and cell wall. The “balls” of low electron density were observed in sections of many cells including those with damaged plasma membranes (
Figure 5A,C).
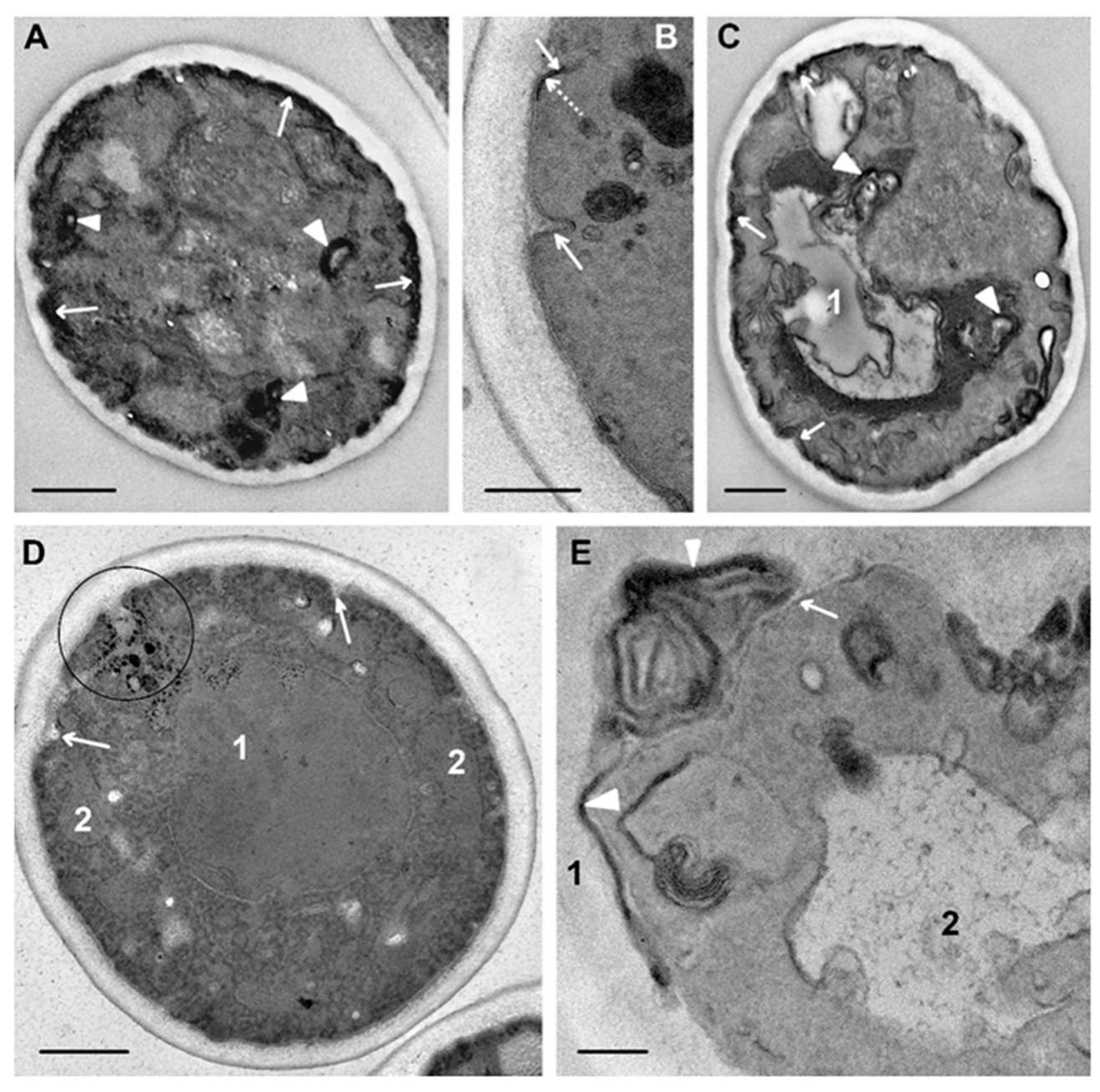
Incubation of
C. albicans with peptide (KFF)3K for 30 min led to distinct changes in the cells compared to 15 min incubation. Vacuoles acquired a bizarre shape; their protrusions extended throughout cytoplasm, often contacting with lipid droplets, and the electron-transparent content was observed only in some cells; mainly vacuoles contained granular and amorphous material of high and/or medium electron density (
Figure 6A). Vacuole changes were better visualized without contrasting, as well as osmiophilic areas in the plasma membrane, the length of which increased. Formation of multilayer flattened structures was observed in areas of visually intact plasma membranes (
Figure 6B,C). The number of layers in these structures varied; sometimes they contained amorphous material, which was apparently a piece of cytoplasm (
Figure 6B). Analysis of sections shows that these multilayer structures were subsequently located in the space between the plasma membrane and cell wall. Apparently, in this way, the cell got rid of membrane portions that were less damaged than in osmiophilic sites. This was also evidenced by a lack of noticeable osmiophilia of the multilayer structures; obviously, the mechanism of their formation differs from the formation of osmiophilic sites in plasma membranes.
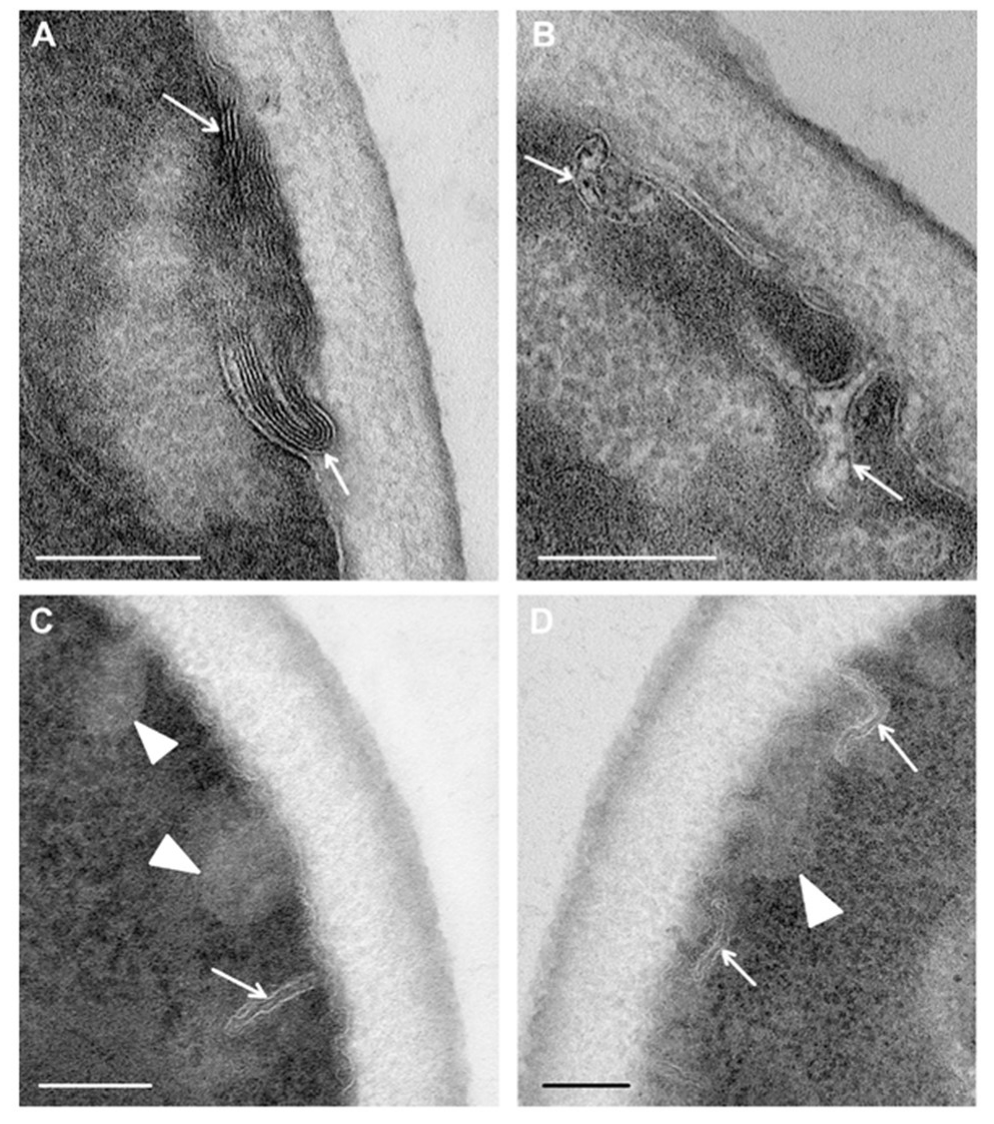
Study of ultrathin sections of
C. albicans cells, incubated for 15 min with R9F2 peptide at the same concentration as (KFF)3K, did not reveal signs of cell alteration; the cell morphology did not differ from intact cells described above. Changes in plasma membranes were detected only after 30 min of incubation with the R9F2 peptide; at the same time, visible changes in other cellular structures were not found. We observed alignment of plasma membranes in most cells, the formation of multilayer folds (
Figure 7A), branched and narrow invaginations protruding into the cytoplasm (
Figure 7B,C), as well as structures resembling foam and formed by thin membranes (
Figure 7D). Obviously, all these structures reflect damage to the plasma membrane by the R9F2 peptide, and their diversity suggests the interaction of the peptide with different components of the plasma membrane. It should be emphasized that there was no formation of osmiophilic sites on plasma membranes when exposed to peptide R9F2.
The observed ultrastructural changes of the C. albicans plasma membrane in response to peptides (KFF)3K and R9F2 indicate different interactions of peptides with membrane components. The peptide (KFF)3K cause a destructive effect on a number of membrane components, resulting in the appearance of osmiophilic sites and holes. In addition, other areas of the plasma membrane formed multilayer structures without signs of osmiophilia, and dynamics of their changes indicate rejection of these structures by the cell. It could be proposed that formation of multilayer structures represent a lesser extent of membrane damage by the peptide (KFF)3K. Thus, the plasma membrane of C. albicans treated with the (KFF)3K peptide show three morphological conditions: visually intact areas, multilayer structures, and osmiophilic sites and holes. Perhaps these features are due to the different molecular composition of the plasma membrane of C. albicans cells, which is at various stages of the life cycle in the logarithmic growth phase.
The peptide R9F2 also led to various changes in ultrastructure of the plasma membrane of
C. albicans, but their character was clearly different from those of peptide (KFF)3K. It can be supposed that formation of multilayer folds and a thin-membrane “foam” occurs similarly to the effect of polyarginine peptides on the plasma membrane of mammalian cells, where the membrane bilayer splits and loosens, and its layers are inverted [
59,
60].
Incubation of
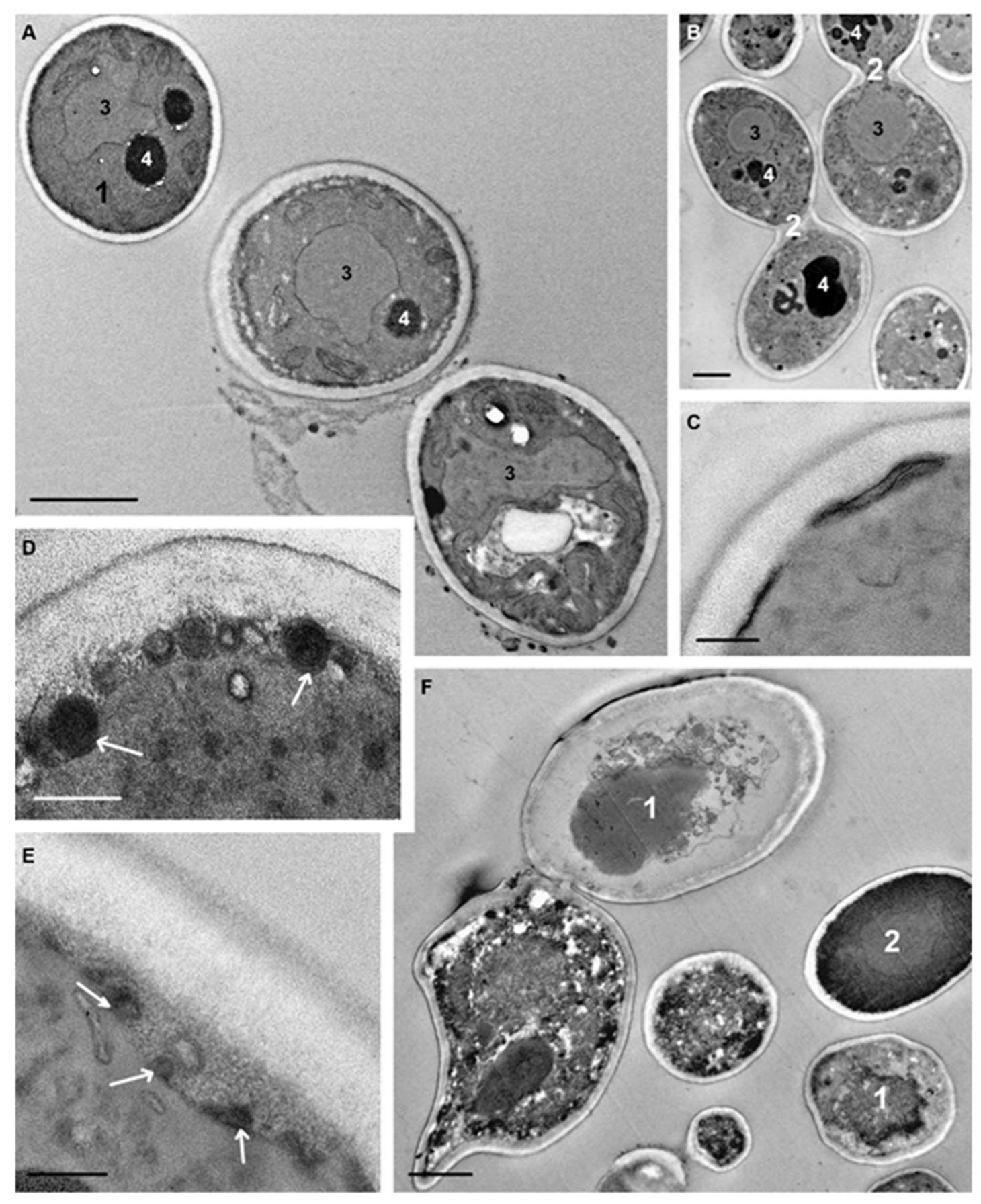
C. albicans cells with peptide (KFF)3K for 75 min resulted in a very high diversity of cell morphology due to the varying degree of destructive changes (
Figure 8A,B). In order not to “drown” in morphological details, we focused on changes in those cellular structures that were the first targets for the peptide. The vacuoles had a diverse shape and basically granular contents; there were some vacuoles with electron-transparent and homogeneous electron-dense contents, and vacuoles of many cells were of a mixed type (
Figure 8A,B).
Osmiophilic sites remained in the plasma membrane, and accumulation of osmiophilic material associated with the plasma membrane was observed (
Figure 8C). The multilayer structures noted above were still present in space between the plasma membrane and cell wall. Expansion of this space (
Figure 8D,E) was a noticeable change in the structure of
C. albicans cells and was observed in many cells. Judging by the changes in contents during observations, at first an unstructured granular material appeared in the space, apparently being an exudate of the cytoplasm. Later, membrane structures and various particles appeared in the space that could enter it upon rupture of the plasma membrane due to damage by peptide (KFF)3K.
The peptide R9F2 also led to diverse morphological patterns of
C. albicans cells during 75 min, similar to peptide (KFF)3K. Both slightly changed cells (
Figure 9D) and cells with destroyed organoids were observed in sections (
Figure 9A,C). In many cells, a vacuole acquired electron-transparent content (
Figure 9C), which, obviously, reflects a violation of the water-ion balance in damaged cells in this period. Deep invaginations of plasma membranes were observed in the cells (
Figure 9B,D), probably reflecting the “development” of invaginations found earlier (
Figure 7). Osmiophilic multimembrane (myelin-like) bodies were present in the cytoplasm (
Figure 9A,C,E). These can be a derivative of plasma membrane folds described above, which became osmiophilic due to developing destructive processes. Formation of the same structures in mammalian cells treated with polyarginine peptides as CPP was described. The authors showed direct translocation of the peptide-transported molecule through plasma membranes and the ability of cells to eject multimembrane bodies outside [
61]. Formation of multimembrane structures as a result of the interaction of polyarginine (R9) CPP with cell membranes and liposomes was reported in another study [
59]. The formation of multimembrane structures from plasma membranes in
C. albicans cells under R9F2 peptide influence observed in our work suggests a similar mechanism for penetration of the peptide into cells, despite differences in the molecular composition of the mammalian and fungal plasma membrane. On the surface of many cells wide osmiophilic regions of high electron density were seen (
Figure 9A,C), the structural details of which were poorly distinguished even without section contrasting. We think that these regions may form from the extended folds (
Figure 7A) during destructive processes resulting in osmiophilia of plasma membranes.
A six-hour incubation of
C. albicans with peptide (KFF)3K led to an increase of dead cells (
Figure 8F), the cytoplasm of which contained shapeless amorphous and granular material, membrane fragments, lipid droplets, and myelin-like structures—the “remains” of organoids. The rigid cell wall provided the visibility of cell integrity even after complete organoid destruction (
Figure S2). Individual cells with a “normal” morphology were noted in sections in this period. Incubation of
C. albicans with peptide R9F2 for 6 h also led to cell destruction; the cell wall limited the accumulation of detritus (
Figure S2). Cells showing “normal” morphology and budding patterns were almost absent in the sections.
To determine the specificity of observed structural changes in
C. albicans when exposed to peptides (KFF)3K and R9F2, we analyzed fungal cells exposed to chlorhexidine. It turned out that the structural changes caused by this antiseptic (
Figure S3) were completely different from the changes induced by the peptides, which once again confirms the specificity and depth of the observed changes in the morphology of fungal cells upon exposure to peptides.