Escherichia coli O157:H7 Curli Fimbriae Promotes Biofilm Formation, Epithelial Cell Invasion, and Persistence in Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria, Plasmids, Primers, and Growth Conditions
2.2. Transposon Mutagenesis
2.3. Crystal Violet Biofilm Assay
2.4. Invasion Assay
2.5. Construction of csgD Promoter Mutants
2.6. Curli Isolation and Detection
2.7. Operon Sequences
2.8. Immunofluorescence Microscopy
2.9. Transmission Electron Microscopy
2.10. Cattle Challenge and O157 Enumeration
2.11. Statistical Analysis
3. Results
3.1. Tn5 Biofilm-Negative Insertions Map to LPS Synthesis or Curli Fimbriae Genes, but Only the Latter Class of Mutant Had Reduced Epithelial Cell Invasion
3.2. A Single Base Pair Change (A to T) in the csgD Promoter of O157 Strains Sakai and 43894, Conferred the Biofilm/Invasive-Positive Phenotype
3.3. Curli Fimbriae Are Expressed in O157 Strains 43895, 43894R, and SakaiR but Not in Wild Type Strains 43894, Sakai, or the Mutant csgB::Tn5
3.4. Deletion of csgB Resulted in the Same Phenotype as the csgB::Tn5
3.5. Curli Was Required for O157 Adherence to and Invasion of MAC-T Cells
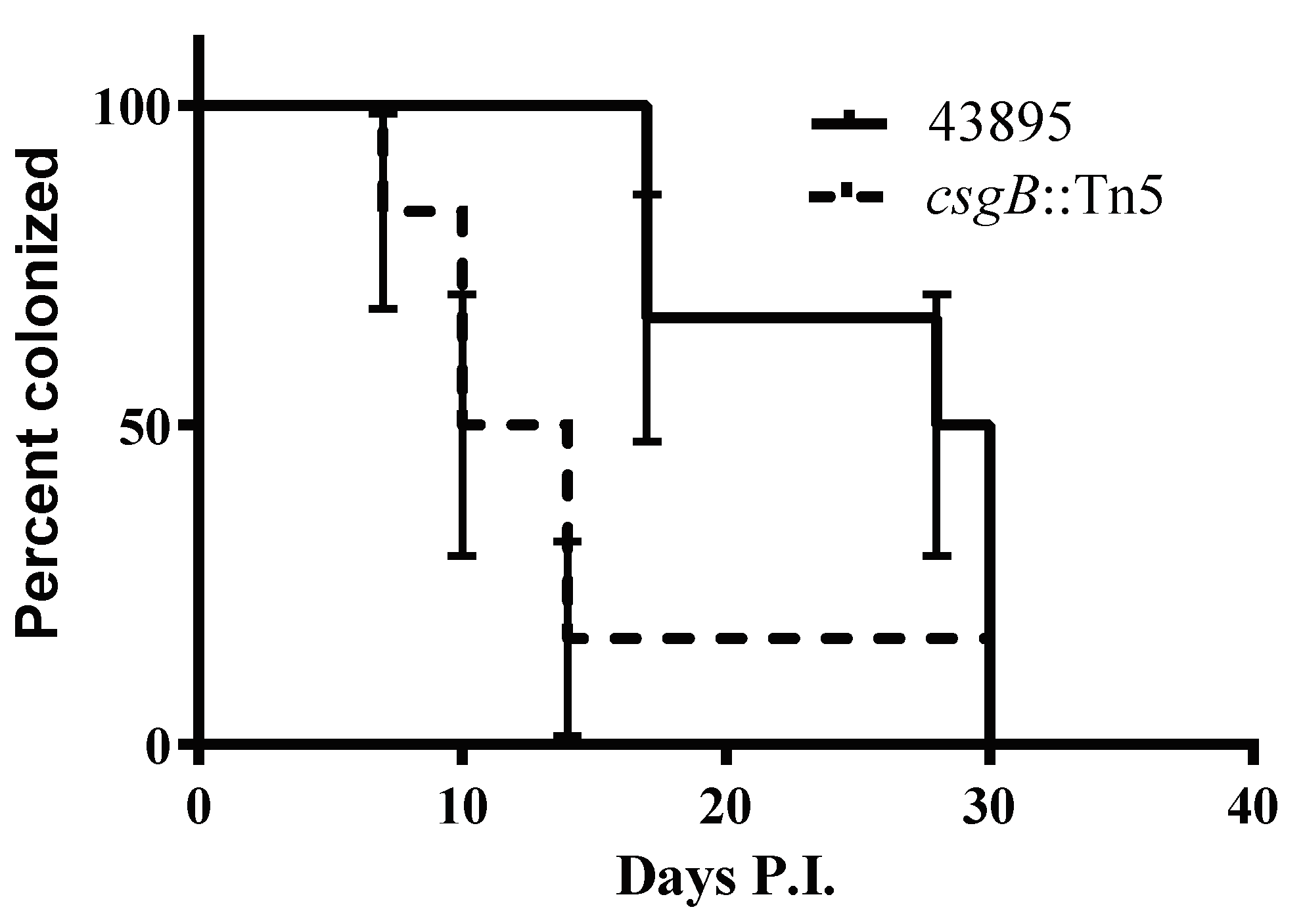
3.6. The csgB::Tn5 Mutation Reduced Persistence of E. coli O157 Strain 43895 in Cattle
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paton, J.C.; Paton, A.W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 1998, 11, 450–479. [Google Scholar] [CrossRef] [PubMed]
- Karmali, M.A.; Steele, B.T.; Petric, M.; Lim, C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1983, 1, 619–620. [Google Scholar] [CrossRef]
- Griffin, P.M.; Tauxe, R.V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 1991, 13, 60–98. [Google Scholar] [CrossRef] [PubMed]
- Michino, H.; Araki, K.; Minami, S.; Takaya, S.; Sakai, N.; Miyazaki, M.; Ono, A.; Yanagawa, H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 1999, 150, 787–796. [Google Scholar] [CrossRef]
- Smith, H.R. Escherichia coli O157 infections: The Scottish experience. Hosp. Med. 1998, 59, 164. [Google Scholar]
- Blanco, M.; Blanco, J.E.; Blanco, J.; Gonzalez, E.A.; Mora, A.; Prado, C.; Fernández, L.; Rio, M.; Ramos, J.; Alonso, M.P. Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol. Infect. 1996, 117, 251–257. [Google Scholar] [CrossRef]
- Sheng, H.; Davis, M.A.; Knecht, H.J.; Hancock, D.D.; Donkersgoed, J.V.; Hovde, C.J. Characterization of a Shiga toxin-, intimin-, and enterotoxin hemolysin-producing Escherichia coli ONT:H25 strain commonly isolated from healthy cattle. J. Clin. Microbiol. 2005, 43, 3213–3220. [Google Scholar] [CrossRef][Green Version]
- Hancock, D.D.; Besser, T.E.; Kinsel, M.L.; Tarr, P.I.; Rice, D.H.; Paros, M.G. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 1994, 113, 199–207. [Google Scholar] [CrossRef]
- Zhao, T.; Doyle, M.P.; Shere, J.; Garber, L. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 1995, 61, 1290–1293. [Google Scholar] [CrossRef]
- Naylor, S.W.; Low, J.C.; Besser, T.E.; Mahajan, A.; Gunn, G.J.; Pearce, M.C.; McKendrick, I.J.; Smith, D.E.; Gallyet, D.L. Lymphoid Follicle-Dense Mucosa at the Terminal Rectum is the Principal Site of Colonization of Enterohemorrhagic Escherichia coli O157:H7 in the Bovine Host. Infect. Immun. 2003, 71, 1505–1512. [Google Scholar] [CrossRef]
- Sheng, H.; Davis, M.A.; Knecht, H.J.; Hovde, C.J. Rectal administration of Escherichia coli O157:H7: Novel model for colonization of ruminants. Appl. Environ. Microbiol. 2004, 70, 4588–4595. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Lim, J.Y.; Watkins, M.K.; Minnich, S.A.; Hovde, C.J. Characterization of an Escherichia coli O157:H7 O-antigen deletion mutant and effect of the deletion on bacterial persistence in the mouse intestine and colonization at the bovine terminal rectal mucosa. Appl. Environ. Microbiol. 2008, 74, 5015–5022. [Google Scholar] [CrossRef] [PubMed]
- Vogeleer, P.; Tremblay, Y.D.N.; Jubelin, G.; Jacques, M.; Harel, J. Biofilm-forming abilities of Shiga toxin-producing Escherichia coli isolates associated with human infections. Appl. Environ. Microbiol. 2015, 82, 1448–1558. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, S.J.; Ritchie, J.M.; Torres, A.G. Fimbriation and curliation in Escherichia coli O157:H7: A paradigm of intestinal and environmental colonization. Gut Microbes 2012, 3, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Wells, T.J.; Sherlock, O.; Rivas, L.; Mahajan, A.; Beatson, S.A.; Torpdahl, M.; Webb, R.I.; Allsopp, L.P.; Gobius, K.S.; Gally, D.L.; et al. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 2008, 10, 589–604. [Google Scholar] [CrossRef]
- Sheng, H.; Wang, J.; Lim, J.Y.; Davitt, C.; Minnich, S.A.; Hovde, C.J. Internalization of Escherichia coli O157:H7 by bovine rectal epithelial cells. Front. Microbiol. 2011, 2, 32. [Google Scholar] [CrossRef]
- Corbishley, A.; Ahmad, N.I.; Hughes, K.; Hutchings, M.R.; McAteer, S.P.; Connelley, T.K.; Helen Brown, H.; Gally, D.L.; Tom, N.; McNeilly, T.N. Strain-dependent cellular immune responses in cattle following Escherichia coli O157:H7 colonization. Infect. Immun. 2014, 82, 5117–5131. [Google Scholar] [CrossRef]
- Smith, D.R.; Price, J.E.; Burby, P.E.; Blanco, L.P.; Chamberlain, J.; Chapman, M.R. The production of curli amyloid fibers is deeply integrated into the biology of Escherichia coli. Biomolecules 2017, 7, 75. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef]
- Olsen, A.; Wick, M.J.; Morgelin, M.; Bjorck, L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 1998, 66, 944–949. [Google Scholar] [CrossRef]
- Tursi, S.A.; Lee, E.Y.; Medeiros, N.J.; Lee, M.H.; Nicastro, L.K.; Buttaro, B.; Gallucci, S.; Wilson, R.P.; Wong, G.C.L.; Tukel, C. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.; Arnqvist, A.; Hammar, M.; Sukupolvi, S.; Normark, S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectinbinding curli in Escherichia coli. Mol. Microbiol. 1993, 7, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Hammar, M.; Arnqvist, A.; Bian, Z.; Olsen, A.; Normark, S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995, 18, 661–670. [Google Scholar] [CrossRef]
- Kikuchi, T.; Mizunoe, Y.; Takade, A.; Naito, S.; Yoshida, S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005, 49, 875–884. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Cooke, P.H.; Solomon, E.B. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 2006, 72, 2564–2572. [Google Scholar] [CrossRef]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [CrossRef]
- Cookson, A.L.; Cooley, W.A.; Woodward, M.J. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int. J. Med. Microbiol. 2002, 292, 195–205. [Google Scholar] [CrossRef]
- Perna, N.T.; Plunkett, G.; Burland, V.; Mau, B.; Glasner, J.D.; Rose, D.J.J.; Mayhew, G.F.; Evans, P.S.; Gregor, J.; Kirkpatrick, H.A.; et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 2001, 409, 529–533. [Google Scholar] [CrossRef]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef]
- Sheng, H.; Duan, M.; Hunter, S.S.; Minnich, S.A.; Settles, M.L.; New, D.D.; Chase, J.R.; Fagnan, M.W.; Hovde, C.J. High-Quality Complete Genome Sequences of Three Bovine Shiga Toxin-Producing Escherichia coli O177:H- (fliCH25) Isolates harboring virulent stx2 and multiple plasmids. Genome Announc. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Uhlich, G.A.; Chen, C.Y.; Cottrell, B.J.; Hofmann, C.S.; Yan, X.; Nguyen, L. Stx1 prophage excision in Escherichia coli strain PA20 confers strong curli and biofilm formation by restoring native mlrA. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bian, Z.; Brauner, A.; Li, Y.; Normark, S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 2000, 181, 602–612. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Keen, J.E.; Elder, R.O. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2001, 67, 2367–2370. [Google Scholar] [CrossRef]
- Luck, S.N.; Bennett-Wood, V.; Poon, R.; Robins-Browne, R.M.; Hartland, E.L. Invasion of epithelial cells by locus of enterocyte effacement-negative enterohemorrhagic Escherichia coli. Infect. Immun. 2005, 73, 3063–3071. [Google Scholar] [CrossRef]
- Lim, J.Y.; La, H.J.; Sheng, H.; Forney, L.J.; Hovde, C.J. Influence of plasmid pO157 on Escherichia coli O157:H7 Sakai biofilm formation. Appl. Environ. Microbiol. 2010, 76, 963–966. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Nat. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Simons, R.W.; Houman, F.; Kleckner, N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 1987, 53, 85–96. [Google Scholar] [CrossRef]
- Kolodziejek, A.M.; Schnider, D.R.; Rohde, H.N.; Wojtowicz, A.J.; Bohach, G.A.; Minnich, S.A.; Hovde, C.J. Outer membrane protein X (Ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infect. Immun. 2010, 78, 5233–5243. [Google Scholar] [CrossRef]
- Matthews, K.R.; Murdough, P.A.; Bramley, A.J. Invasion of bovine epithelial cells by verocytotoxin-producing Escherichia coli O157:H7. J. Appl. Microbiol. 1997, 82, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Small, P.L.; Isberg, R.R.; Falkow, S. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within HEp-2 cells. Infect. Immun. 1987, 55, 1674–1679. [Google Scholar] [CrossRef]
- Sheng, H.; Lim, J.Y.; Knecht, H.J.; Li, J.; Hovde, C.J. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 2006, 74, 4685–4693. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.H.; Sheng, H.; Wynia, S.A.; Hovde, C.J. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 2003, 41, 4924–4929. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Blanco, L.P.; Smith, D.R.; Chapman, M.R. Bacterial amyloids. Methods Mol. Biol. 2012, 849, 303–320. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Chen, C.Y.; Cottrell, B.J.; Andreozzi, E.; Irwin, P.L.; Nguyen, L.H. Genome amplification and promoter mutation expand the range of csgD-dependent biofilm responses in an STEC population. Microbiology 2017, 163, 611–621. [Google Scholar] [CrossRef]
- National Advisory Committee on Microbiological Criteria for Foods. Response to questions posed by the Food and Drug Administration regarding virulence factors and attributes that define foodborne Shiga toxin-producing Escherichia coli (STEC) as severe human Pathogens (dagger). J. Food Prot. 2019, 82, 724–767. [Google Scholar] [CrossRef]
- Romling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 1998, 28, 249–264. [Google Scholar] [CrossRef]
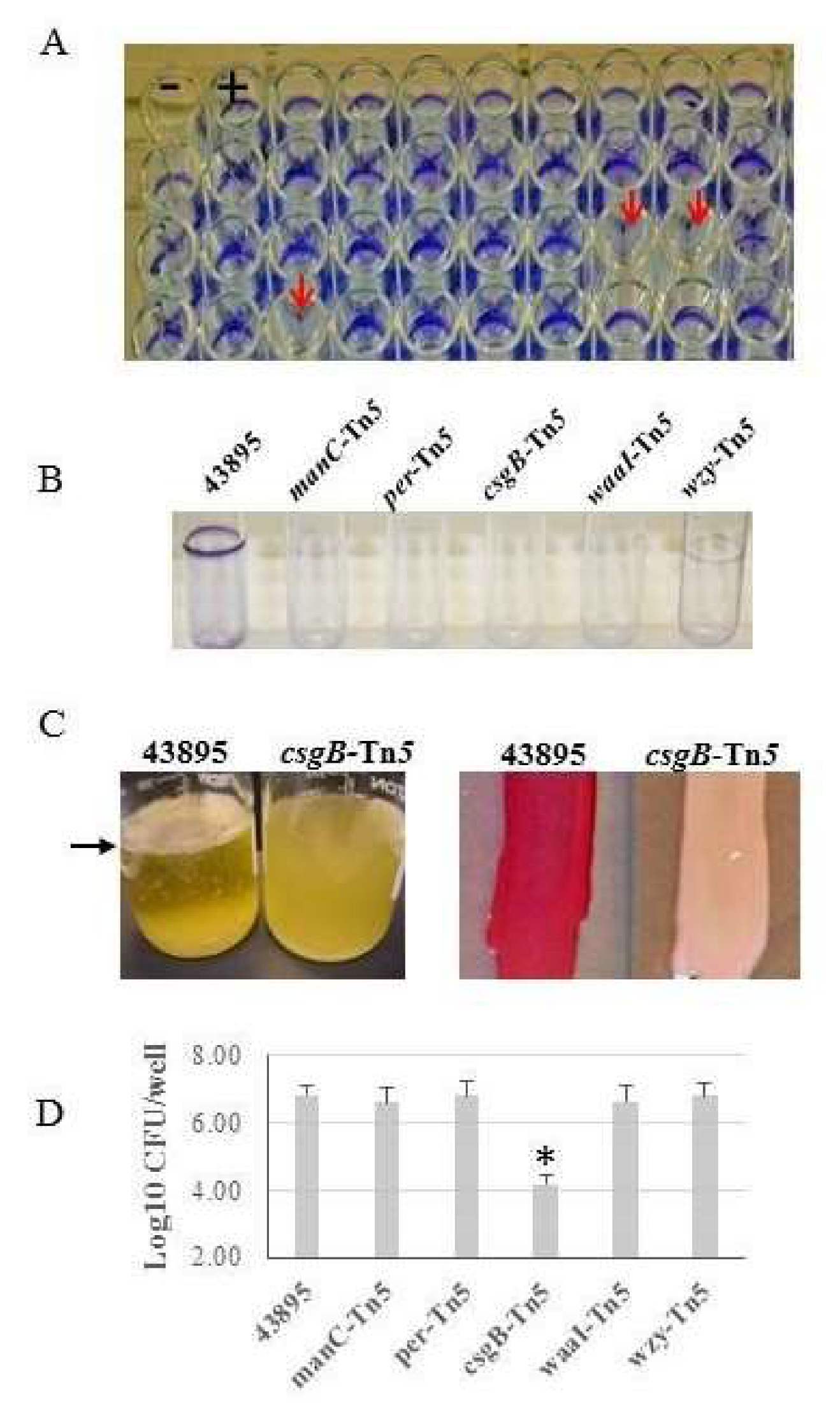
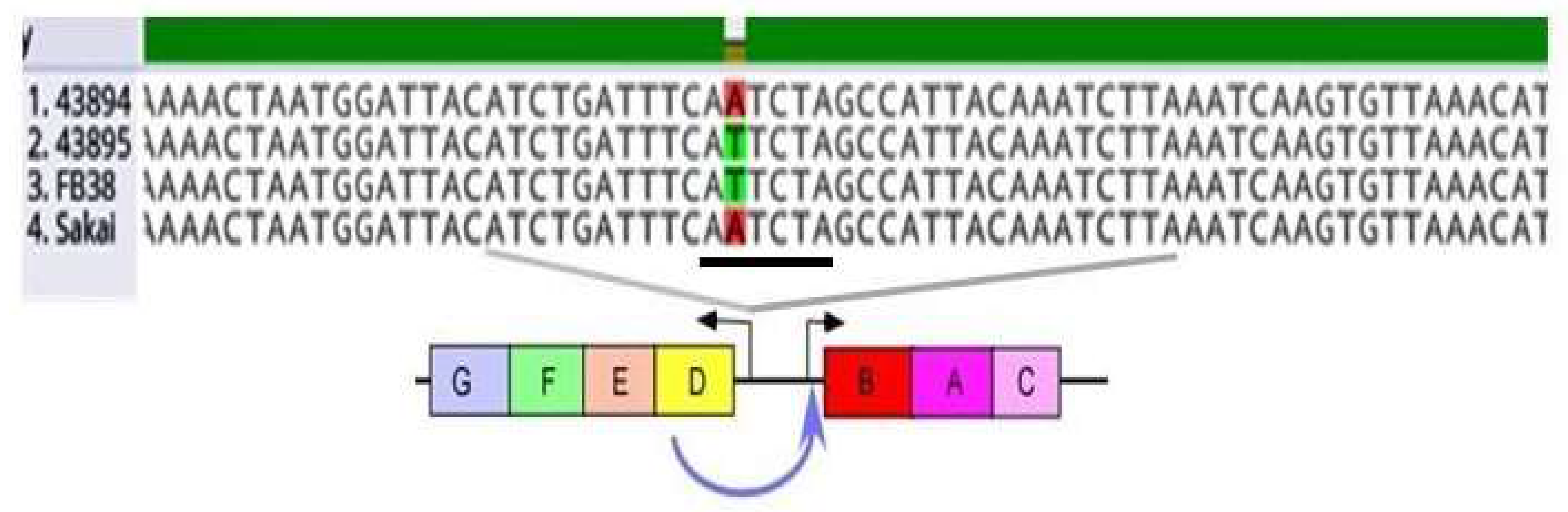

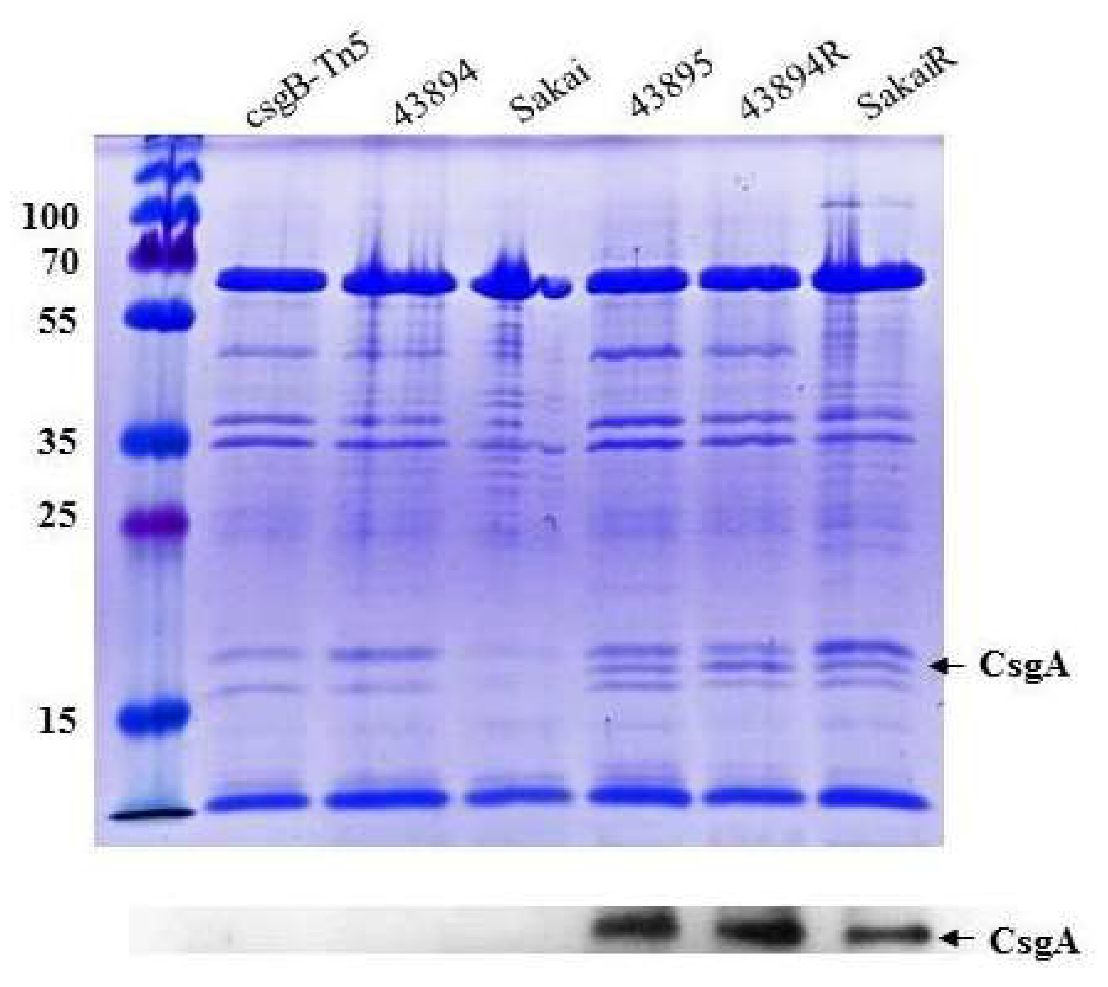

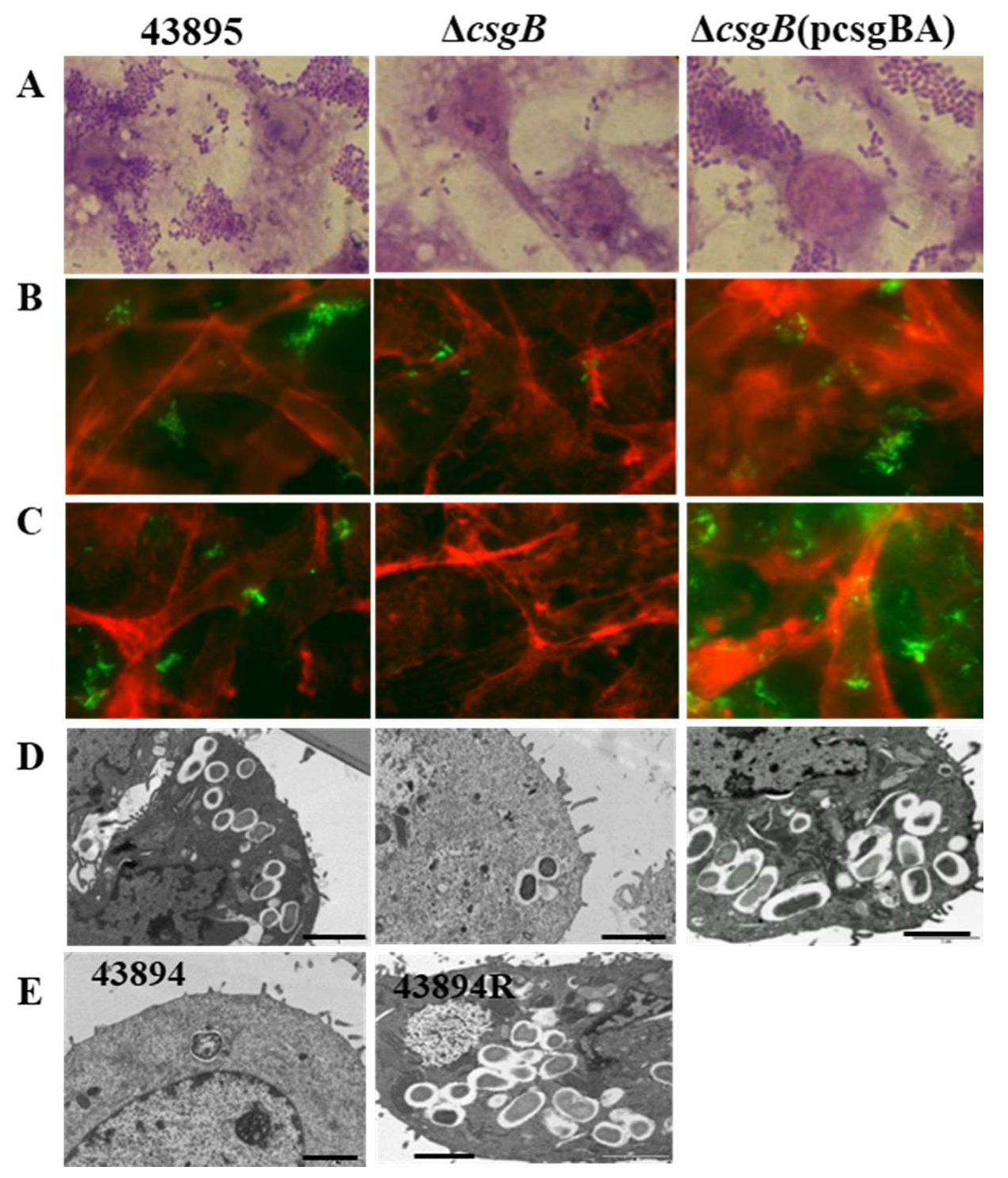
| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| 43895 | E. coli O157:H7 ATCC 43895, a clinical isolate, stx1+/stx2+, curli+ | ATCC * |
| manC-Tn5 | Tn5 inserted in manC of 43895 | This work |
| per-Tn5 | Tn5 inserted in per of 43895 | This work |
| csgB-Tn5 | Tn5 inserted in csgB of 43895 | This work |
| waaI-Tn5 | Tn5 inserted into waaI of 43895 | This work |
| wzy-Tn5 | Tn5 inserted into wzy of 43895 | This work |
| 43895ΔcsgB | 43895 with csgB deletion | This work |
| 43895ΔcsgB(pcsgBA) | 43895ΔcsgB complemented csgB | This work |
| 43894 | E. coli O157:H7 ATCC 43894, a clinical isolate, stx1+/stx2+, curli- | ATCC |
| 43894R | 43894 mutant with A to T transversion in csgD promoter, curli+ | This work |
| Sakai SakaiR | E. coli O157:H7, a clinical isolate, stx1+/stx2+, curli- Sakai mutant with A to T transversion in csgD promoter, curli+ | [16] This work |
| FB38 | E. coli O157:H7, a bovine isolate, stx1+/stx2-, curli positive | Laboratory stock |
| S17-1 λ pir | pro recA thi hsdR Hfr RP4-2 (Tc::Mu) (Km::Tn7, SmR, λ pir lysogen | [38] |
| K-12 | E. coli MG1655 strain | Laboratory stock |
| pKD4 | Template plasmid for mutagenesis (AmpR KanR) | [37] |
| pKD3 | Template plasmid for mutagenesis (AmpR CmR) | [37] |
| pKD46 | Red recombinase helper plasmid, RepA101(Ts), AmpR | [37] |
| pACYC177 | Cloning vector, AmpR, KanR | ATCC |
| pGP704.L | Suicide plasmid, mob+, sacBR+, pir-dependent oriR6K, AmpR | [39] |
| pGP704csg1 | pGP704.L containing csgBA genes of 43895; allelic exchange plasmid | This work |
| pcsgBA | csgBA and intergenic region of 43895 cloned into pACYC177 | This work |
| Primer | * Sequence (5′–3′) |
|---|---|
| KAN-2 FP-1 | ACCTACAACAAAGCTCTCATCAACC |
| KAN-2 RP-1 | GCAATGTAACATCAGAGATTTTGAG |
| IBA-F | GTTTGGATCCAAACCCCGCTTTTTTTATTGATC (BamHI) |
| IBA-R | GTTTCTGCAGTTAGTACTGATGAGCGGTCGCGT (PstI) |
| csgDBign-F | GAGCCTGAAGAGATATCGTCCA |
| csgDBign-R | GCGCACCCAGTATTGTTAAC |
| csgB-LF | ATGAAAAACAAATTGTTATTTATGATGTTAACAATACTGGGTGTGTAGGCTGGAGCTGCTTCG |
| csgB-LR | TTAACGTTGTGTCACGCGAATAGCCATTTGCGACTGTCTCTGCATATGAATATCCTCCTTA |
| Dele-1F | AGAAGTACTGACAGATGTTGCACTGCTGTGTGTAGTAATAAATGTGTAGGCTGGAGCTGCTTCG |
| Dele-1R | AACTTAATAAAACCTTAAGGTTAACATTTTAATATAACCAGTCATATGAATATCCTCCTTA |
| DBC1bF | TATACCCGGGTTCTTGATCCTCCATGGCATAAAA (SmaI) |
| DBC1bR | ATATAGATATCCTGCGTTACGATGGAAAGTATGTC(EcoRV) |
| Strains a | Cell-Associated b (CFU × 107/Well) | Internalized c (CFU × 104/Well) |
|---|---|---|
| 43895 | 8.78 ± 0.73 | 467 ± 190 |
| ΔcsgB | * 0.71 ± 0.12 | ** 4.58 ± 0.89 |
| ΔcsgB(pcsgBA) | 7.66 ± 0.67 | 389 ± 110 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, H.; Xue, Y.; Zhao, W.; Hovde, C.J.; Minnich, S.A. Escherichia coli O157:H7 Curli Fimbriae Promotes Biofilm Formation, Epithelial Cell Invasion, and Persistence in Cattle. Microorganisms 2020, 8, 580. https://doi.org/10.3390/microorganisms8040580
Sheng H, Xue Y, Zhao W, Hovde CJ, Minnich SA. Escherichia coli O157:H7 Curli Fimbriae Promotes Biofilm Formation, Epithelial Cell Invasion, and Persistence in Cattle. Microorganisms. 2020; 8(4):580. https://doi.org/10.3390/microorganisms8040580
Chicago/Turabian StyleSheng, Haiqing, Yansong Xue, Wei Zhao, Carolyn J. Hovde, and Scott A. Minnich. 2020. "Escherichia coli O157:H7 Curli Fimbriae Promotes Biofilm Formation, Epithelial Cell Invasion, and Persistence in Cattle" Microorganisms 8, no. 4: 580. https://doi.org/10.3390/microorganisms8040580
APA StyleSheng, H., Xue, Y., Zhao, W., Hovde, C. J., & Minnich, S. A. (2020). Escherichia coli O157:H7 Curli Fimbriae Promotes Biofilm Formation, Epithelial Cell Invasion, and Persistence in Cattle. Microorganisms, 8(4), 580. https://doi.org/10.3390/microorganisms8040580





